PMG Exam 3
1/19
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
20 Terms
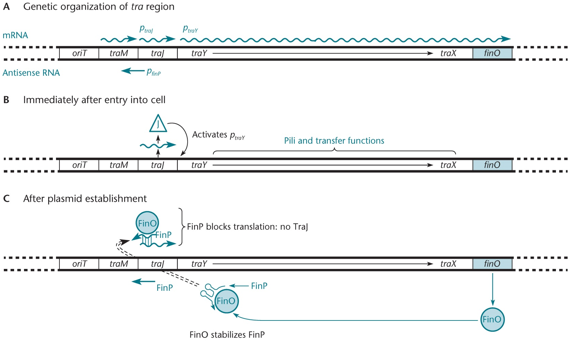
Fertility inhibition of the F plasmid involves the traJ gene, a transcriptional activator needed for traY, traX, and finO expression from the promoter ptraY. Antisense RNA FinP, which is stabilized by FinO, a protein, blocks TraJ translation. This process prevents the transfer of the F plasmid during conjugation.
"What is finP?"
"Antisense RNA that transcribes in the opposite direction of mRNA. Since it is easy to degrade, it needs FinO, a protein that blocks translation, resulting in no TraJ."
"Mutation in FinP sequence?"
"traJ will also have that mutation since they're complimentary; everything will be ok."
"Mutation in RNA binding region of FinO?"
"This will cause FinP to degrade, and it will not be able to be annealed anymore. FinO can also bind tighter and stop the work of traJ sooner; translation."
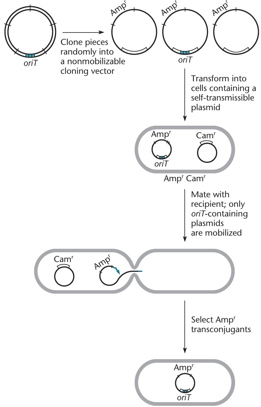
The oriT site is identified by cloning random plasmid fragments into a nonmobilizable vector, transforming cells with a self-transmissible plasmid, and testing for mobilization. Mobilized vectors contain the oriT site.
"What does the cloning experiment need?"
"Plasmid needs oriT to be mobilized and cloned. It also needs restriction enzymes to cut the oriT site."
"What should you look for in a cloning experiment product?"
"You should look for the clones that have growth on ampicillin after doing a passage with ampicillin."
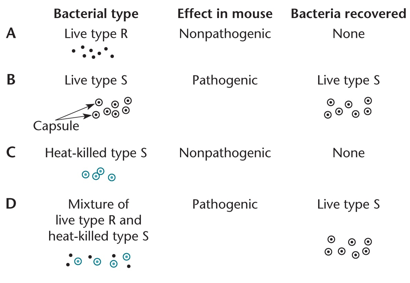
"Griffith Experiment"
"The Griffith experiment shows bacterial transformation. Live type R bacteria are nonpathogenic and can NOT replicate/recover bacteria, while Live type S bacteria are pathogenic and can replicate due to the capsule. Heat-killed type S bacteria are killed by heat alone. Still, when mixed with live type R bacteria, they transform to pathogenic bacteria due to bacteria that may have released their DNA extracellularly and been uptaken by live type R."
"What is the Griffith Experiment used for and how?"
"It is used to improve transformation by uptaking extracellular DNA."
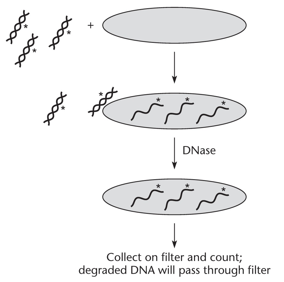
Only double-stranded DNA binds to initiate transformation. Once inside, it becomes single-stranded and can only transform new cells after integrating into the chromosome by recombination. The presence of transformants in step 4 confirms that the DNA was double-stranded during DNase treatment.
"Why is the time of DNase insertion necessary?"
"By adding DNase much later, intracellular DNA is recombined with the chromosome and is thus double-stranded, allowing Arg+ transformants. Transformation can NOT occur with single-stranded DNA. Using DNase too soon could degrade the cell."
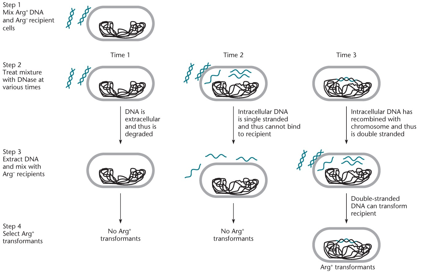
Determining the efficiency of DNA uptake during transformation. DNA in the cell is insensitive to DNase. DNA that did not enter the cell is degraded and passes through a filter. The asterisks indicate radioactively labeled DNA.
"Why is a filter needed for cell suspension?"
"The filter captures the bacteria, while the damaged DNA passes right through. At the end of the experiment, the numbers should add up."
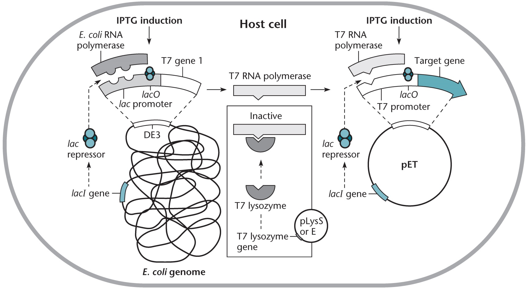
"pET System"
In pET vectors, T7 RNA polymerase is chromosomally encoded in E. coli under the lac promoter and only expressed with IPTG, which micics lactose. It transcribes the cloned gene downstream of the T7 promoter. If the gene is toxic, transcription is further reduced by T7 lysozyme from pLysS, which inactivates residual polymerase, and by lac operators between the T7 promoter and the gene.
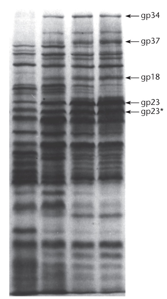
Phage T4-infected bacteria were given radioactive amino acids (Cysteine and Methionine) at different times to label phage (not host) proteins. These proteins, made only during labeling periods, were denatured with SDS, separated by SDS-PAGE, and visualized by autoradiography. Gel lanes show proteins made at 5–10 min (lane 1), 10–15 min (lane 2), 15–20 min (lane 3), and 20–30 min (lane 4) after infection.
"Does T4 have its own RNA Polymerase?"
"No, it uses the one from E. Coli."
"What does 7.7 represent?"
"The lanes in Figure 7.7 represent different times and when the T7 proteins are produced. They are radioactive and contain Cysteine and Methionine. The bands also belong to E. Coli."
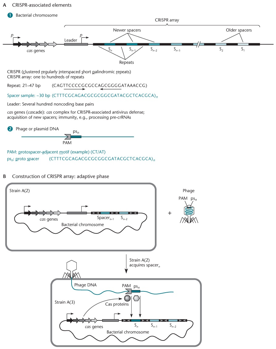
A CRISPR array consists of repeated sequences and unique spacers (sn to s1), with a leader region containing a promoter and cas genes. Spacers nearest the leader are the most recent. When phage DNA with a matching protospacer (psn) and adjacent PAM is detected, Cas proteins recognize the PAM and insert the psn as a new spacer (sn) at the leader end. Existing spacers shift down, and the oldest is discarded.
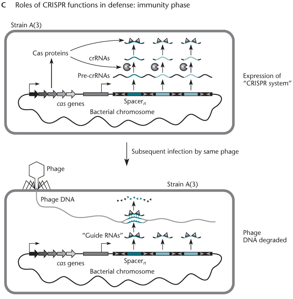
In the immunity phase, the CRISPR array is transcribed and cut into crRNAs, each matching a spacer. If the same phage returns, a crRNA pairs with its matching protospacer (psn), guiding Cas proteins to cut and inactivate the phage DNA.
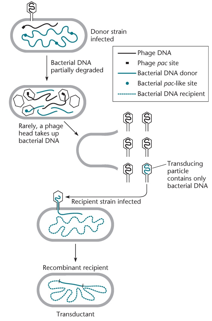
In generalized transduction, a phage mistakenly packages bacterial DNA instead of its own by recognizing a pac site in the host DNA. In the subsequent infection, it injects this bacterial DNA into another cell, where it may replicate (if a plasmid) or recombine with the chromosome (if chromosomal DNA), creating a recombinant.