BIO 120.11 | M4 Genetic Engineering
1/66
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
67 Terms
Genetic engineering is also known as _
Gene modification
Uses recombinant DNA (rDNA) and lab-based technologies to alter the genetic makeup of an organism
Genetic engineering (modification)
Enumerate 3 fields through which genetic engineering is usually applied
iam
Industrial epfatb
Enzymes, plastics, food, additives, textiles, bioenergy
Agriculture mps
Modified food, pest control, stress resistance
Medical avt
Antibiotics, vaccines, therapeutics
3 other terms for Genetically Engineered Organisms (GEOs)
rgt
Recombinant
Genetically modified organisms (GMO)
Transgenic
Other ways through which genetic engineering can be done apart from recombinant DNA
mcat
Mutagenesis
CRISPR/Cas9
Allelic exchange
Transposons
Earliest step for developing genetically modified organisms (GMOs)
Gene cloning
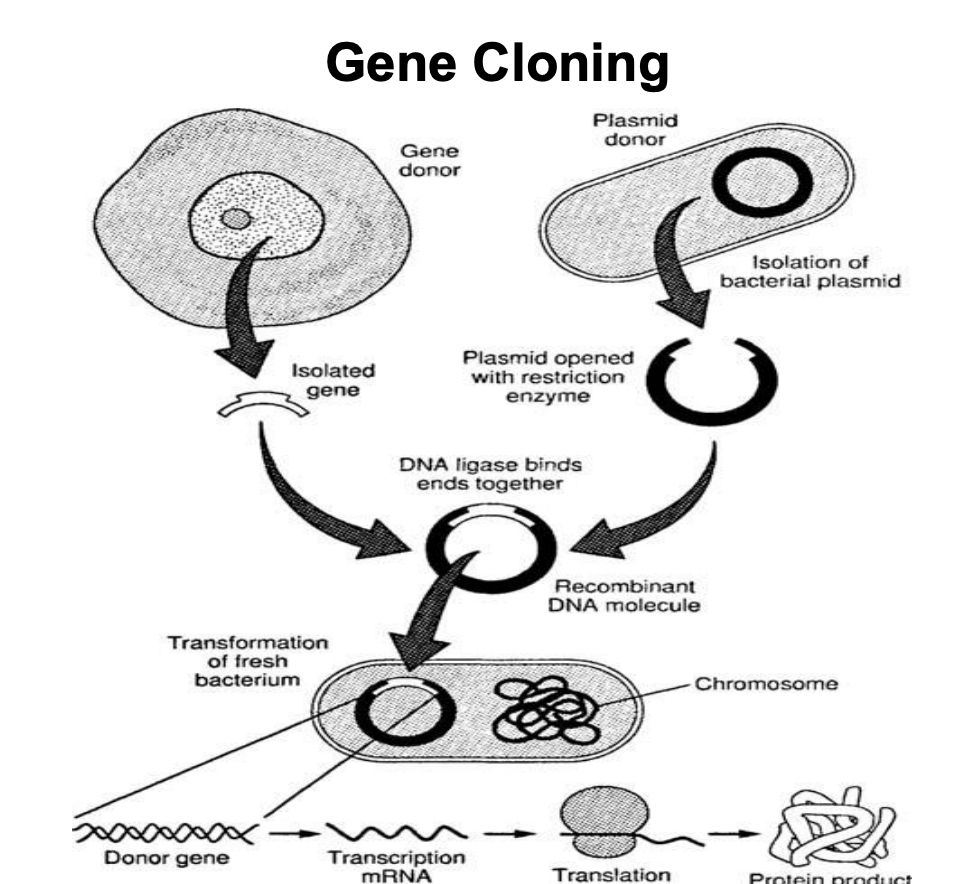
Explain 4 general steps in gene cloning
ilts
Isolation of the gene to be cloned
Ligation of the DNA or gene-of-interest into a vector
Transformation of host cell with recombinant DNA
Selection or screening of host cell with recombinant DNA
Host carrying the recombinant DNA
Recombinant organism
Contains foreign gene / gene-of-interest
Recombinant DNA
T/F: Any organism can be a source of recombinant DNA as long as the sequence of GOI is known
TRUE
3 methods through which isolation of gene to be cloned can be done
rpa
Restriction enzymes
PCR
Artificial gene synthesis
Molecular scissors that accurately and reproducibly cut genomic DNA into fragments called _
Restriction enzymes
Restriction fragments
Has unique recognition sequences; gene (to be cloned/isolated) must be near or between restriction sites
Restriction enzymes
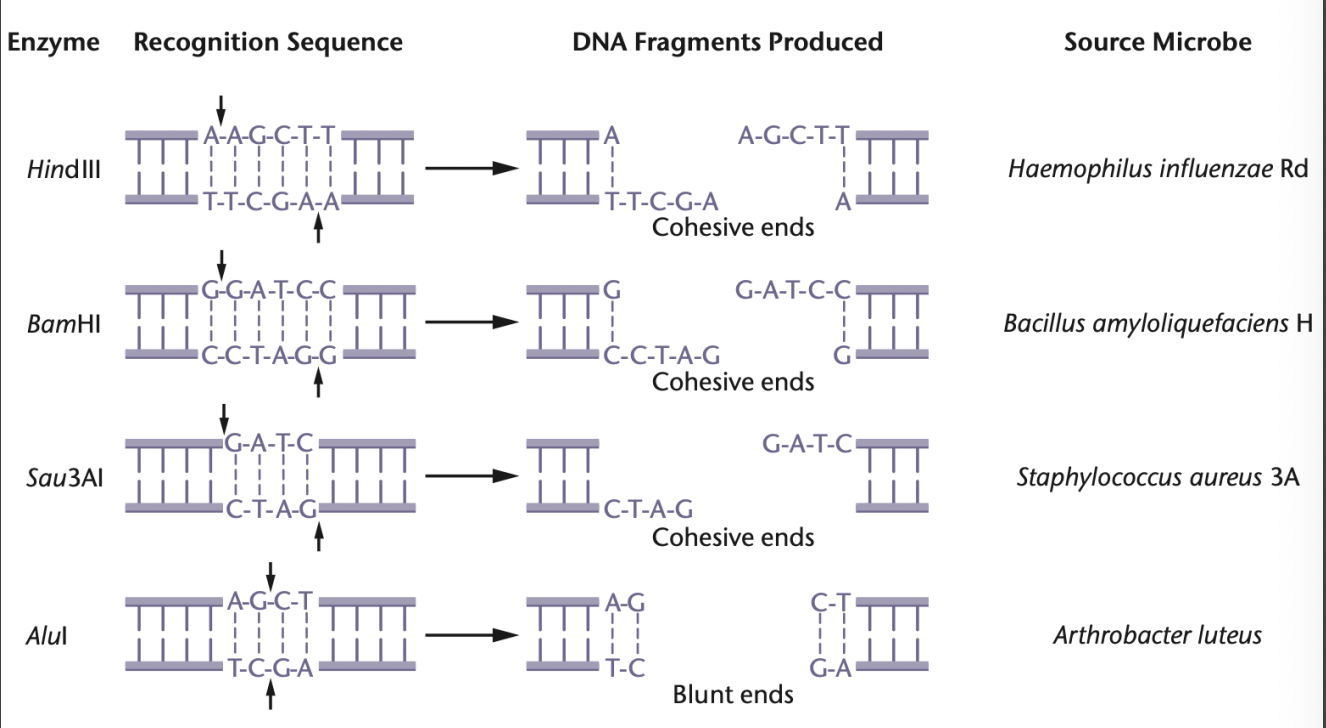
4 examples of restriction enzymes & source microbes
habs
(cohesive ends)
HindIII = Haemophilus influenzae Rd
AluI = Arthrobacter luteus
BamHI = Bacillus amyloliquefaciens H
(blunt end)
Sau3AI = Staphylococcus aureus 3A
Polymerase Chain Reaction (PCR) is a rapid method of _
isolating and copying a gene
Can amplify specific DNA sequences that are present in very small quantities even when they are mixed with many other DNA molecules
PCR
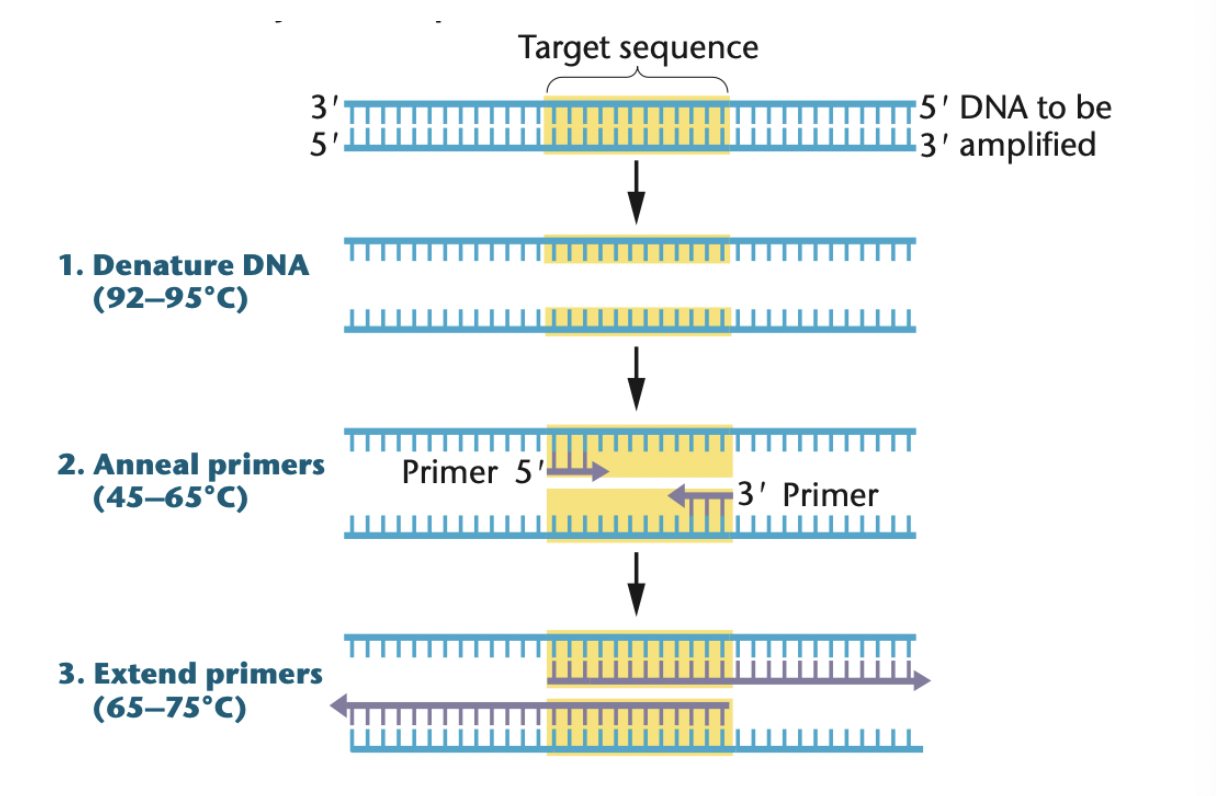
3 steps in PCR amplification
dae
Denature DNA (92 - 95C)
Anneal primers (45 - 65C)
Extend primers (65 - 75C)
_-nucleases are used for restriction enzymes
Endo
If you have a linear DNA with 2 restriction sites, how many restriction fragments will be produced after introduction of RE?
3 RFs
If you have a circular DNA with 2 restriction sites, how many restriction fragments will be produced after introduction of RE?
2 RFs
During PCR denaturation, what type of bonds are broken?
Hydrogen bonds
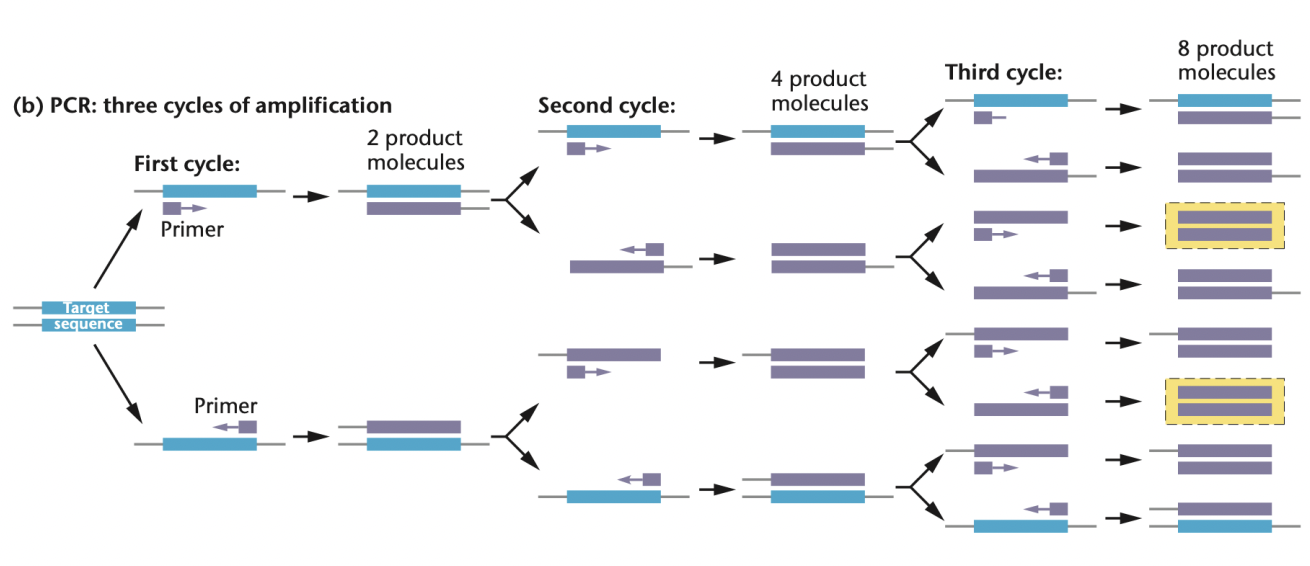
T/F: In PCR amplification, DNA copies increase exponentially
TRUE
T/F: Artificial gene synthesis is also highly reliant on PCR
TRUE
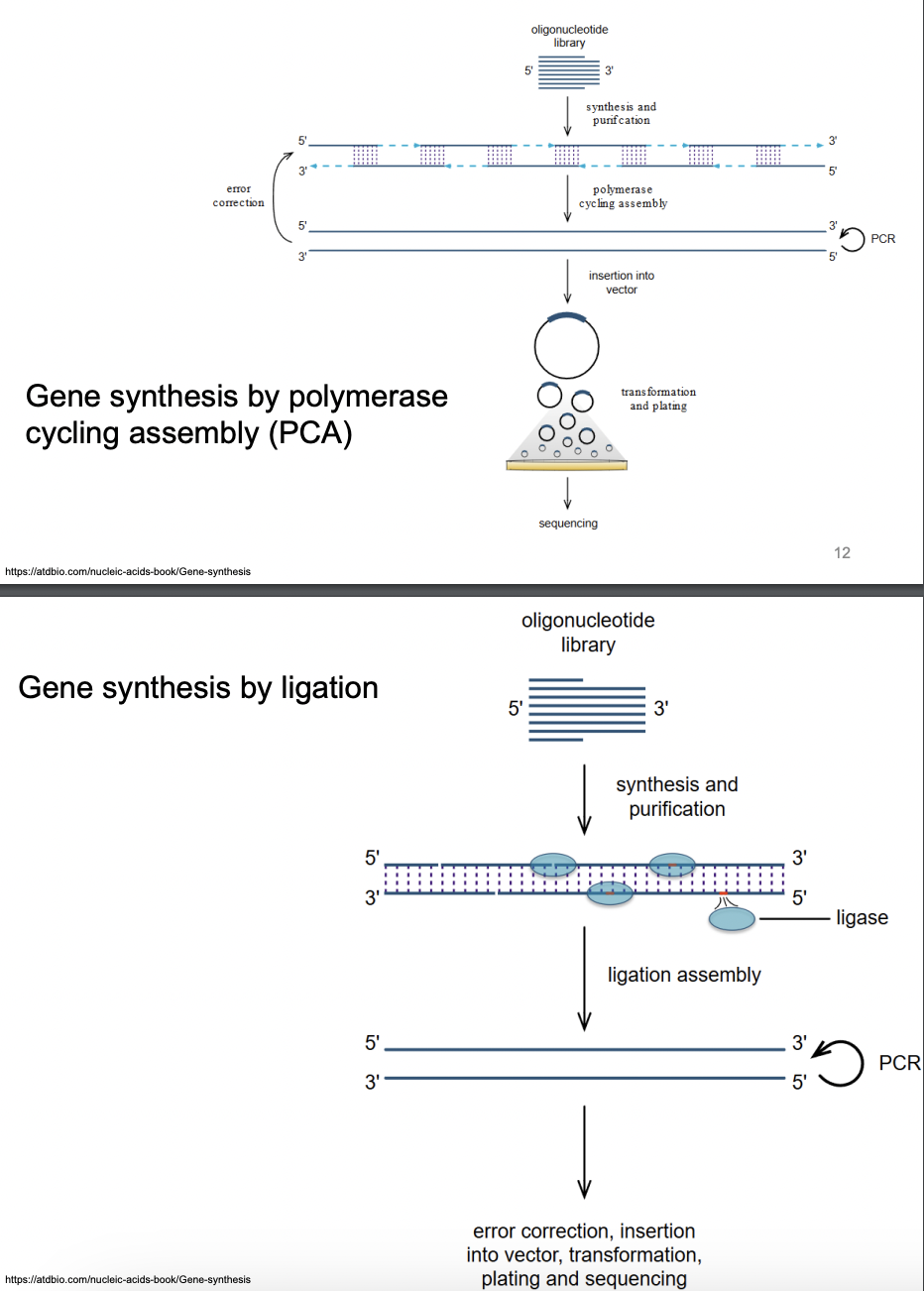
Enumerate, explain 2 ways through which artificial gene synthesis can be done
Polymerase Cycling Assembly (PCA)
Fragment preparation: GOI is cut into smaller fragments with overlapping regions to allow annealing during PCR
PCR: Fragments are amplified in a series of PCR cycles, allowing overlapping regions between fragments to fuse in the process
Assembly: As cycles progress, fragments join together in stepwise fashion, forming full-length genome
Ligation
Short synthetic oligonucleotides are chemically synthesized based on desired genetic sequence
Oligonucleotides are then assembled into larger fragments, which are then ligated together to form full-length genome
Oligonucleotides synthesized are designed to have overlapping regions that can anneal to each other
T/F: DNA ligase creates covalent bonds between sugar and phosphate of adjacent nucleotides
TRUE
What type of bond is formed by ligase?
Phosphodiester bond
Stage where DNA/GOI is inserted into vector
Ligation of DNA into vector
5 kinds of vectors used for DNA ligation into vector
pbcyb
Plasmids
Bacteriophages
Cosmids
Yeast Artificial Chromosomes (YACs)
Bacterial Artificial Chromosomes (BACs)
Vector consisting of cos site of phage + plasmid
Cosmid
Carriers of GOI into host cell; make numerous copies of the gene
Cloning vector
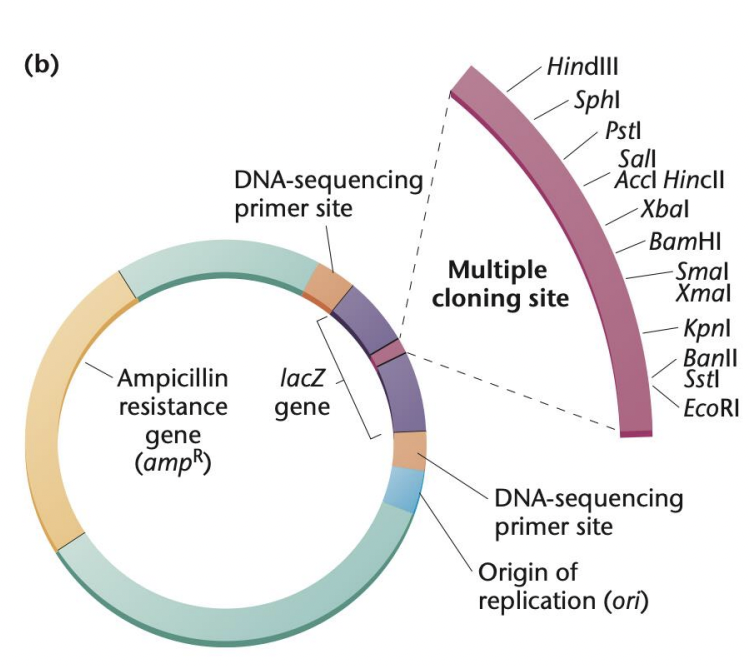
Enumerate parts of cloning vector
ossr
Origin of replication (ori)
Ensures that vector replicates in the host cell
Several restriction enzyme sites
Provides flexibility in inserting GOI at specific sites
Selectable marker gene (antibiotic resistance genes)
Allows identification of successfully transformed host cells
Reporter gene (GFP or lacZ for blue/white screening)
Allows visual or measurable confirmation of GOI expression
This is inserted to observe actual mRNA expression of GOI into mRNA and protein in target organism
Expression vector
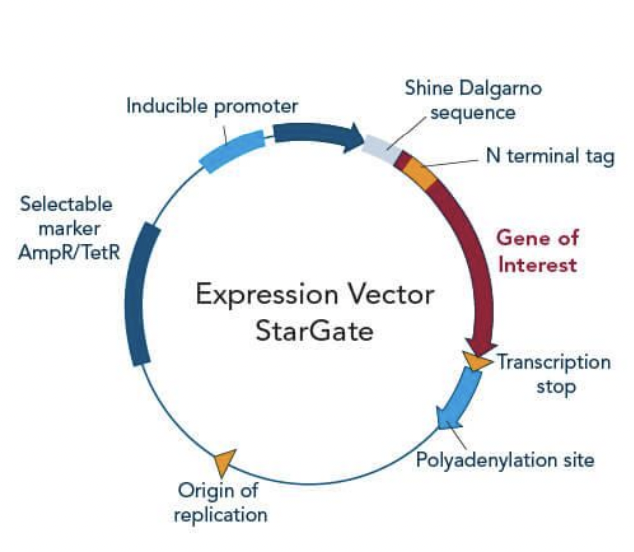
Enumerate parts of expression vectors
Origin of replication
Several restriction enzyme sites
Selectable marker gene
Reporter genes
Regulatory sequences
Drives expression → transcription/translation of GOI in host cells
e.g., Promoters, Shine Dalgarno sequence (RBS)
“Molecular paste” joining DNA fragments with complementary sticky or blunt ends
DNA ligase
DNA ligase creates _ bonds
Phosphodiester bonds
Covalent bonds between sugar and phosphate of adjacent nucleotides

Explain general process for producing recombinant DNA
Restriction enzyme cuts sugar-phosphate backbones in specific restriction sites, producing complementary sticky/blunt ends
DNA/GOI cut by the same RE from another molecule is then added
Base pairing occurs
DNA ligase seals strands
Recombinant DNA is formed by _
Cutting plasmid vector with RE
Cutting and isolating GOI with same RE from donor cell
Ligating those 2 DNAs to form recombinant molecule
Stage where recombinant DNA/plasmid is inserted into host cells; replicates within host cells, producing dozens of identical copies/clones
Transformation of host cell with recombinant DNA
Enumerate and explain pros & cons of molecular cloning hosts
Bacteria
Escherichia coli wmm pp
Pros
Well-developed genetics
Many strains available
Most studied bacterium
Cons
Pathogenic
Periplasm traps proteins G(-)
Bacillus subtilis enne gg
Pros
Easily transformed
Nonpathogenic
Naturally secretes proteins G(+)
Endospore formation simplifies culture
Cons
Genetically unstable
Genetics is less developed than E. coli
Eukaryotes: Saccharomyces cerevisiae wnce pw
Pros
Well-developed genetics
Nonpathogenic
Can process eukaryotic mRNA
Easy to grow
Cons
Plasmids unstable
Will not replicate most bacterial plasmids
Enumerate and explain 6 common gene transfer methods
tbmect
Transformation: uptake of environmental naked DNA by competent cells
Biolistics: direct delivery of DNA-coated microprojectiles using high-velocity gene guns
Microinjection: direct delivery of DNA using fine needles
Electroporation: use of electrical pulses to create temporary pores in host cell membrane, allowing DNA entry
Conjugation (e.g., allelic exchange): transfer of DNA between cells via direct contact (pilus)
Transduction: transfer of DNA between cells via bacteriophage

Process where recombinant DNA within host cells are passed onto their progeny, creating population of host cells carrying cloned DNA sequence
Replication of host cells
Stage where colonies that were successfully genetically transformed are identified/determined
Screening or selection of host cells with recombinant DNA
T/F: Genetic transformation is a ‘hit or miss’ strategy
TRUE
Screening process could either be:
Detection/isolation of the GOI via PCR, RE digestion
Expression of selectable marker and reporter gene
e.g., lacZ gene for blue/white screening
lacZ gene encodes B-galactosidase, which leaves lactose analogs, including X-gal (chromogenic)
When X-gal is cleaved = blue colonies are expressed, which means
In case of successful transformation (insertion of GOI into vector, disrupting lacZ) = white colonies must be observed
Hence, failure to transform and thus insert GOI into vector, leaves lacZ functional (ultimately cleaving X-gal) = blue colonies
Corrosions accelerated by microorganisms
Microbially influenced corrosions (MICs)
2 classifications of MICs
Metal corrosions
Crown corrosions
Type of MIC
Iron, steel
Microbes implicated sffm
Sulfate-reducing bacteria
Ferrous iron (Fe2+) -oxidizing bacteria
Ferric iron (Fe3+) -reducing bacteria
Methanogens (CO2 + H → CH4 + H2O)
Metal corrosion
Rapid form of microbial biodeterioration
Crown corrosion
Crown corrosions are often observed in _
concrete sewer tiles
A consequence of interactions between sulfate-reducing and sulfur-oxidizing bacteria
Crown corrosion
Can lead to collapse of underground wastewater transmission pipe systems
Crown corrosions
T/F: In observation of transformation, E. coli needs to be lysed to express proteins
TRUE
T/F: Proteins expressed using B. subtilis as host will be observed in cells
FALSE
Proteins expressed using B. subtilis as host will be observed in medium
T/F: All antibiotic-resistant bacteria observed are successfully transformed
FALSE
Possible that vector plasmid was empty, i.e., did not contain GOI
Explain MIC mechanism of iron corrosion by SRB (via increasing acidity/lowering pH)
SRB produce H2S through sulfate-reduction (SO42-)
This produced H2S then reacts with Fe2+ in the iron metal, producing FeS + H+
H2S + Fe2+ → FeS + H+
Increase in H+ leads to more acidic environment that initiates and accelerates iron corrosion

Explain MIC mechanism of iron corrosion by SRB (accelerated via bacterial consumption of H2)
When iron Fe0 oxidizes in anodic region, it forms Fe2+, leaving behind a corrosion pit
Electrons produced from oxidation then migrates to cathodic region, where water dissociates into protons + hydroxide ions
Protons are then utilized by SRB to reduce sulfate, producing H2S, which then again reacts with Fe, producing more H+ that further acidifies environment, accelerating iron metal corrosion
H2 + SO42- → HS-
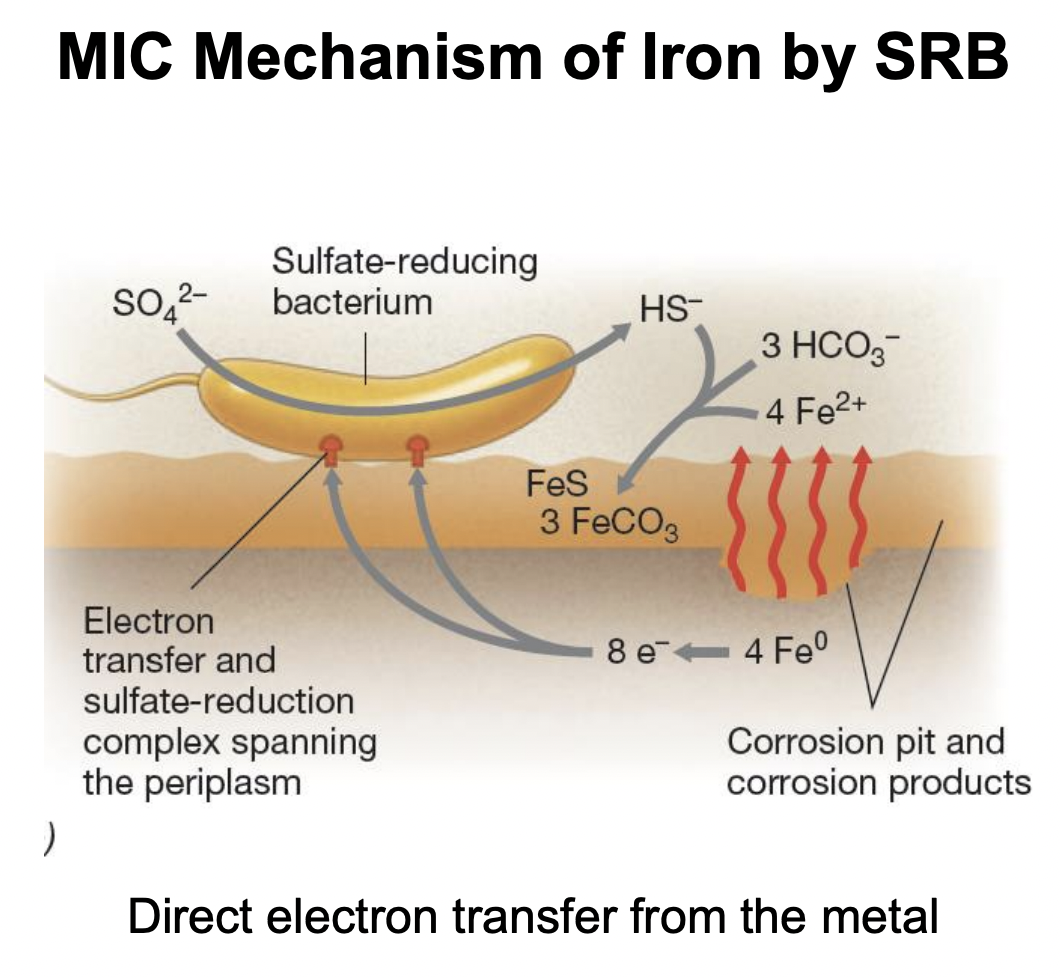
What compounds indicate iron metal corrosion?
Ferrous iron (Fe2+)
Bicarbonate (HCO3-)
Hydrosulfide (HS-)
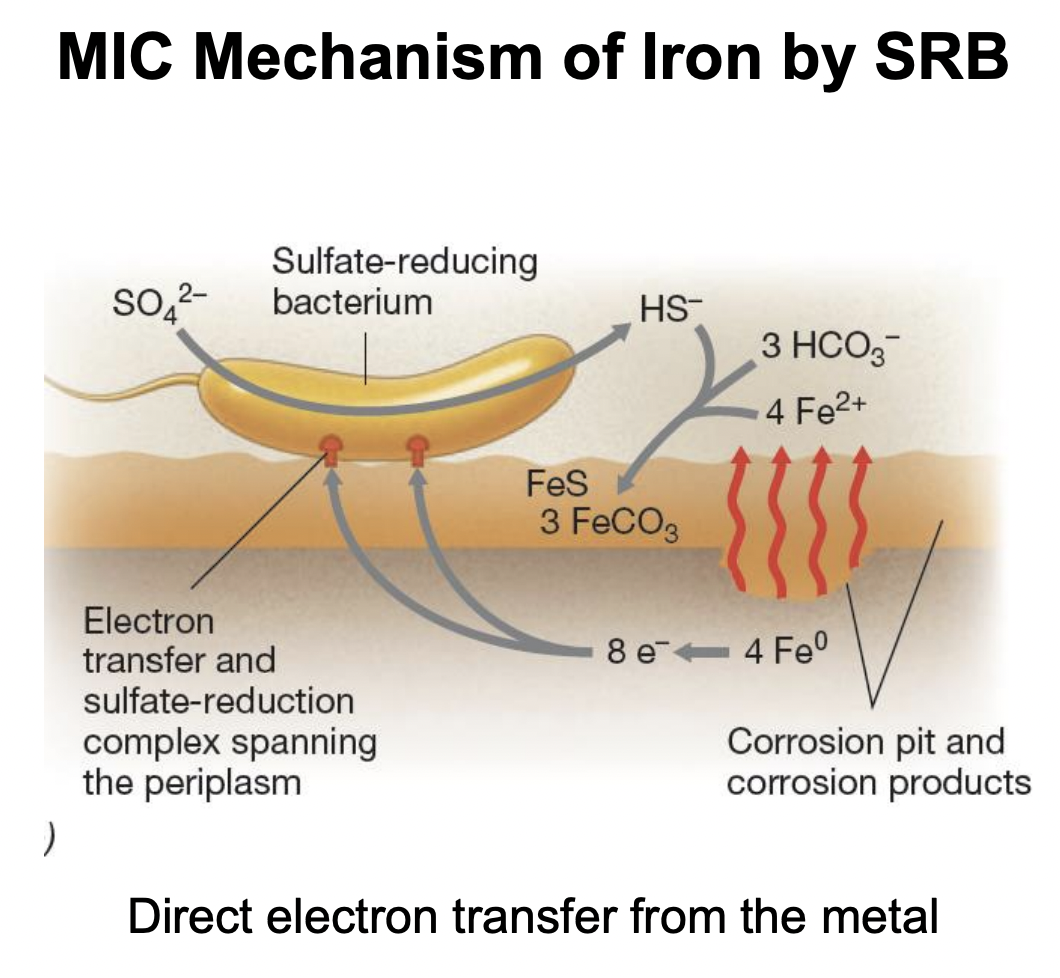
Explain MIC mechanism of iron corrosion by SRB (accelerated via direct electron transfer from metal)
Fe0 oxidizes into Fe2+, forming corrosion pits
Fe0 directly donates electrons to SRB via periplasmic electron transfer complexes
Electrons are then used by SRB to reduce sulfate SO42- → HS-
HS- reacts with Fe2+ → FeS + H, accelerating corrosion
T/F: Corrosion only happens anaerobically
FALSE
Iron-oxidizing bacteria (IOB) do it aerobically
T/F: Iron-oxiding bacteria are anaerobes
FALSE
Aerobes

Explain MIC mechanism of iron by IOB
(A) Role of IOB in iron corrosion
Fe0 oxidizes into Fe2+
IOB catalyzes oxidation of Fe2+ → Fe3+
IOB utilize O2 + H2O → OH-, which increase alkalinity → accelerating iron corrosion (in oxic environments)
(B) Interaction between SRB & IOB
IOB → Fe2+
SRB (via direct electron transfer from iron, consumption of H+ from cathodic region) reduces SO42- into HS-, which can then react with Fe2+ → FeS, accelerating corrosion
(C) Cathodic, anodic regions
Fe0 oxidizes into Fe2+ in anodic regions
SRB reduces SO42- → HS- in cathodic regions
Both combine to create localized corrosion pits
Explain MIC mechanism of iron corrosion by IRB
IRB reduces Fe3+ → Fe2+ under anaerobic conditions
IRB reducing Fe3+ → Fe2+ ,which can then react with H2S → FeS + H+, further accelerating corrosion
T/F: IRB are anaerobes, IOB are aerobes
TRUE
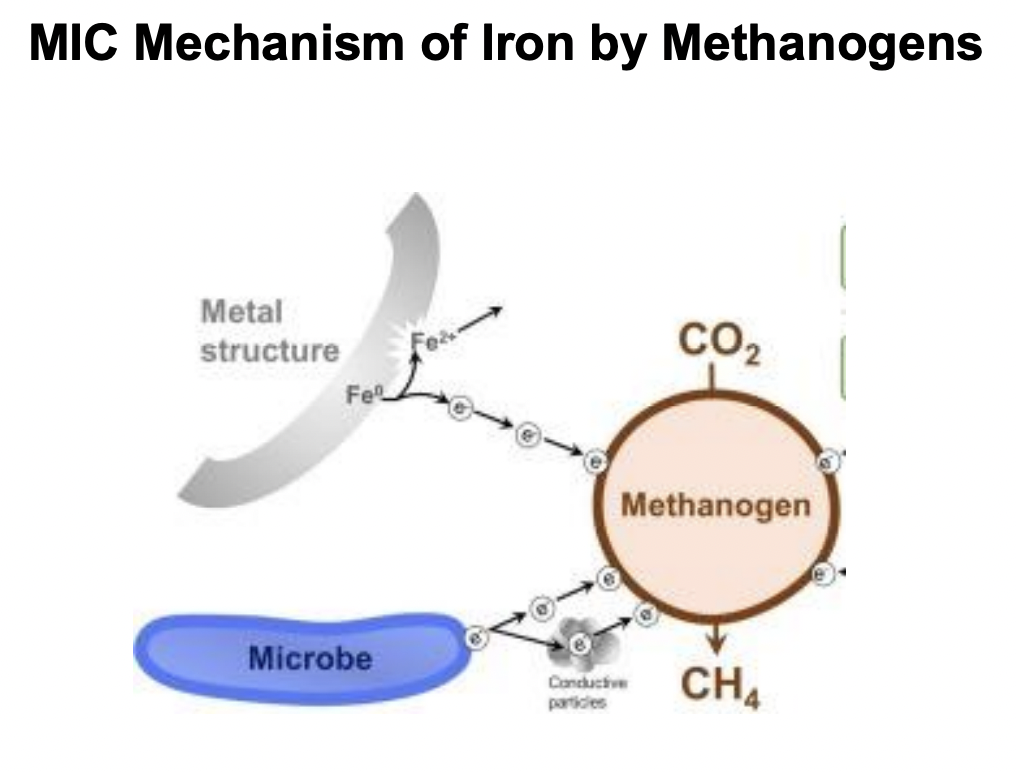
Explain MIC mechanism of iron corrosion by methanogens
Electrons released from iron oxidation is used by methanogens to reduce CO2 → CH4
Fe0 oxidation → Fe2+, which can then react with H2S → FeS + H+, further accelerating corrosion
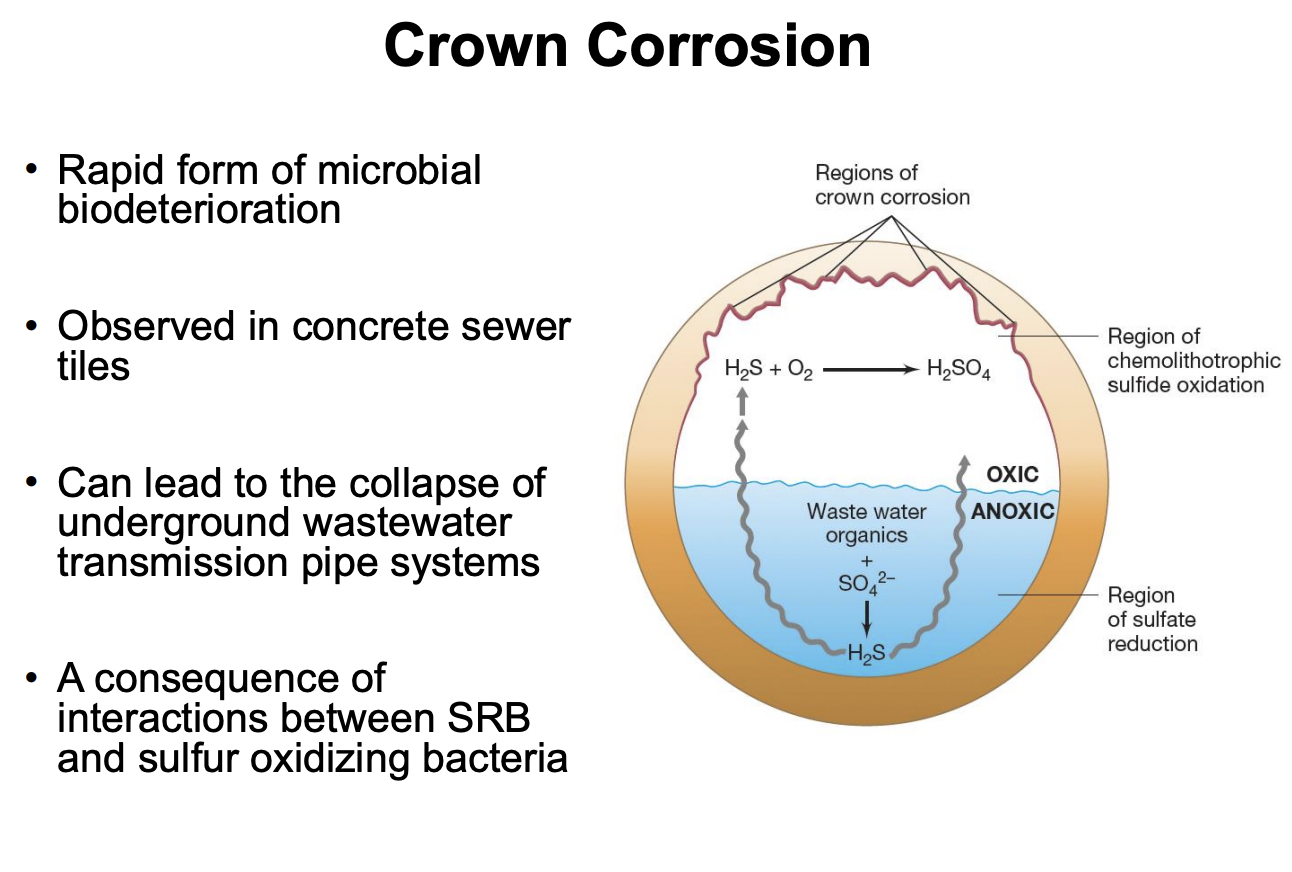
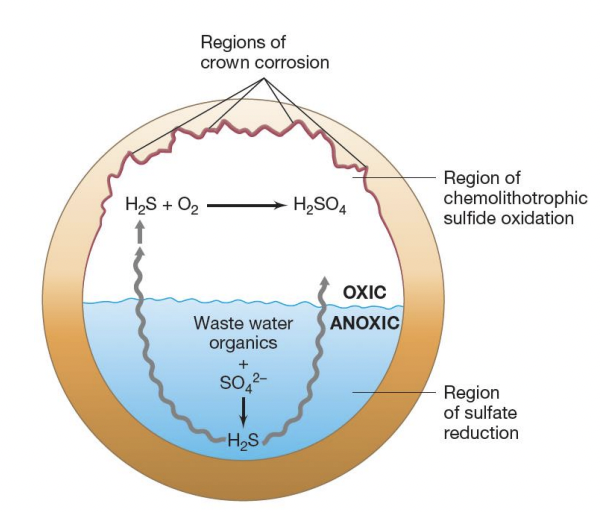
Explain crown corrosion
In anoxic region (submerged in water), SRB reduces SO42- → H2S, which is then released up towards crown
In oxic region, SOB then oxidizes H2S, forming sulfuric acid
H2S + O2 → H2SO4
Sulfuric acid → lowers pH, further accelerating corrosion
T/F: SOB alone can cause crown corrosion
FALSE
SRB produces H2S through sulfate reduction. H2S is then oxidized by SOB, producing H2SO4 that acidifes environment
T/F: SRB is aerobic, SOB is anaerobic
FALSE
SRB is anaerobic, SOB is aerobic