BS3013 - Week 8: Enzymes and Drug Targets
1/55
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
56 Terms
What is the main function of an enzyme?
to lower the activation energy — thereby accelerating the reaction rate
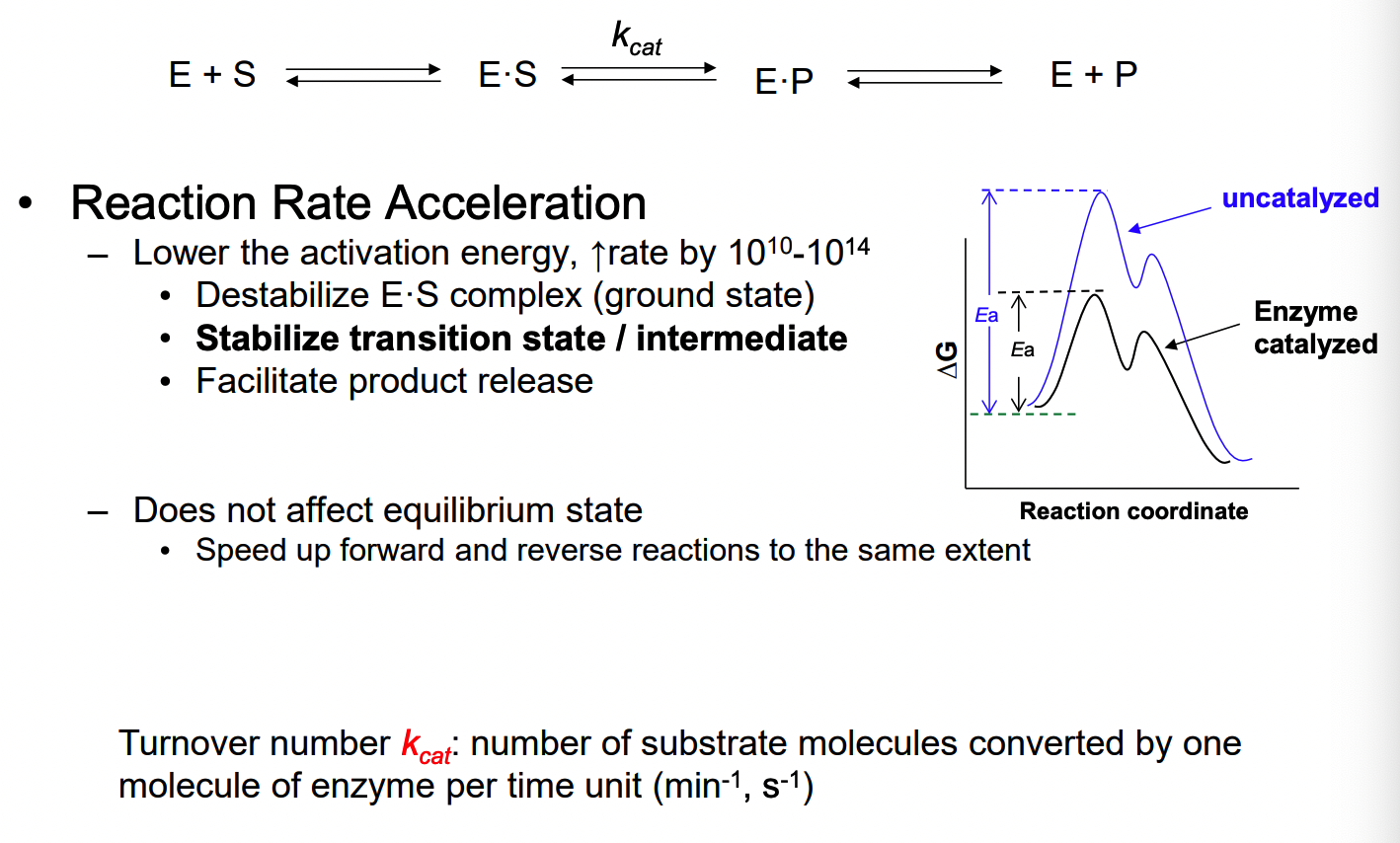
black curve = enzyme catalyzed reaction
blue = uncatalyzed: high activation energy (transition state with the highest energy)
What 2 specificities are involved in an enzymatic reaction?
binding and reaction specificity
binding: recognizes certain substrates only
reaction: catalyze certain type of reaction only
ex. trypsin = cleaves peptide bone after basic amino acid
Protease = hydrolyzing peptise
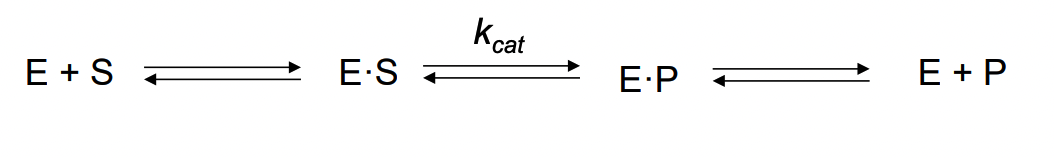
What is involved in binding specificity?
certain constituents at active site are involved in binding interactions
binding to substrate, transition state, intermediates, product
max binding interactions with transition state: up to ~ 10^12 times more tightly than with substrate or product
What does reaction specificity include?
rxn: arises from specific acid, base and nucleophilic groups of amino acids
What are the 6 components of enzyme catalysis?
approximation
bring reacting partners to proximity, increasing EM
Covalent catalysis
nucleophilic catalysis: active site amino acid side chain functional groups attach substrate and form covalent bond to it
general acid-base catalysis
active site: acid/base
electrostatic catalysis
ionic charge or dipole interacting with opposite charge developing on the substrate at the transition state of the reaction
Desolvation:
removal of water molecules from charged groups at the active site on substrate binding → ground state destabilization/transition state stabilization
strain or distortion:
binding of substrate to the enzyme inducing a conformational change in the active site - deformation of the enzyme and/or the substrate, leading to strain (destabilization, higher ground energy)
What are the 4 types of proteases?
serine protease
systeine protease
aspartyl protease
metalloprotease
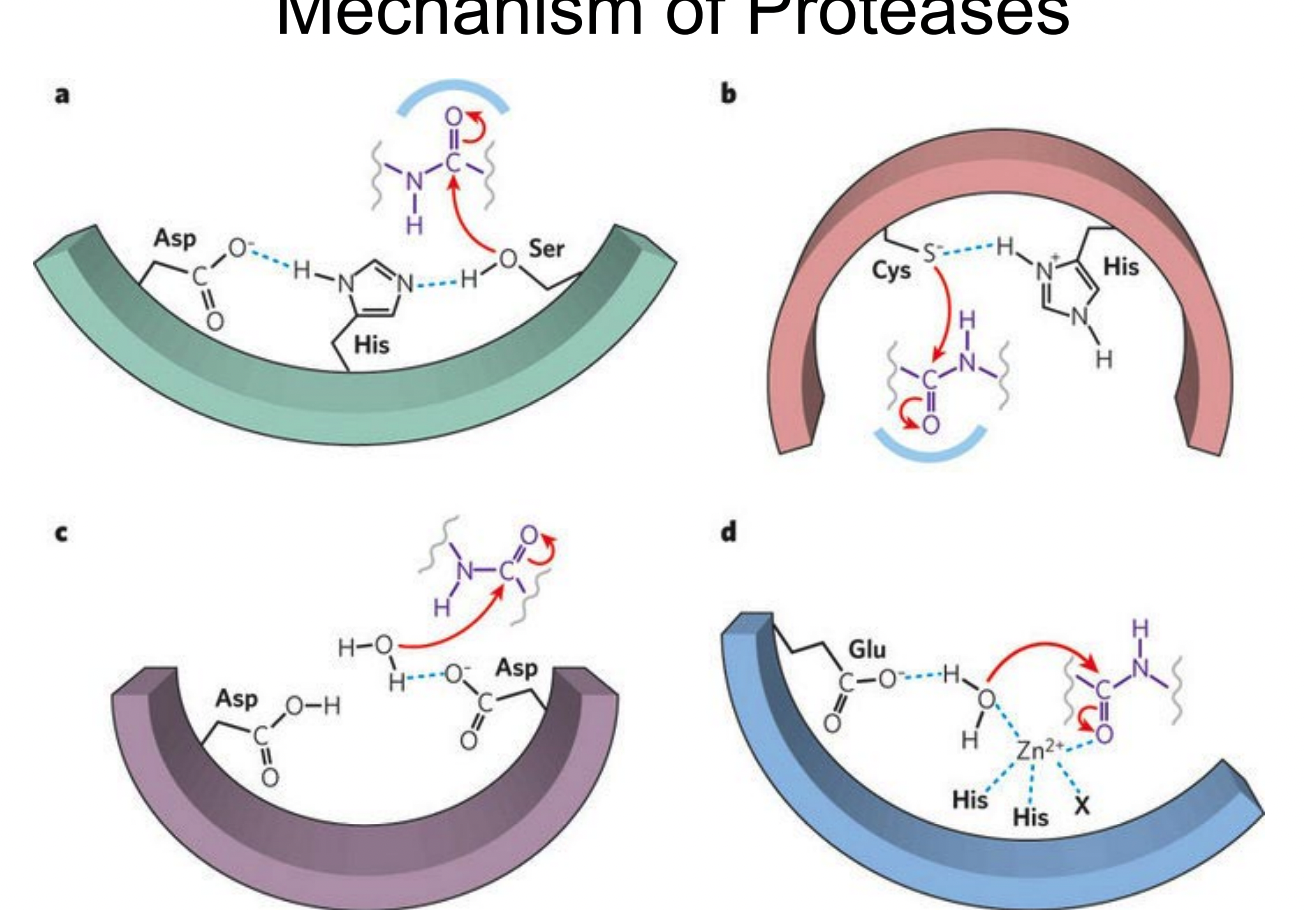
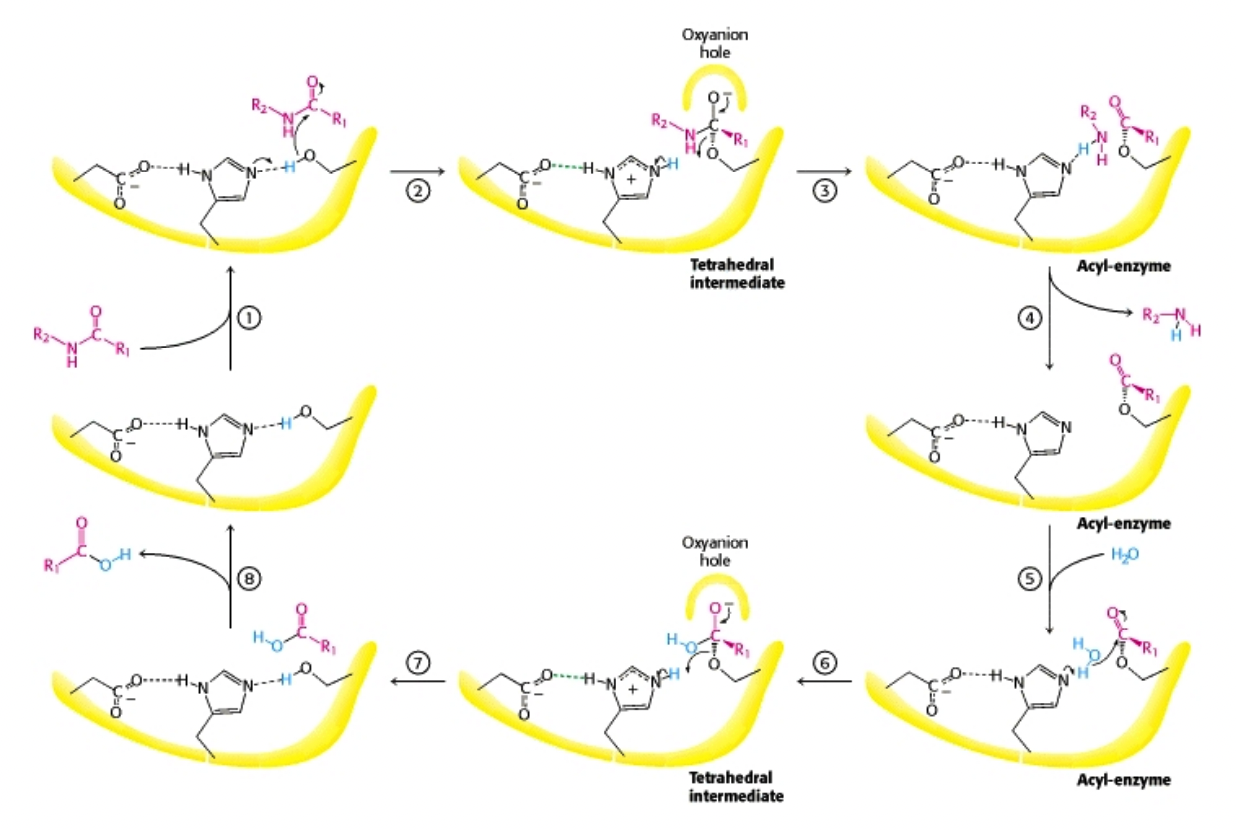
What is the mechanism of serine protease-catalyzed hydrolysis?
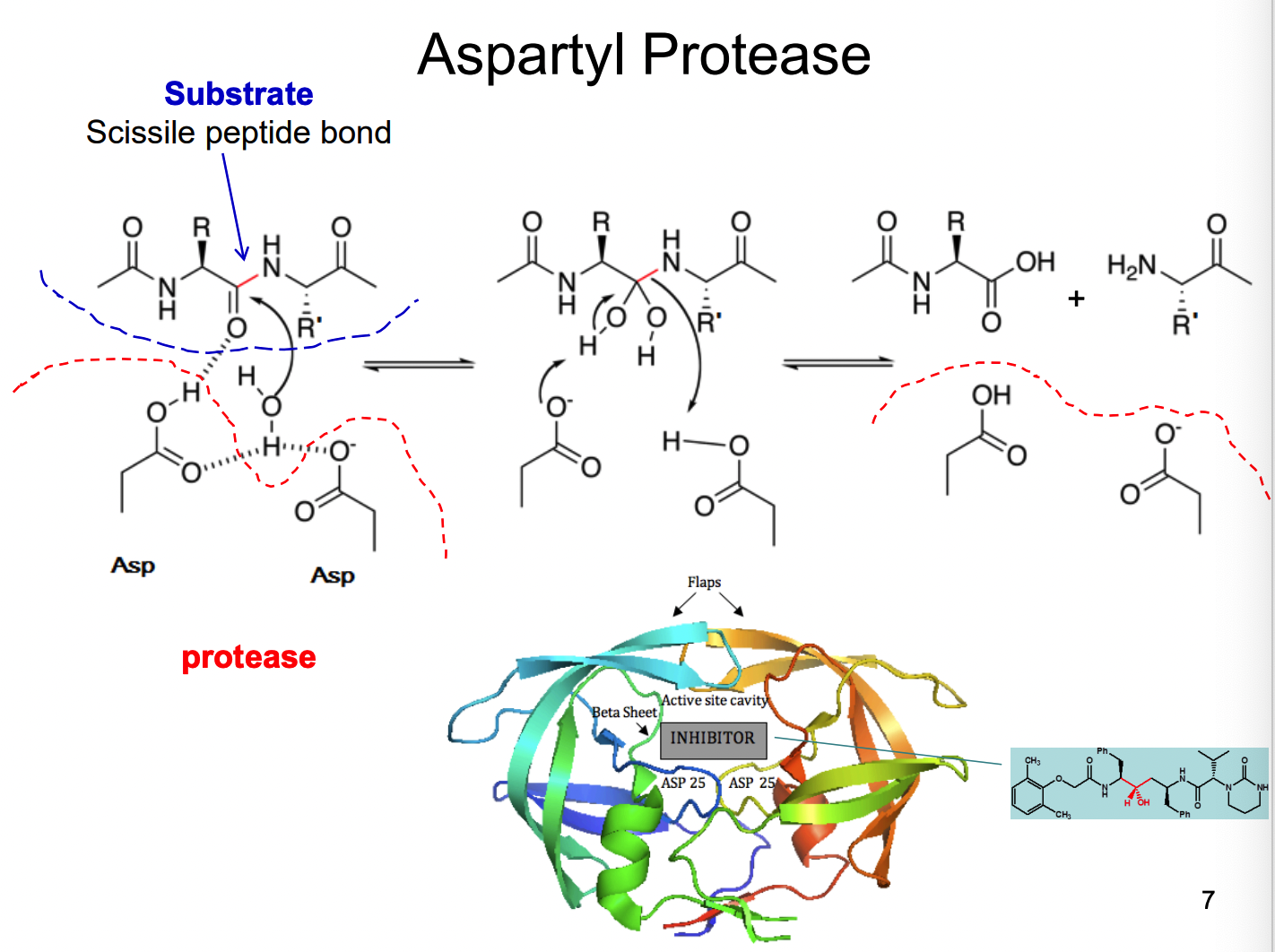
How does Aspartyl protease work?
What is an enzyme Commission Number system?
depending on the type of enzyme they start with a number

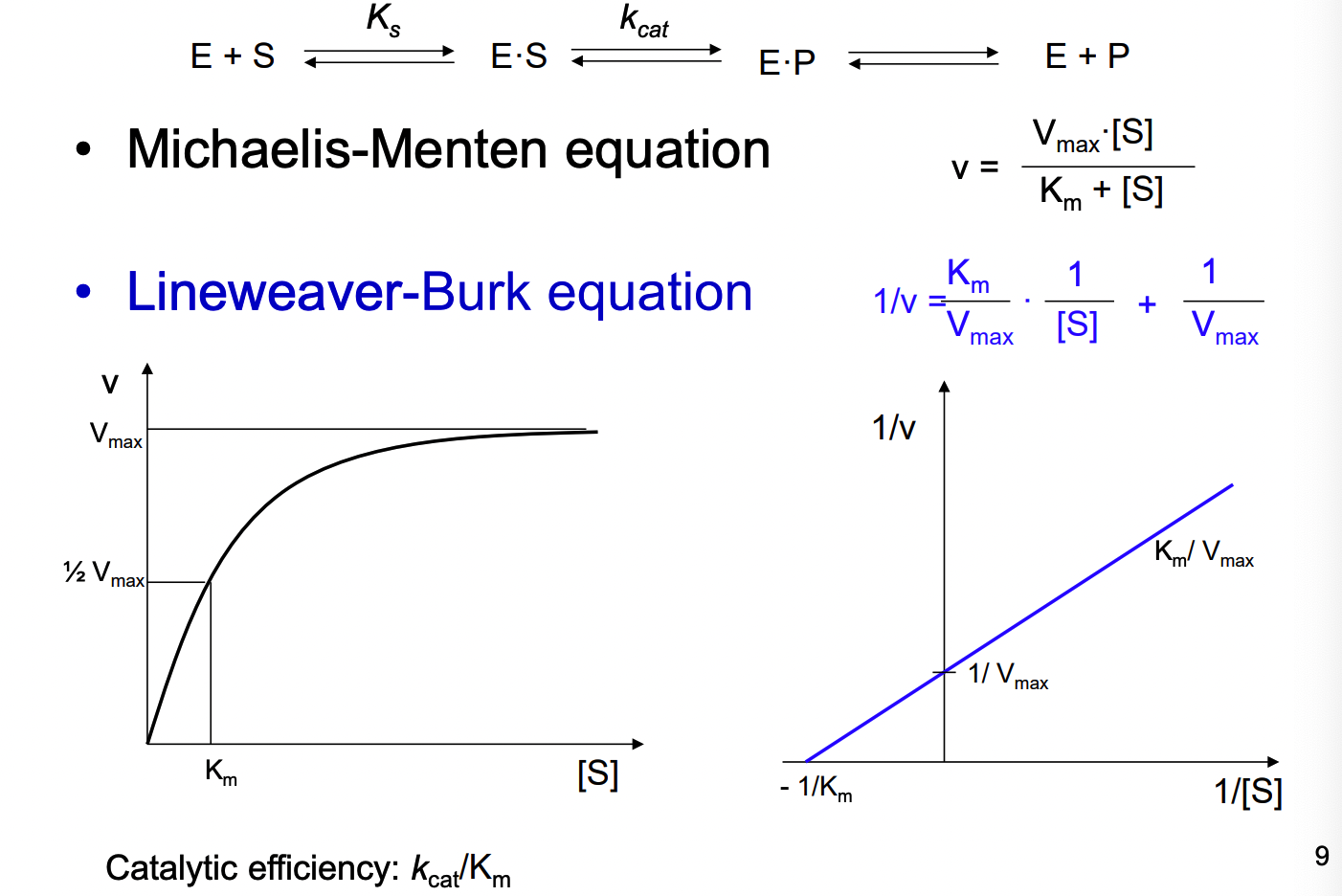
What 2 equations are used relating to enzyme kinetics
michaelis-menten equation
lineweaver-burk equation
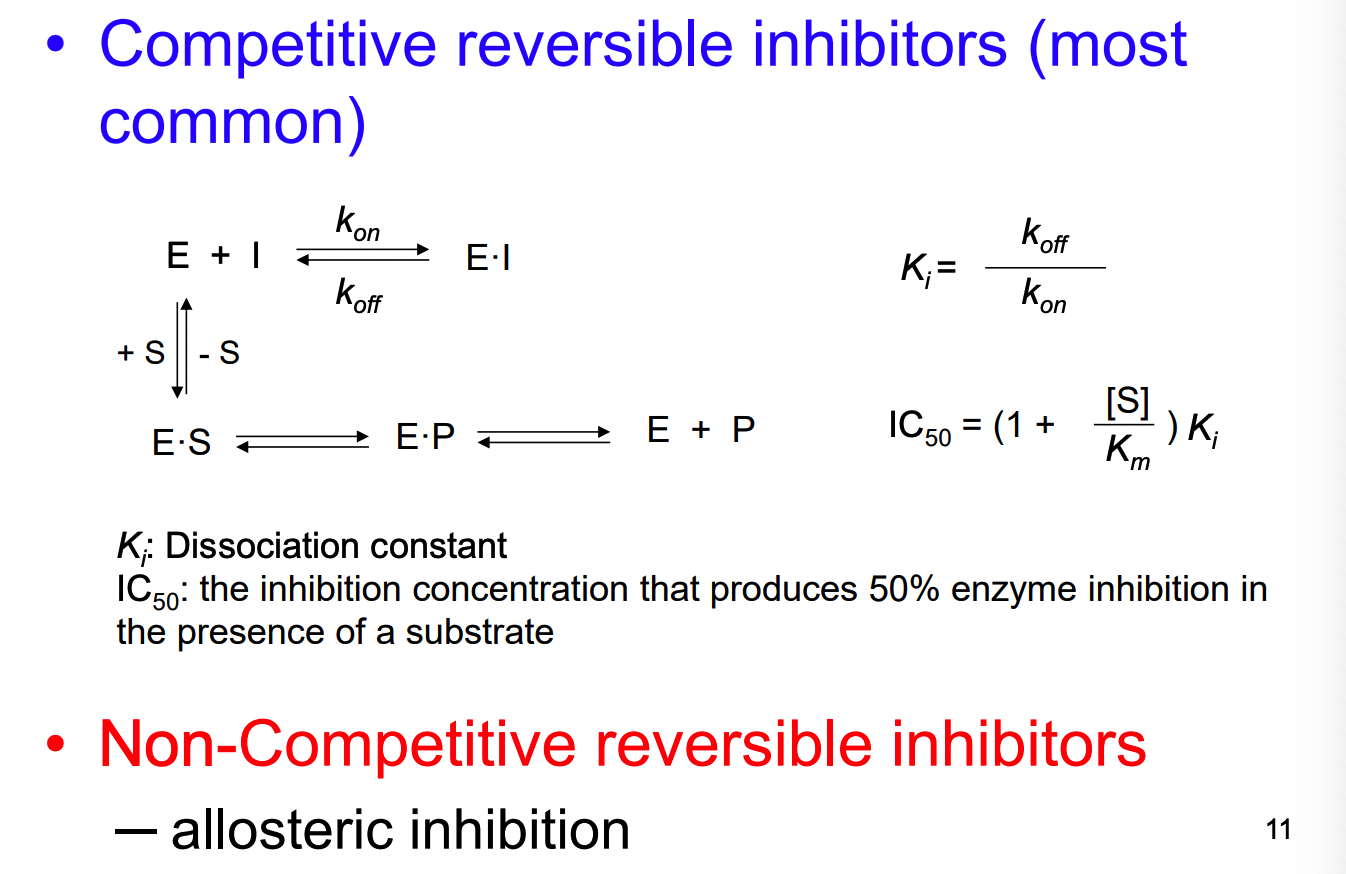
What are the 2 main types of enzyme inhibition?
competitive - same site
non-competitive - allosteric site
** both reversible or irreversible
When enzymes are reversible what does this mean?
hat are the 4 types of inhibition of a competitive reversible inhibitor?
simple competitive: designed structure resembles substrate
alternative substrate: binds to enzyme and acts as substrate
products are useless to the organism
transition state analog: design molecule to resemble substrate at transition state of the reaction (this type would bind much more tightly to the enzyme)
slow, tight-binding: compound with tight binding to the enzyme and equilibrium of binding between it and the enzyme = reached slowly
inhibition time dependent (reminiscent of the kinetics of irreversible inhibitors)
What is an example of simple competitive inhibition?
inhibitors of angiotension-converting enzyme (ACE)
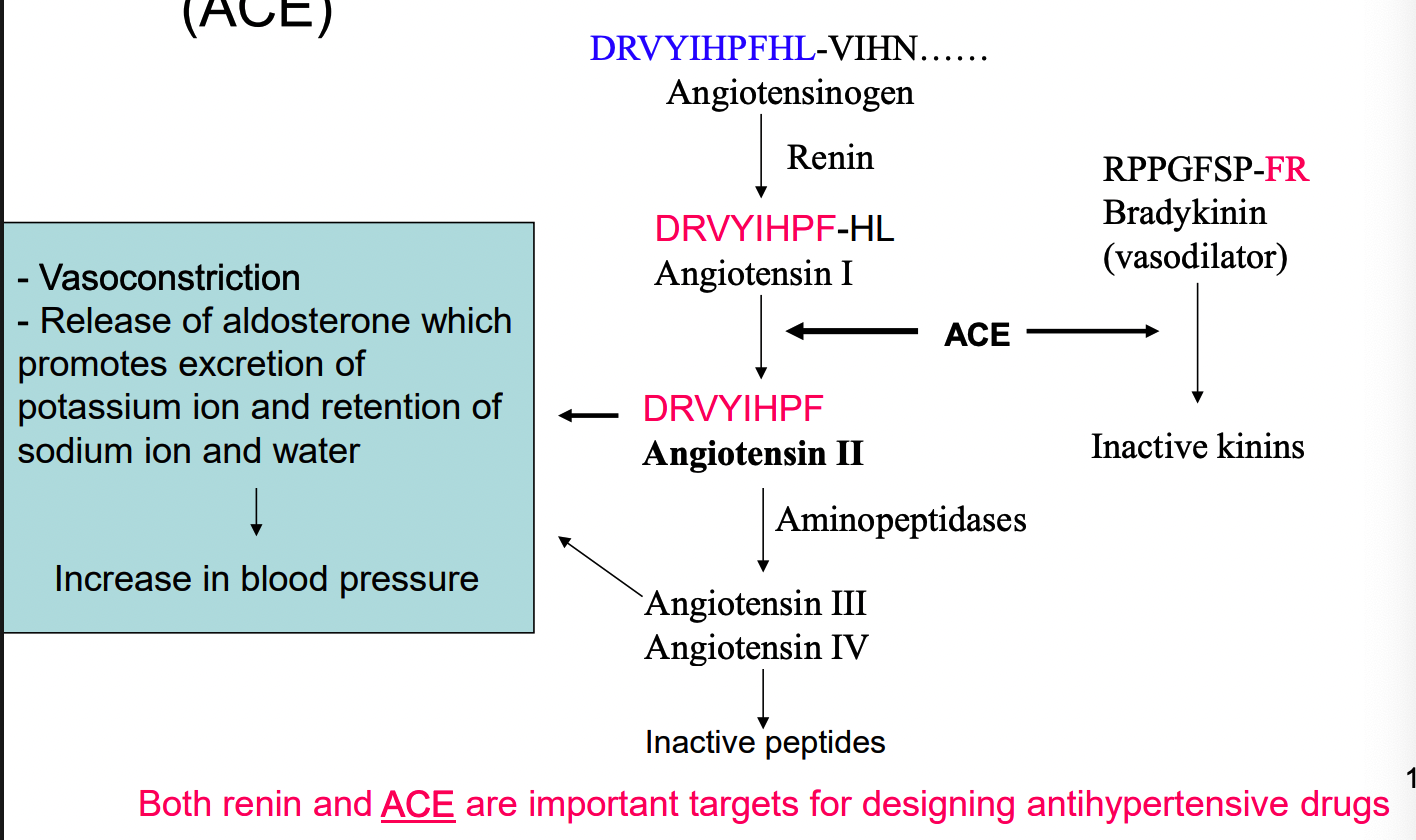
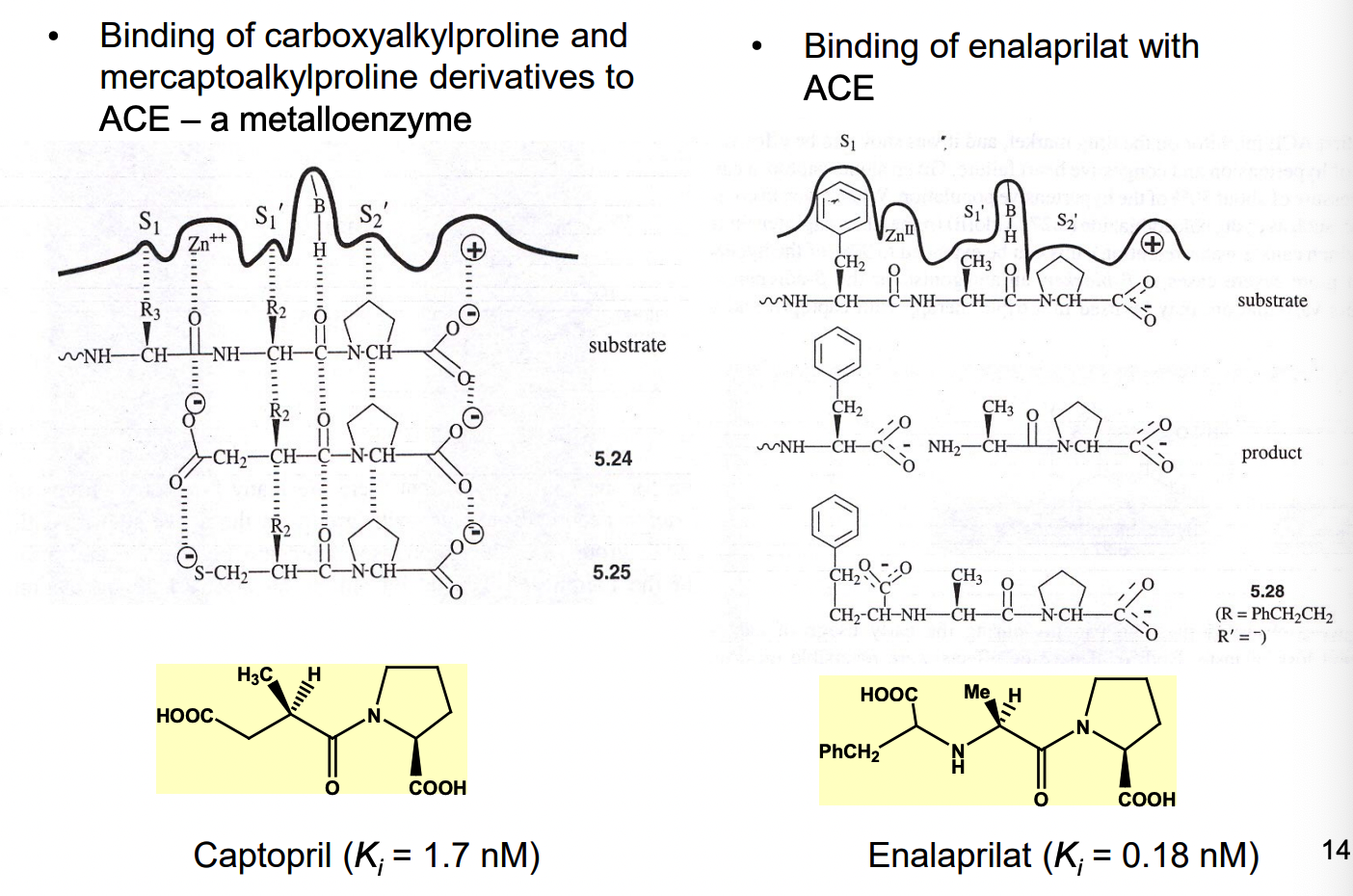
what are 2 examples of substrate analogs as ACE inhibitors?
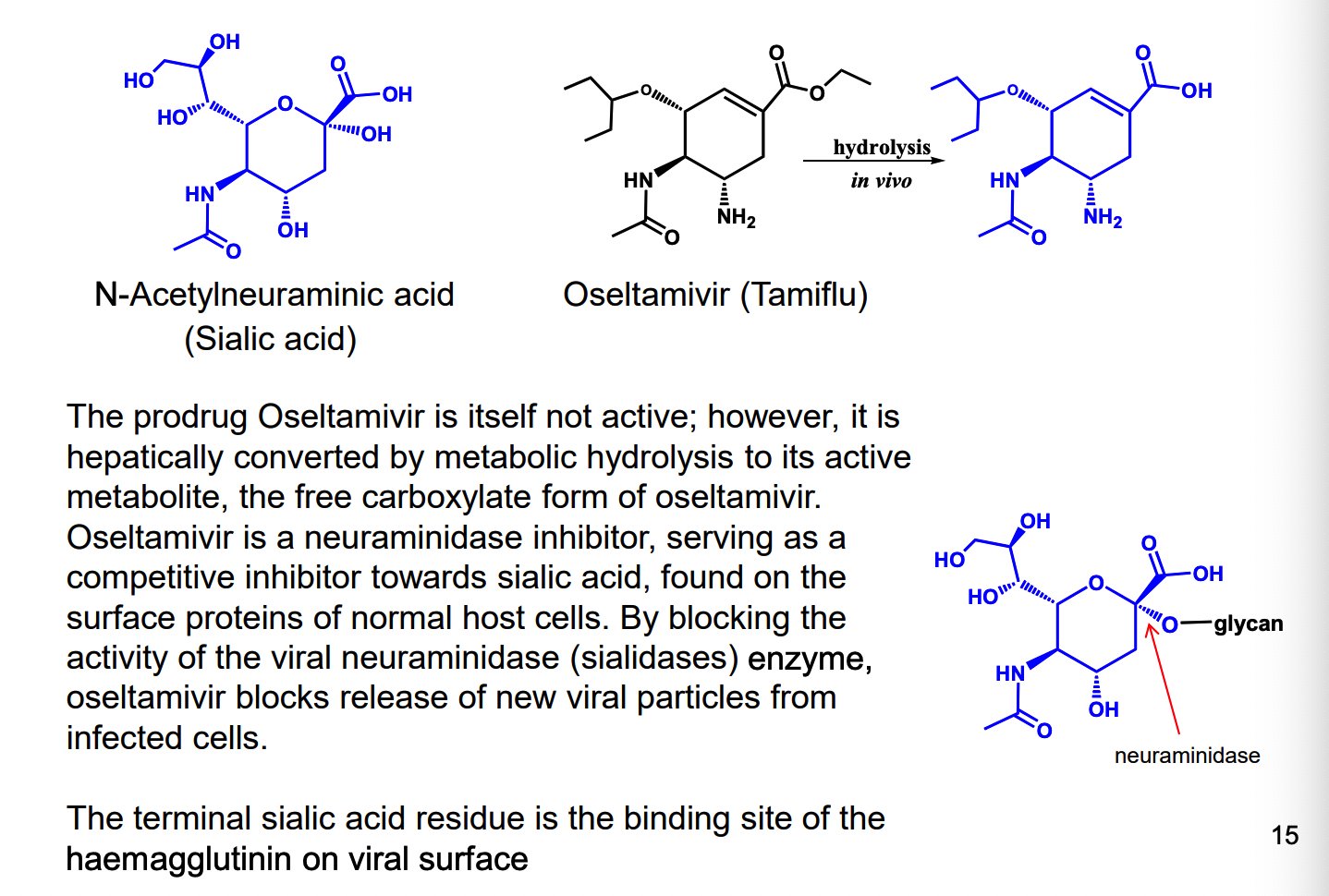
What are examples of neuraminidase inhibitor?
Oseltamivir (the free carboxylate form)
competitive inhibitor of sialic acid on surface proteins of normal host cells
block viral neuraminidase enzyme, oseltamivir blocks release new viral particles form infected cells
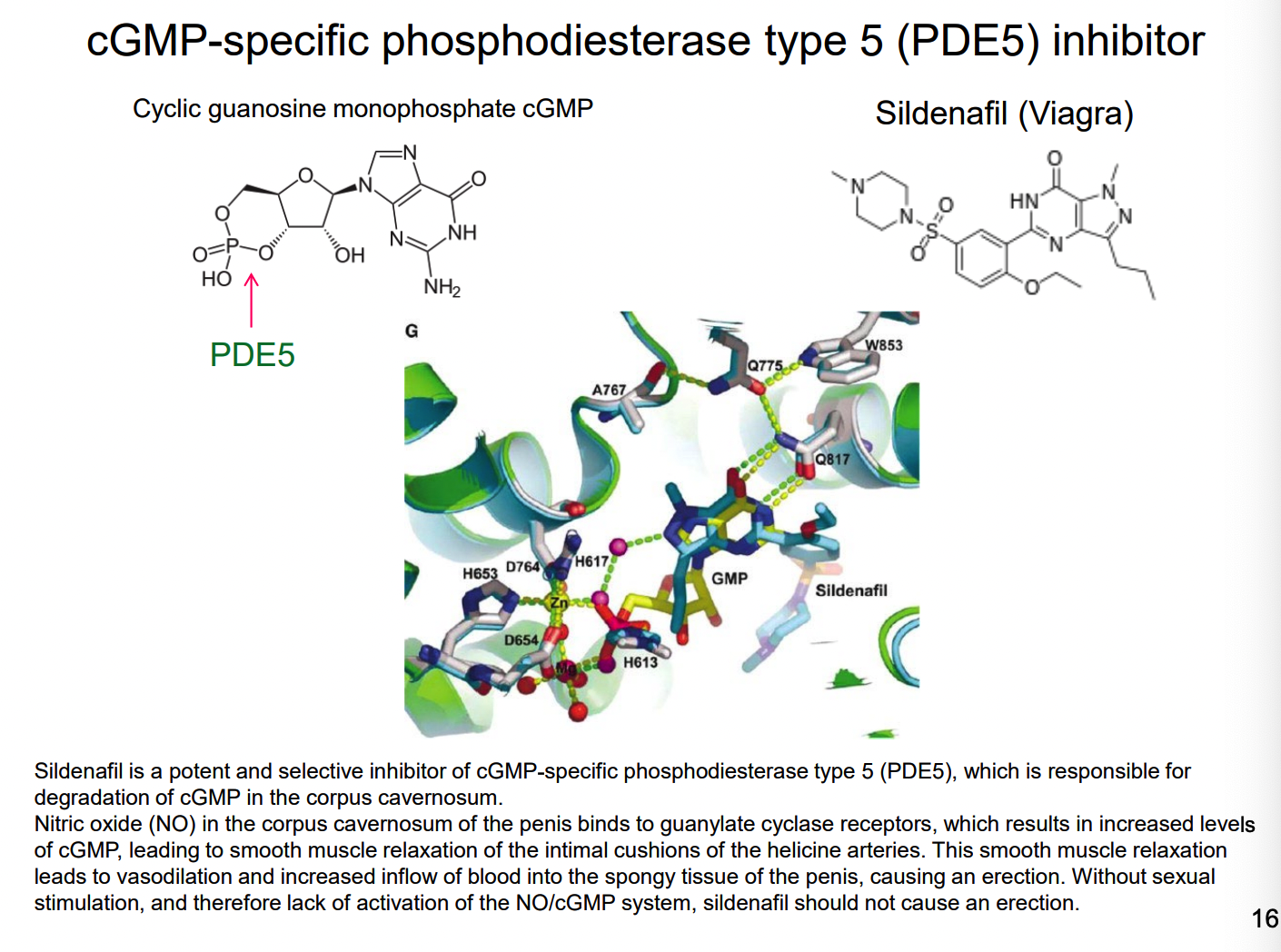
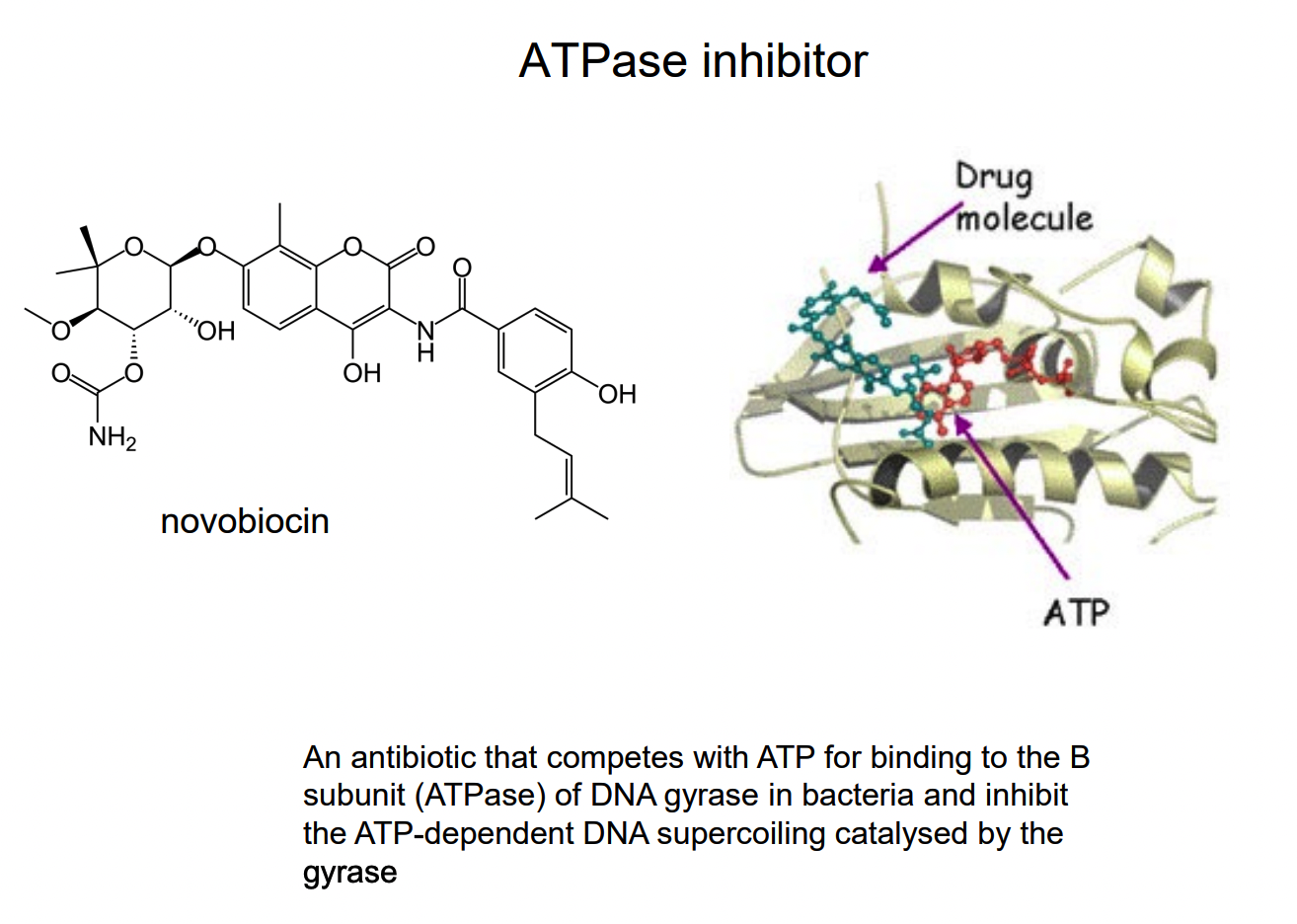
What is an alternative substrate inhibitor?
where the inhibitor is a substrate itself, but the products produced = useless
preventing the natural substrate from being converted to the product that the organism needs
con: product produced could be toxic/side effects
drug is “consumed” constantly
ex. sulfonamide antibacterial agents (sulfa drugs)

What is Prontosil? How does it work?
1st sulfonamide antibacterial agent (sulfa drug)
active principle generated by reductive metabolism reaction
bacteriostatic agent — inhibits further growth of the bacteria
many sulfa drugs = developed based on the p-aminobenzenesulfamide structure
How does sulfa drugs inhibit folic acid biosynthesis?
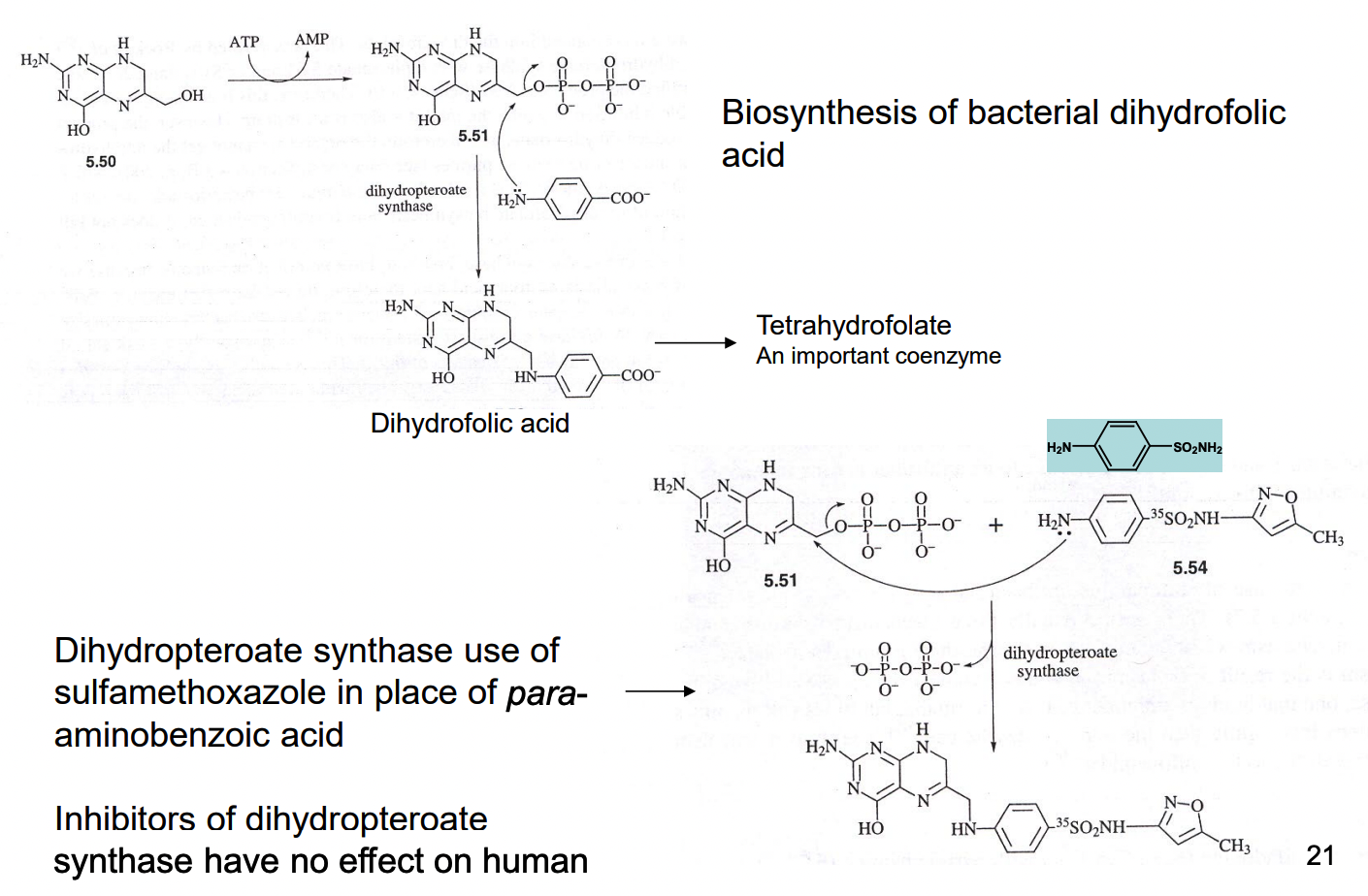
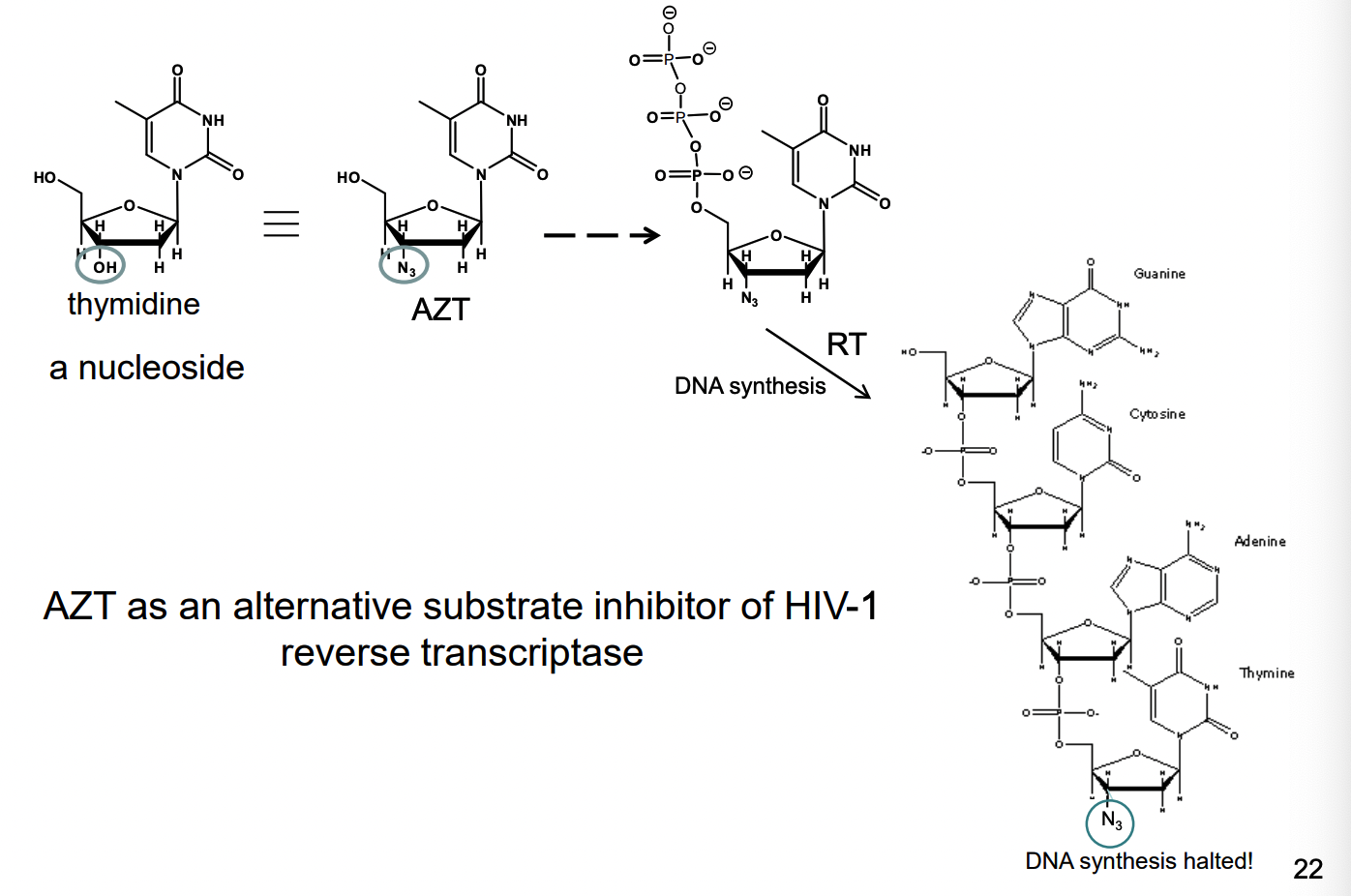
How do nucleoside analogs work as an alternative substrate inhibitor of reverse transcriptase
How do nucleotide analogs work as inhibitor alternative substrate inhibitors of viral RNA polymerase?
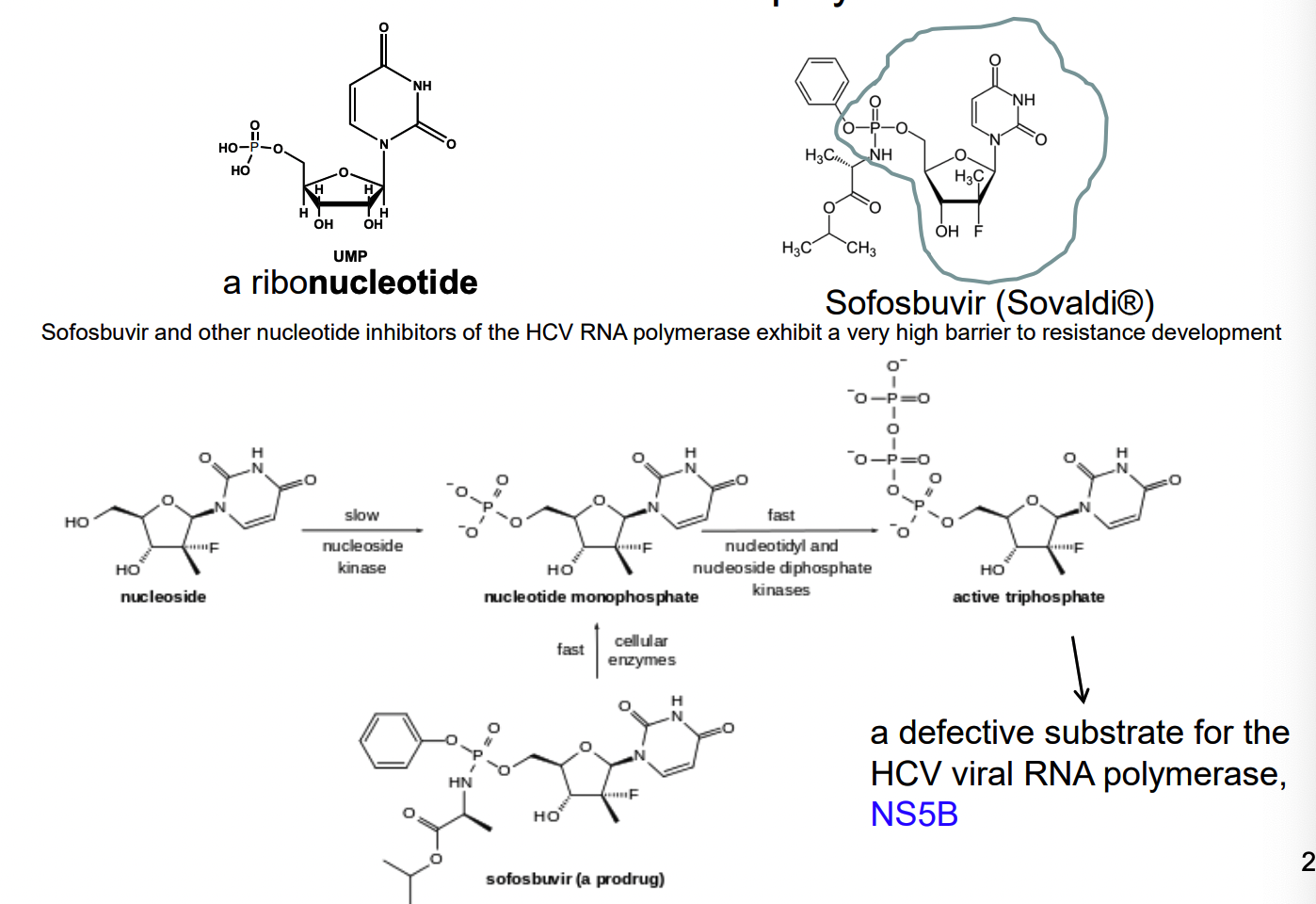
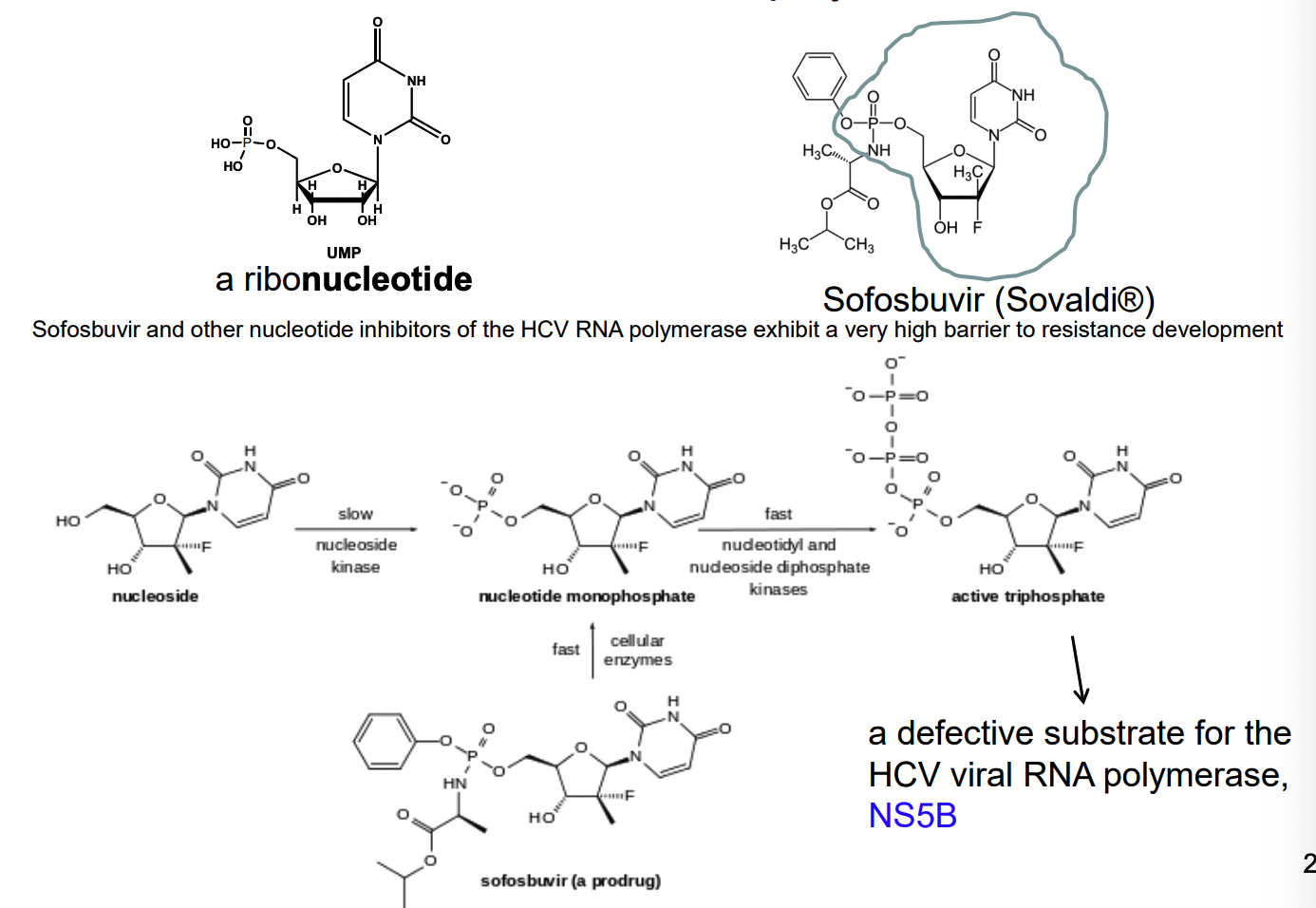
Nucleotide analog reverse transcriptase inhibitors
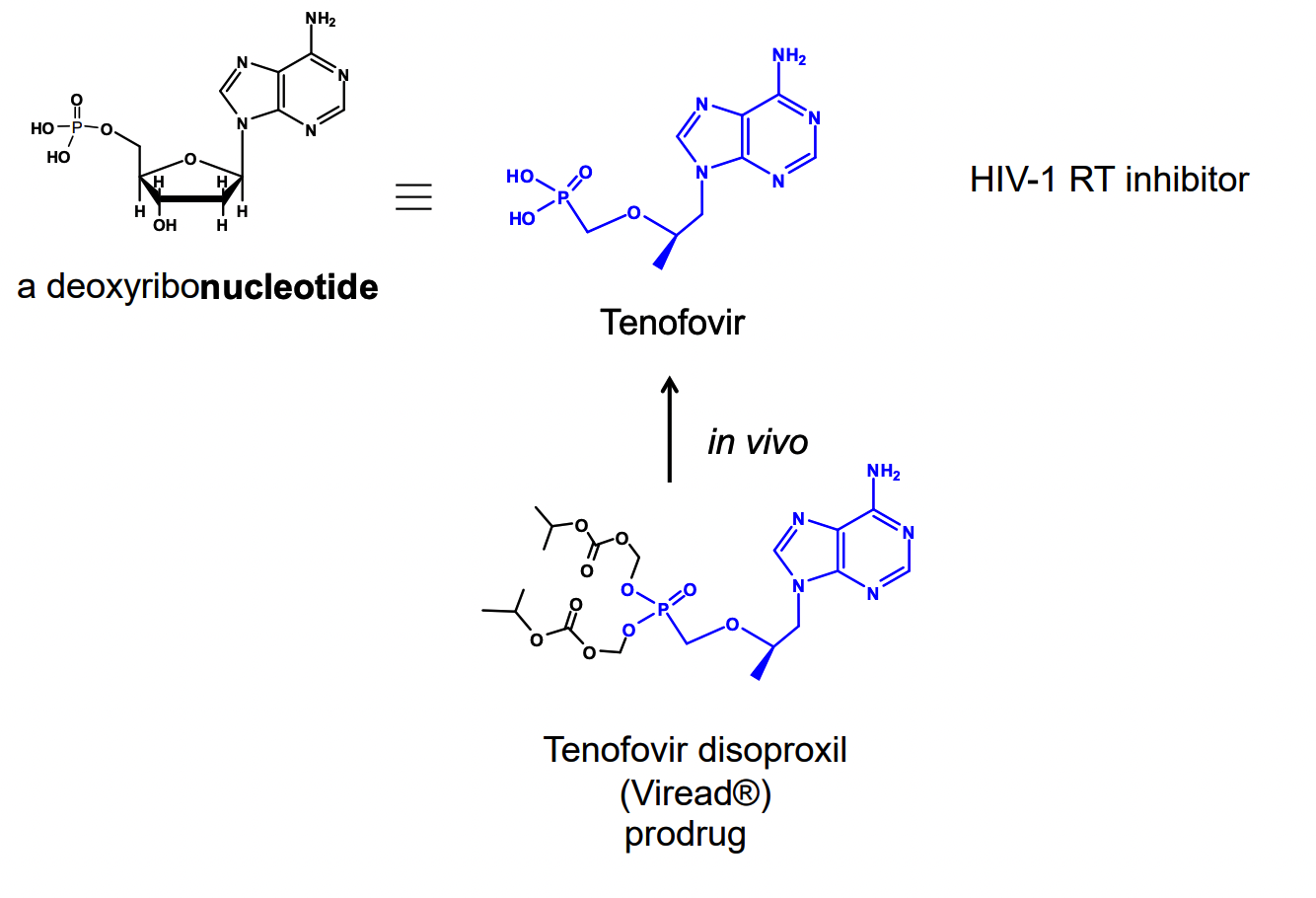
What is Remdesivir used for?
treatment of Ebola virus & Marburg virus infections
a prodrug that metabolizes into active form
acts as a nucleotide analog inhibitor of RNA polymerase
developed by Gilease Sciences
subsequently found to show antiviral activity against other single stranded RNA viruses
respiratory syncytial virus
junin virus
lassa fever
nipah virus
hendra virus
coronaviruses
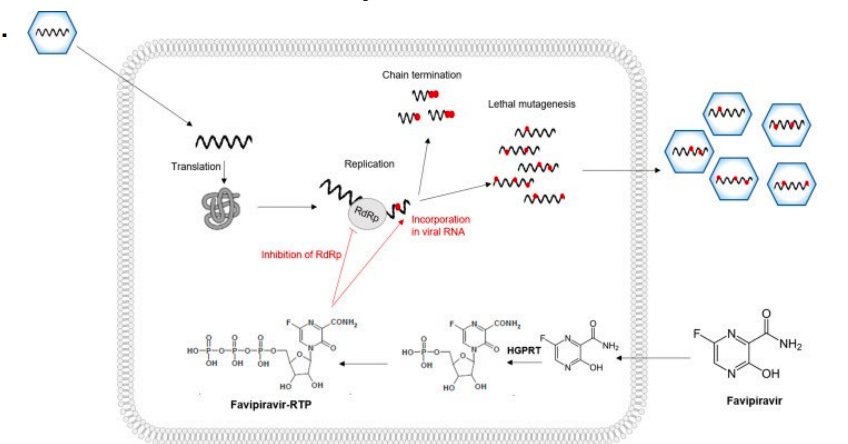
What is Favipiravir used for? What kind of inhibitor it is?
approved to treat influenza in Japan
activity against many RNA viruses: west nile virus, yellow fever, foot-and-mouth disease
flaviviruses arenaviruses, bunyaviruses and alphaviruses (possible covid-19)
doesn’t inhibit RNA or DNA synthesis in mammalian cells (nontoxic to humans)
Favipiravir = prodrug (metabolized into active form — favipiravir-ribofuranosyl-5’-triphosphate
What is the strongest interaction between the substrate and enzyme?
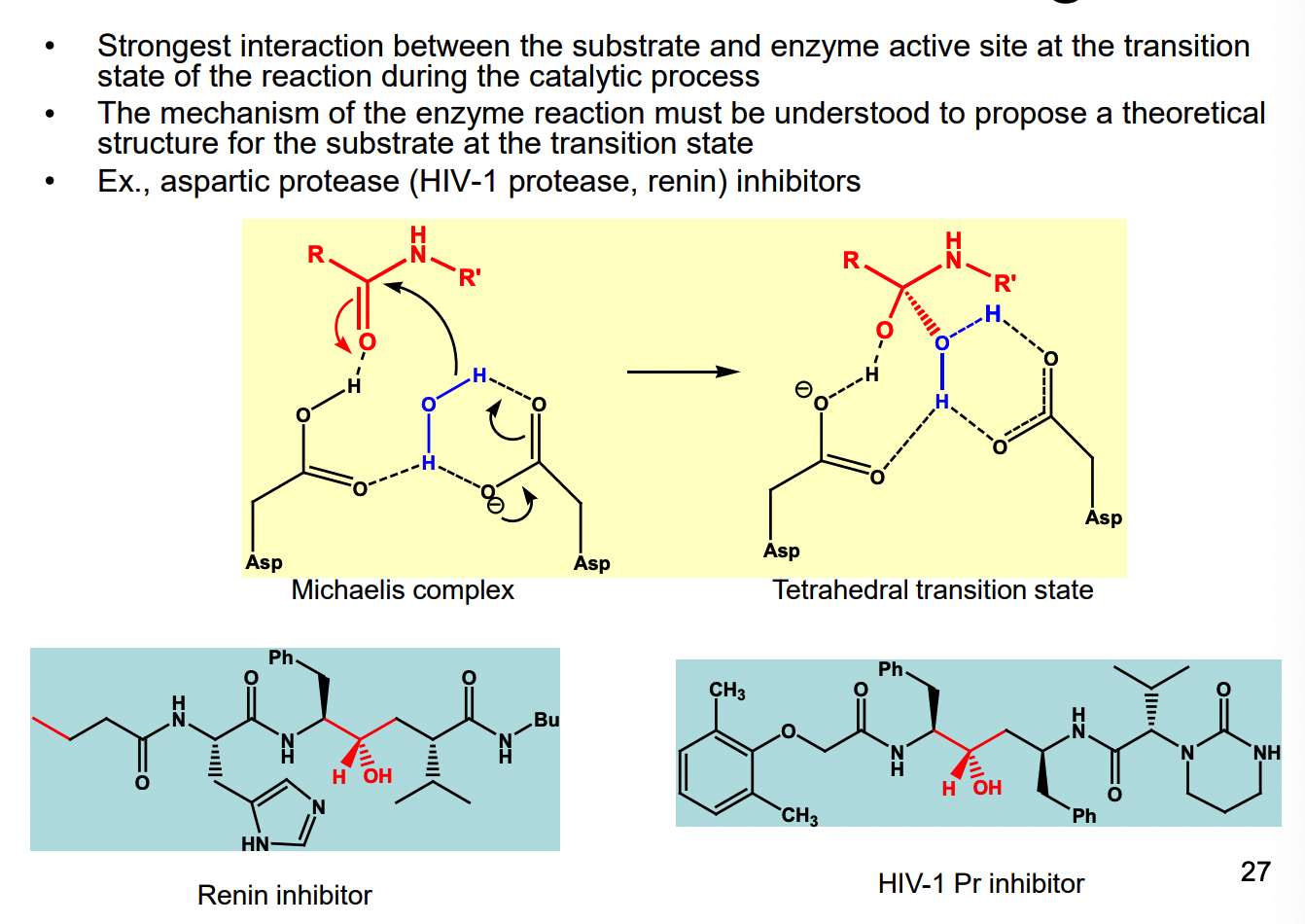
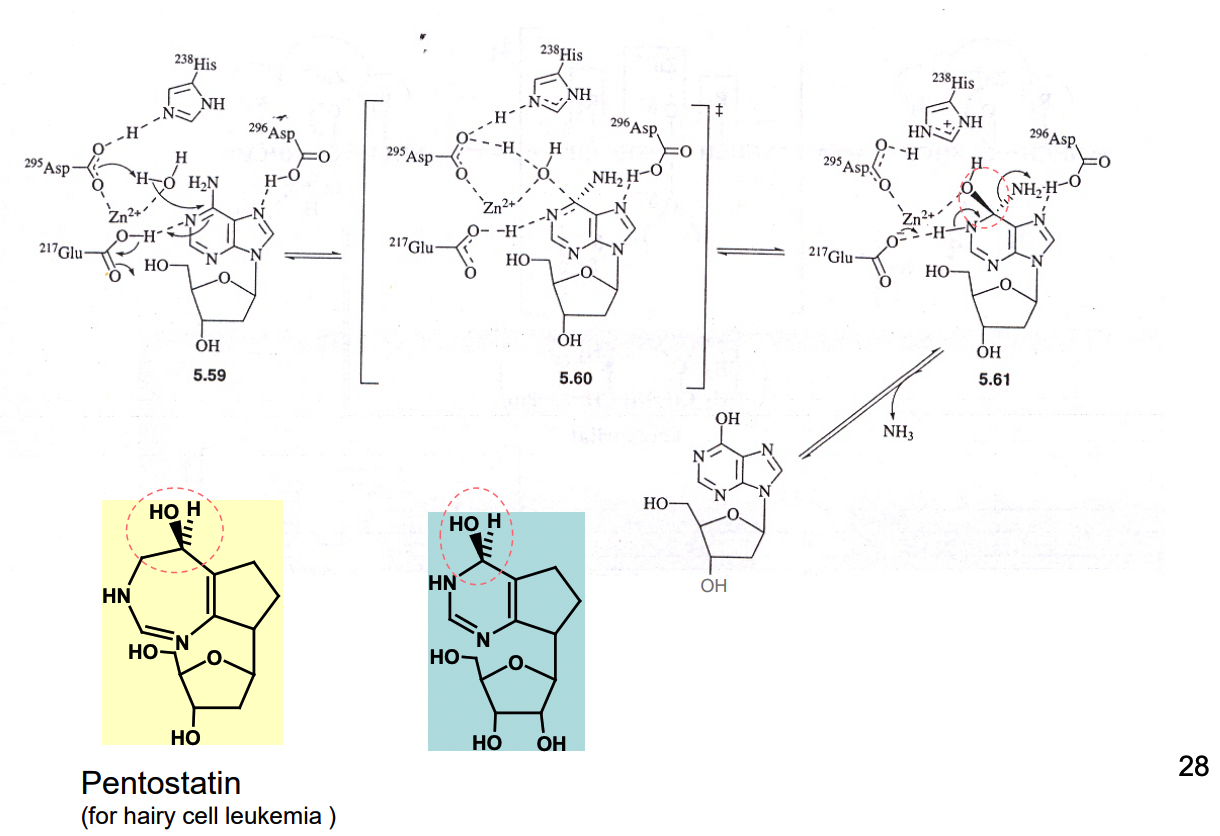
What is Pentostatin a potent inhibitor of?
it is a transition state analog — inhibitor of adenosine deaminase
Ki = 2.5 × 10^-12 M, Km of adenosine ~ 10^-5 M
What kind of analogs are slow, tight-binding analogs?
slow to reach equilibrium
off rate v low, very tight binding
functionally equivalent to irreversible inhibitors
What are irreversible enzyme inhibitors?
reversible enzyme inhibitor = need to maintain a high enough conc of inhibitor to sustain E•I complex
competitive irreversible enzyme inhibitor are active-site directed irreversible inhibitor or enzyme inactivator
compound structure is similar to the substrate, generally forms a covalent bond to an active site residue
not necessary to sustain the inhibitor concentration to retain the enzyme-inhibitor interaction (since enzyme has reacted with irreversible inhibitor)
complex can’t dissociate and enzyme remains inactive (even in absence of additional inhibitor)
What are the 2 types of enzyme inactivators?
affinity labeling agents
reactive compounds
mechanism-based enzyme inactivators
unreactive compounds that are substrates (suicide substrates) of the target enzyme and are activated by the target enzyme
What are affinity labeling agents
reactive compound
similar structure to substrate of target enzyme
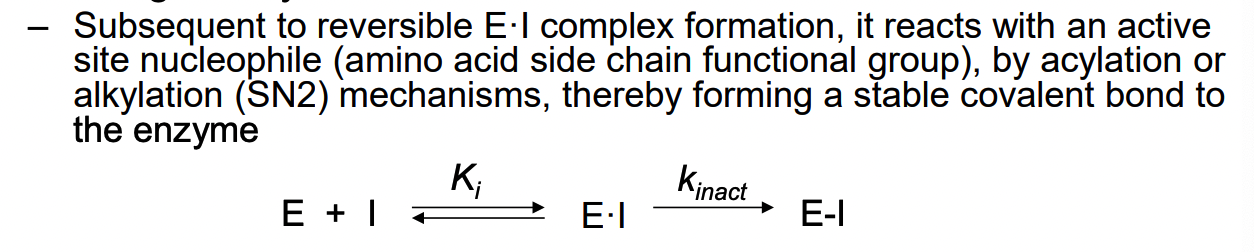
most cases: equilibrium for reversible EI complex formation = rapid and rate of dissociation is fast
k inact = rate-determining step
time-dependent loss of enzyme activity
rate of inactivation = proportional to low conc of inhibitor (becomes independent at high concs due to enzyme saturation
What are some concerns with affinity labeling agents?
potentially toxic — can react with nucleophiles from other biomolecules
key to effective design = specificity of binding
enhance selective reactivity of the subsequent enzyme inactivation step
lower reactivity of the reactive group = can also increase selectivity
What are examples of B-lactam antibiotics?
penicillin & cephalosporins = bacteriocidal
inactivate an enzyme needed for bacterial growth
this doesn’t exist in animals (making it an ideal drug)
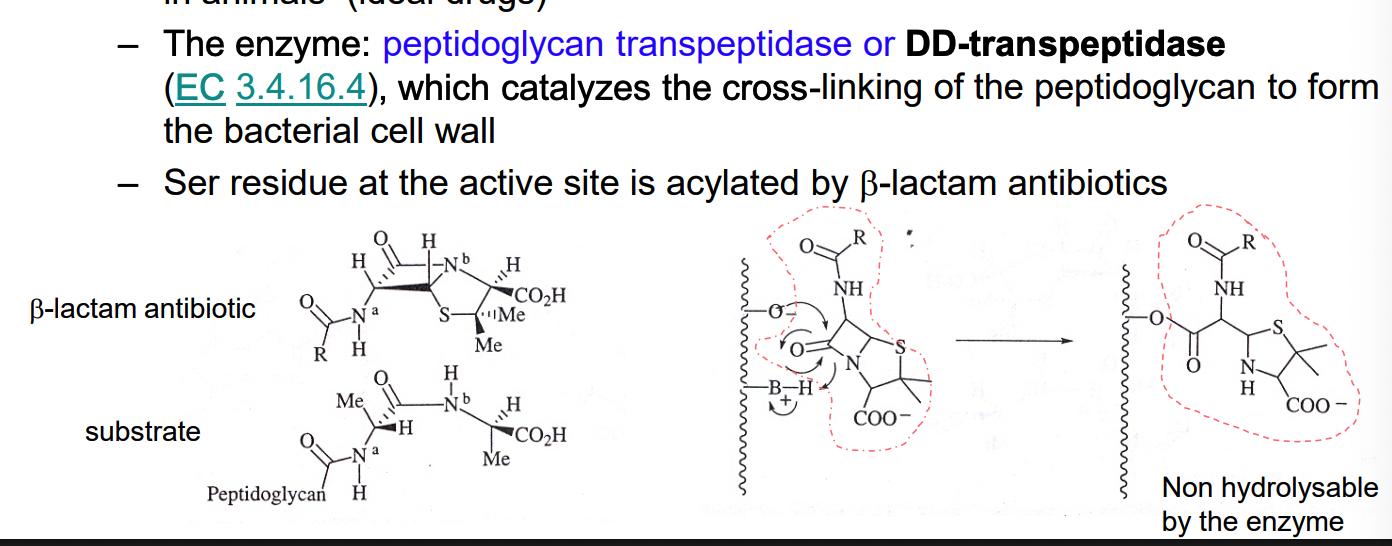
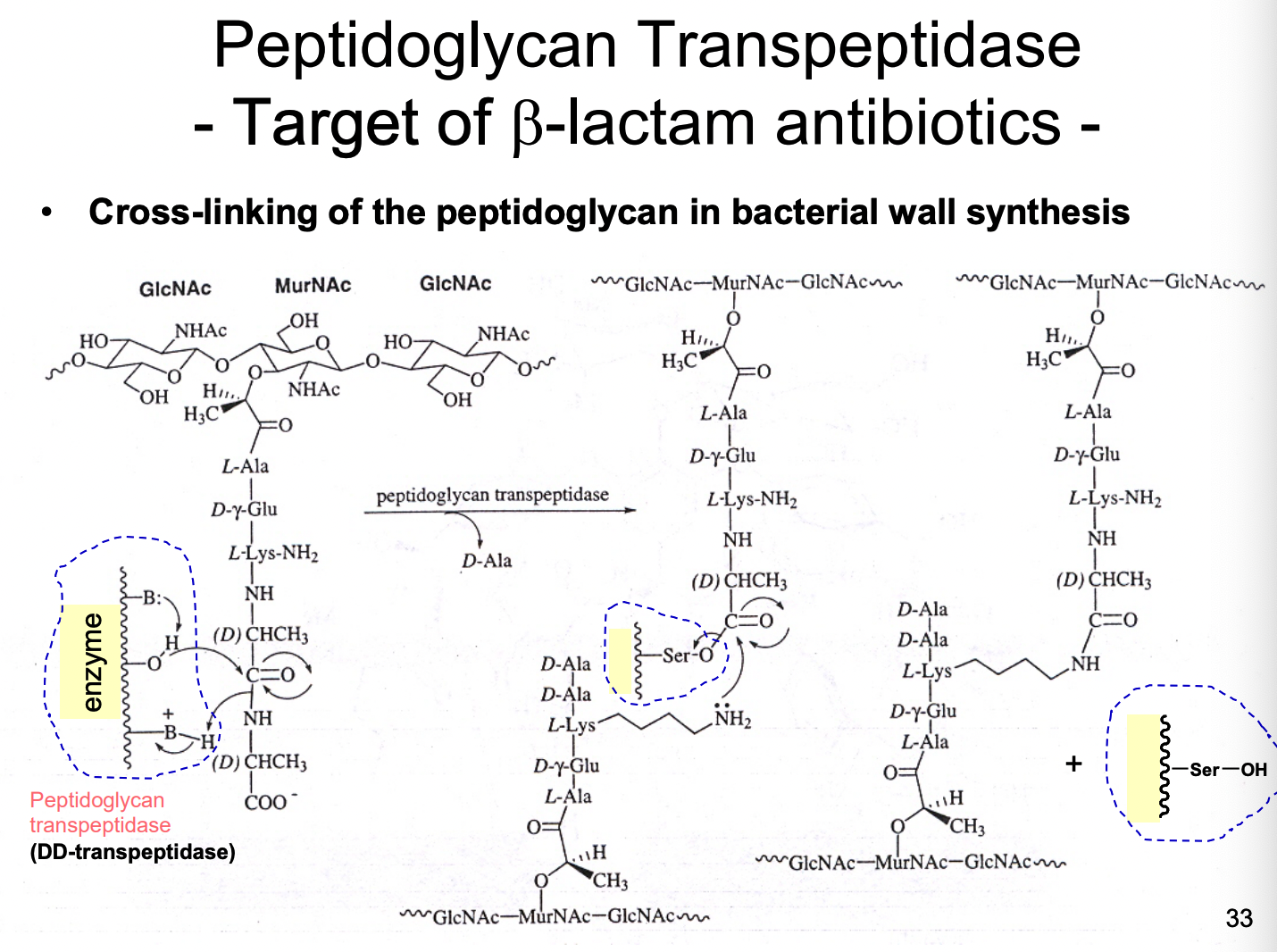
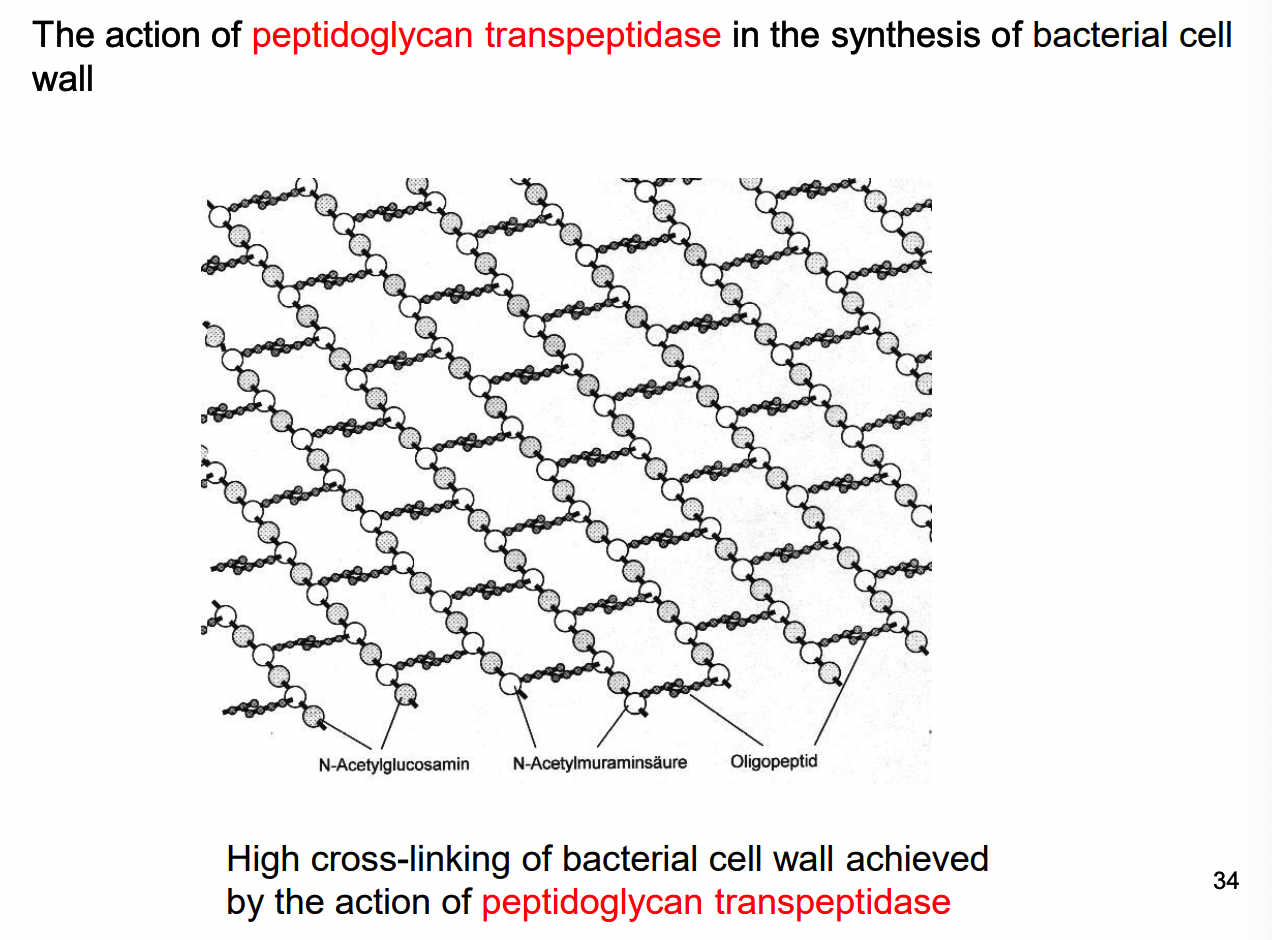
How does bacterial resistance develop for penicillins and cephalosporins?
penicillins and cephalosporins = not exceeedingly reactive as acylating agents
few nonspecific acylation reactions occur — non toxicity, once considered “wonder drugs”
resistance:
half of the strains of staphylococcus aureus = resistant w/n 10 yrs of introduction of penicillin
1990, 90% of strains = resistant
Principal cause = resistant strains synthesize and excrete enzyme B-lactamase which catalyze hydrolysis of B-lactam
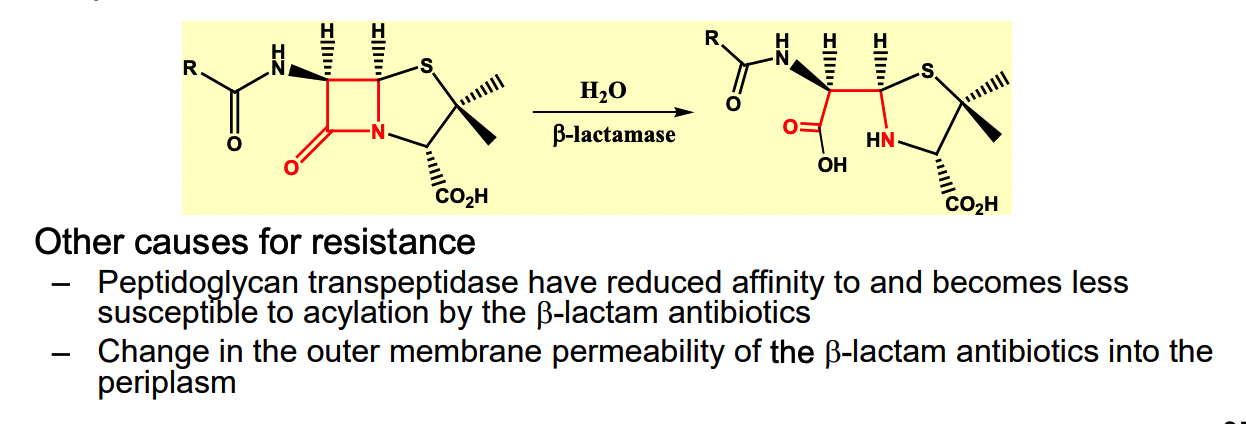
How do we overcome resistance?
1) pair pencillin with B-lactamase inhibitor → killing resistant strains
** ex of drug synergism
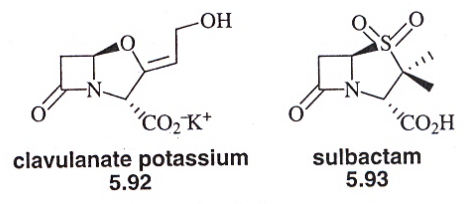
2) new analogs of B-lactam antibiotics can be made
analogs are poorly recognized by B-lactamase, but active against peptidoglycan transpeptidase
3) switch to other antibiotics (vancomycin) with diff mech of action
4) prevent antibiotic abuse
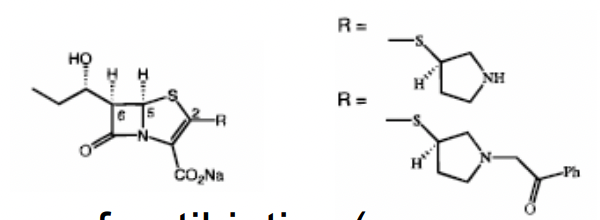
How does Nirmatrelvir work as an irreversible inhibitor of 3C-like protease of SARS-CoV-2?
3C-like protease process viral polyprotein at multiple sites (usually after glutamine residues
cysteine proteases
drug forms covalent bond with thiol group of cys residue in the enzyme
What is a mechanism-base enzyme inhibitor?
inactive compound, structurally similar to the substrate or product of an enzyme
initially serves as a substrate for the enzyme and converted to a product (reactive)
inactivates the enzyme prior to escape from the active site
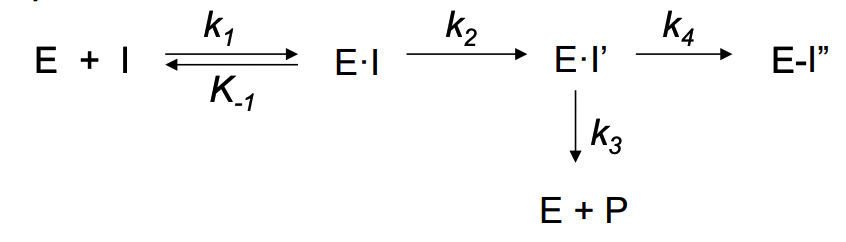
Affinity labeling agents vs mechanism based enzyme inhibitors (2 key features)
1) initial inactivity
2) requirement for the enzyme to catalyze the conversion
Advantages of mechanism-based enzyme inactivators
initially inactive
no problem of nonspecific alkylation or acylation of other proteins
converted reactive product = already in active site of the enzyme and will react within
not many ex: most determined ex post facto rather than designed that way
(not easy to design)
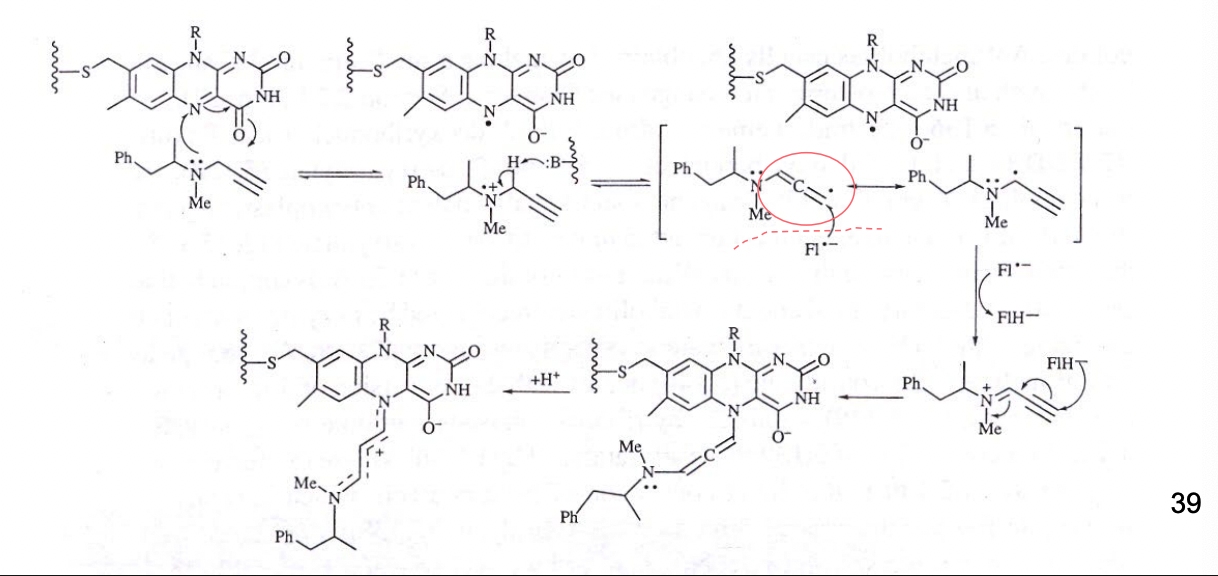
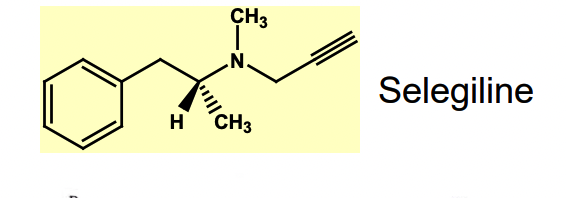
What is Selegiline (L-Deprenyl)?
antiparkinsonian drug
parkinson’s disease = reduction in dopamine conc in the brain
Selegiline deactivates monoamine oxidase B (MAO B) = involved in metabolism of the inhibitory neurotransmitter dopamine
What are new enzyme targets for epigenetic therapy
enzymes responsible for modulating epigenetic makeup of cells
DNA methyltransferase
HATS - histone acetyltransferases
HDACS - histone deacetylases
2 approved drugs as inhibitors of HDAC for the treatment of cutaneous T-cell lymphoma.
histone lysine methyltransferase and lysine demethylase
RNA -modifying enzymes
What is drug resistance mean? how does it happen
formerly effective drug dose no longed effective
related to microorganisms or cancer cell growth
resistance arises mainly by natural selection
evolution and adaptations: 1 in 10^7 microorgs (or cancer cells) in a colony have 1+ mutations making it resistant to that drug
susceptible cells in population get killed → few resistance ones replicate → become predominant population
resistance = seldom caused by drug induced mutation
What is drug tolerance the cause of?
pt’s body adapts to a particular drug in chronic use → more drug needed to attain initial effect
lead to decrease in the TI
What is the mechanism of drug resistance? (10)
1) altered drug uptake
2) overproduction of target enzyme
3) altered target enzyme
4) production of drug-destroying enzyme
5) deletion of a prodrug-activating enzyme
6) overproduction of substrate for the target enzyme
7) new pathways for formation of the product of the target enzyme
8) efflux pumps
What is altered drug uptake in drug resistance?
ability of the organism to exclude the drug from the site of action by preventing the uptake of the drug
how does overproduction of the target enzyme lead to drug resistance?
increases target enzyme production though induction of extra copies of the gene encoding the enzyme
How does altering the target enzyme work for drug resistance
mutations of aa residues in the active site can result in poor binding of the drug to active site
important strat = minimize effect of target enzyme mutation in drug resistance → design a drug similar structure to that of substrate
mutation would also cause auto-inhibition
production of similar enzyme that can do the same fx
does the same fx as og enzyme, but drug can’t bind
ex. methicillin-resistant Staphylococcus aureus (MRSA) - responsible for several difficult to treat infections in humans
Altered active site of action — vancomycin resistance is an ex
vancomycin = last defence against streptococcal or staphylococcal bacteria
forms complex with terminal D-alanyl-D-alanine of peptidoglycan, blocking transpeptidation
Vancomycin resistance example (inhibits bacterial wall synthesis — binds to the substrate of the enzyme
last residue = becomes D-lactate → ester bond formed instead of amide bond
amide can form H-bond with vancomycin
How does drug resistance occur through production of drug-destroying enzyme?
induction of genes that produce new enzymes to degrade drug
B-lactamase
How does deletion of a prodrug-activating enzyme work for drug resistance
no active form = no working drug
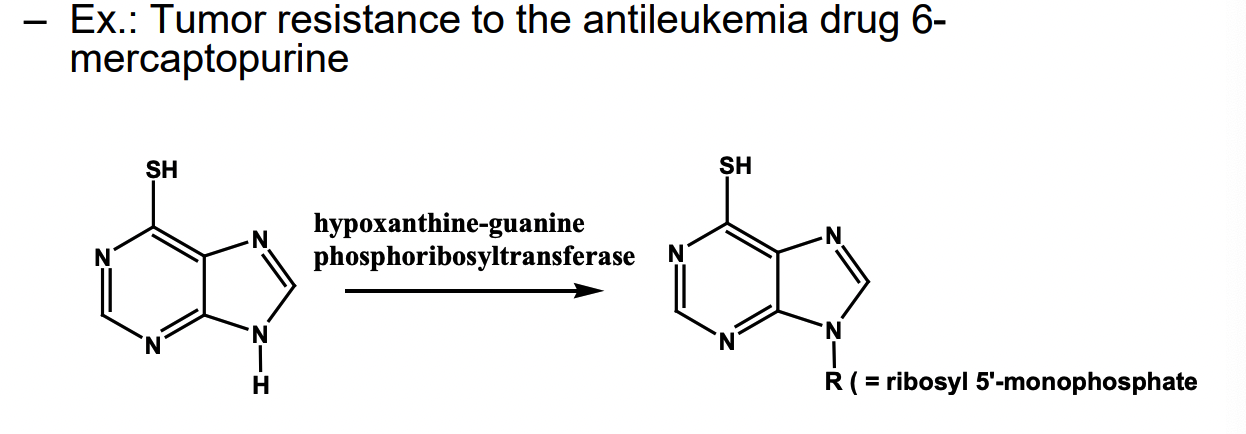
NH → ribose (so it’s like a nucleoside)
in some tumor drugs, the enzyme is deleted
How does overproduction of the substrate for the enzyme work for drug resistance?
overproduction of natural substrate = competitively block ability of the drug to bind at the active site of the target enzyme
how do new pathways form for product of target enzyme?
drug is used to block production of a metabolite using enzyme inhibition
organism can bypass this by creating a new pathway to produce that same metabolite
how do efflux pumps work against drugs → resistance?
tumor cells and microorganisms can develop protein transporters that bind to drug that carry the drugs out of the cell
How can we fight against drug resistance? what is the main strat?
drug synergism
therapeutic effect of 2+ drugs used in combo
combo is greater than the sum of the effects of the drugs administered individually
improve use of antibiotics
avoid overuse and abuse
What are the approaches to drug synergism?
inhibition of drug-destroying enzyme
sequential blocking: inhibition of 2+ consecutive steps in metabolic pathway
inhibition of enzymes in different metabolic pathways
if the cause of drug resistance = production of a new metabolic pathway
efflux pump inhibitors
use of multiple drugs for same target