DNA to RNA
1/49
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
50 Terms
What is transcription?
DNA to RNA
DNA doesn’t direct protein synthesis directly, instead it copies into RNA
Only uses ONE strand to make different transcripts based on promoter sequence
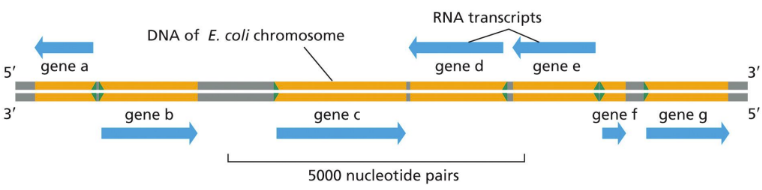
What is consensus nucleotide sequence?
Derived by comparing many sequences with the same basic function and tallying up the most common nucleotides found at each position
The nucleotide size graph
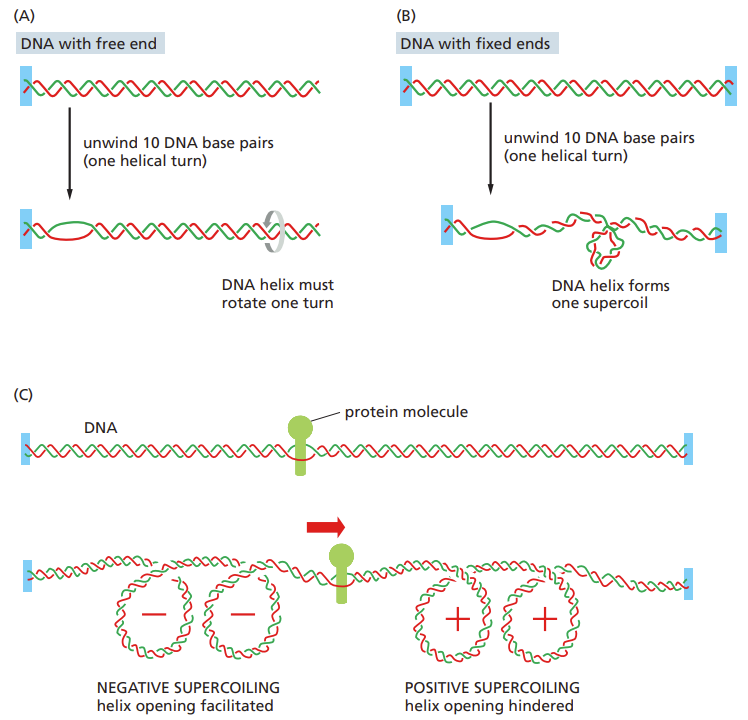
Define DNA supercoiling?
Produced by elongation factors
Because of the need to unwind the helix
For every 10 open nucleotides = 1 supercoil
Topoisomerase helps remove tension during transcription
May aid in unwinding DNA from nucleosomes in eukaryotes
Define general transcription factor?
The transcription factors that eukaryotic RNA polymerases
require for initiate transcription
Are proteins
Help position RNA polymerase II at promoter
The general TF initiate transcription because DNA packaging via nucleosomes present a problem
TFIID, B, A, F, E, & H
What does TFIID do?
Recognizes TATA box (& other DNA sequences) near the transcription start point
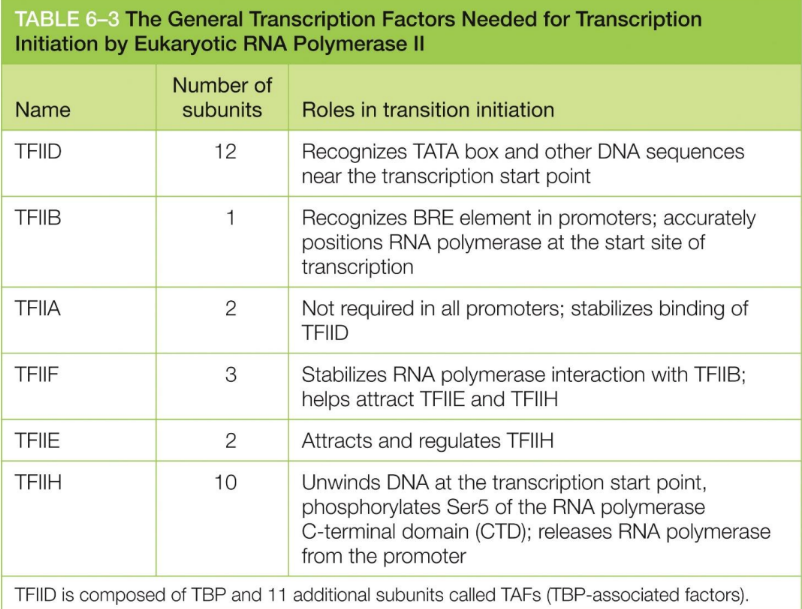
What does TFIIB do?
Recognizes BRE element in promoters
accurately positions RNA polymerase at the start site of transcription
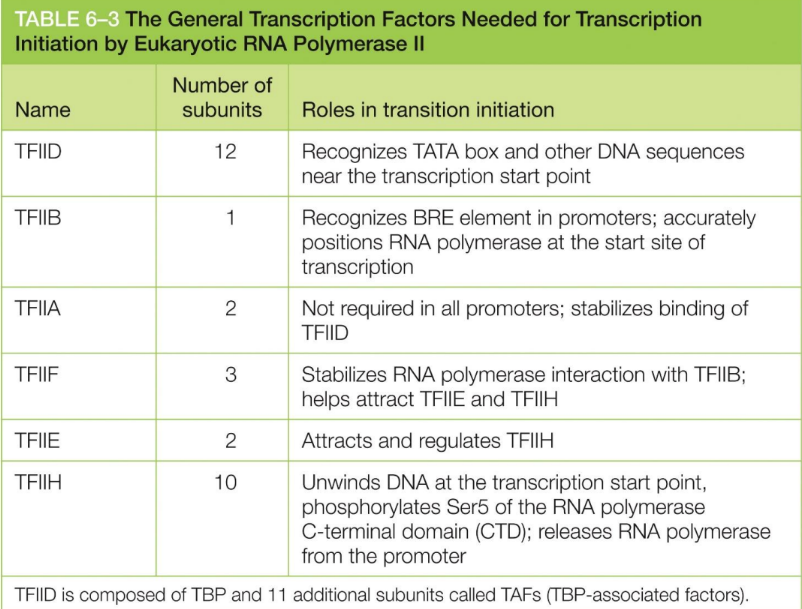
What does TFIIA do?
Stabilize TFIID binding
not required in all promoters
What does TFIIF do?
Stabilizes RNA polymerase interaction with TFIIB
helps attract TFIIE and TFIIH
What does TFIIE do?
Attracts and regulates TFIIH
What does TFIIH do?
Unwinds DNA at the transcription start point
phosphorylates Ser5 of CTD of RNA polymerase
releases RNA polymerase from the promoter
What is an exon?
part of a sequence that is used/transcribed into mRNA or eventually proteins
Expressed coding sequences
Transcribed into RNA
Spliced together to form a functional gene
SR proteins (special serine & arginine rich proteins) assemble on exon to call spliceosomes
What is an intron
Intervening sequences
Transcribed into RNA
Removed via splicing
What is a promoter
a sequence of DNA nucleotides that signal the starting point for RNA synthesis
All promoters need general TF for RNA pol. II to begin
What is RNA polymerase (simple definition)
a group of enzymes that form phosphodiester bonds between ribonucleotides
What is the central dogma
DNA = (DNA synthesis) > DNA = (Transcription) > RNA = (Translation) > Protein
* sometimes RNA is the final product and NOT protein
For structure/regulation
What are the 3 RNA polymerases
RNA polymerase I (r)= transcribes 5.8S, 18S, and 28S rRNA genes
RNA polymerase II (m)= Transcribes ALL protein-coding genes & snoRNA, miRNA, siRNA, lncRNA, and most snRNA genes
Has the D, B, A, F, E, & H transcription factors (required)
RNA polymerase III (t)= Transcribes tRNA, 5S rRNA, and some snRNA genes
What does RNA pol. I transcribe
transcribes 5.8S, 18S, and 28S rRNA genes
What does RNA pol. II transcribe?
Transcribes ALL protein-coding genes & snoRNA, miRNA, siRNA, lncRNA, and most snRNA genes
Has the D, B, A, F, E, & H transcription factors (required)
What does RNA pol. III transcribe
Transcribes tRNA, 5S rRNA, and some snRNA genes
What is snRNA
Small nuclear RNAs
Function in nuclear processes (splicing of pre-mRNA)
Join exons together (after being spliced)
What is snoRNA
Small nucleolar RNAs
Help process and chemically modify rRNAs
What is miRNA
microRNAs (gene expression regulation)
Regulate gene expression by blocking translation of specific mRNAs and causing degradation
What is mRNA
messenger RNAs
codes for proteins
What is rRNA
Ribosomal RNAs (80% of RNAs)
Form the basic structure of ribosome and catalyze protein synthesis
What is tRNA
transfer RNAs
Central to protein synthesis as the adaptors between mRNA and amino acids (at APE site in ribosome)
What is telomerase RNA
Serves as the template for the telomerase enzyme that extends the ends of chromosomes
What is lncRNA
long noncoding RNAs
Some serve as scaffolds (link non-continuous strands) and regulate cell processes (like X-inactivation)
What is siRNA
Small interfering RNAs
Turn off gene expression by directing the degradation of selective mRNAs and helping to establish repressive chromatin structures
What are the steps of transcription? (General)
TFIID binds to TATA box (double stranded state)
binding causes a distortion in the helix (indicating activity is going on)
TFIIB binds to the TBP subunit on TFIID (sometimes TFIIA also binds)
TFIIF binds to the RNA pol. II to stabilize TFIIB (attracts E and H)
TFIIE & TFIIH bind to RNA pol. II and form pre initiation complex
TFIIH uses ATP to open the DNA & phosphorylates RNA pol. II tail
releasing it from the TF & making a conformational change => Can now begin transcription
mRNA processing begins after ~20 nucleotides have been transcribed
5’ cap is added to modify the pre-mRNA transcript (before transcription)
RNA Splicing (during transcription)
MRNA 3’ poly A tail (after transcription/splicing)
mature mRNA
What is the initiation process of creating mRNA (TFs)
TFIID subunit (TBP) binds to DNA double strand at TATA box (Tata Binding Protein = TFIID subunit)
DNA helix distortion occurs (triggered by TFIID binding) signaling activity is going on
More TF gather (F, E, & H) to form the complete transcription initiation complex
TFIIH does the most work
Uses ATP to open DNA (double strands broken apart)
Next, phosphorylates RNA pol. II tail (CTD) -> conformational change that allows it to transcribe
CTD lengthens, allowing more proteins to bind until they “hop off” to the new RNA strand (to begin 5’ capping, RNA splicing, and poly A tail)
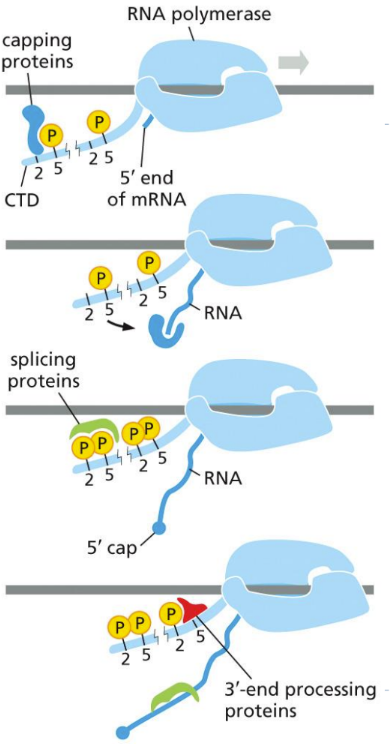
What is the 5’ capping process?
5’ cap is added to modify the pre-mRNA transcript (before transcription)
Cap = methylated guanine
Acts as marker to let cell know what is being made is mRNA
CBC (Cap-Binding Complex)
Essential for translation
Protects mRNA from degradation
Added by 3 enzymes bound to RNA pol. Tail
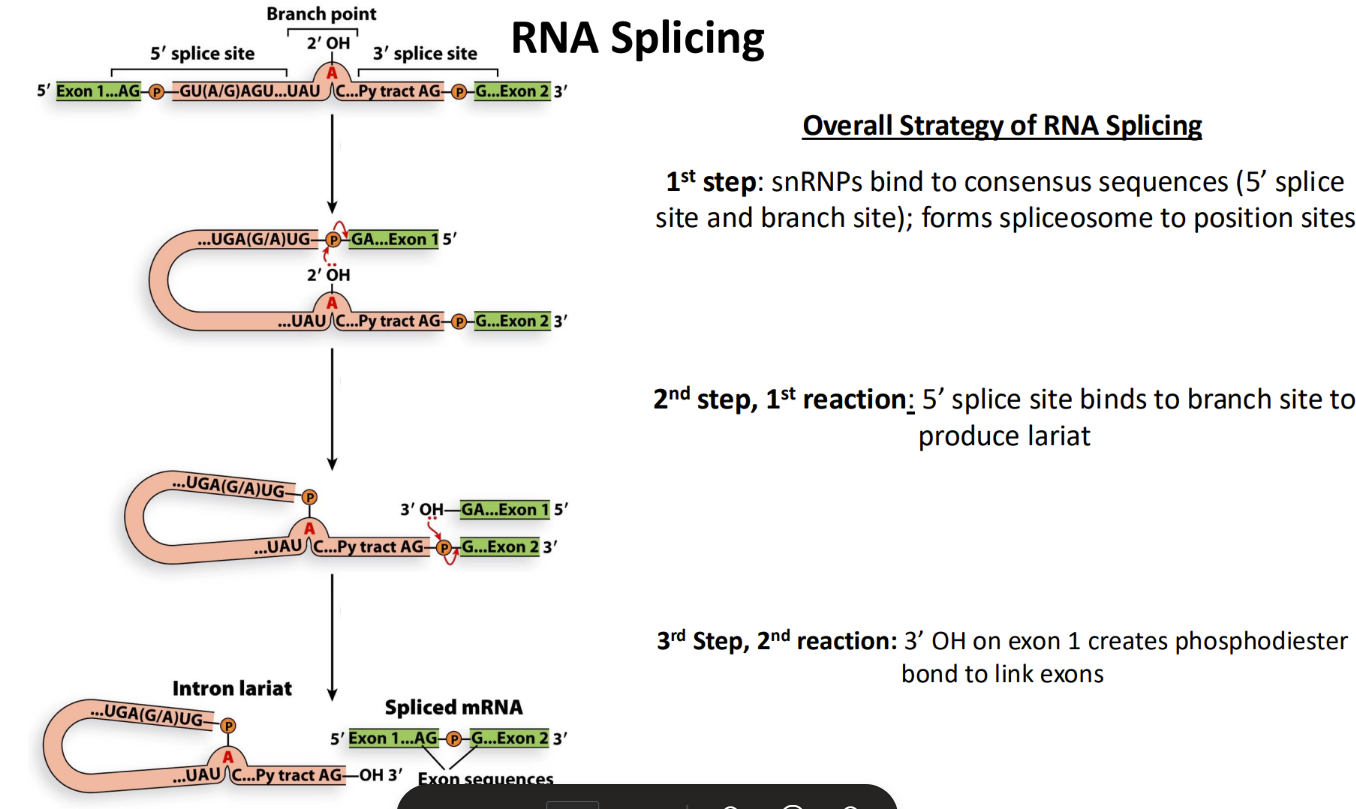
What is the RNA splicing process?
Spliceosome forms (6snRNPs) (called by SR proteins on exons)
2-step enzymatic reaction to remove introns
The 2 neighboring exons are joined together
Exon junction complex protein binds where the intron use to be to indicate that the RNA has undergone splicing successfully
Both exons & introns are transcribed into RNA but introns are removed with splicing
Splicing proteins wait at C terminal domain
snRNA join exons together
OR
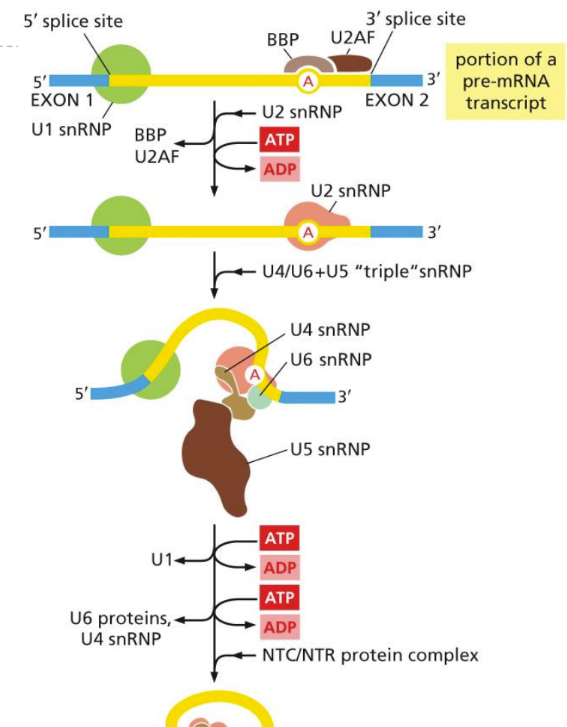
1. snRNPs bind to consensus sequences (form spliceosome to sites)
2. (1st rxn) 5’ splice site binds to branch point (lariat form) (exon 1 released)
3. (2nd rxn) 3’ OH in exon 1 piece binds to phosphate in 3’ splice site (releasing intron)
What is the Poly A tail process?
Consensus sequences (AAUAAAA..CA..~30nucl...GU/U rich region) are bound by binding proteins/enzymes that travel on RNA pol. CTD (tail)
Poly-A polymerase adds 200 As to 3’ of mRNA transcript
Tail length determined by pol-A binding proteins (stabilize tail and count to 200)
When finished, RNA pol. releases the template
How is mature mRNA determined?
look at pick
What are the similarities and differences between transcription and replication
Similarities | Differences |
|
|
Define RNA polymerase (general)
Enzyme that catalyzes the synthesis of an RNA molecule on a DNA template from ribonucleoside triphosphate precursors
Can't dissociate from strand until finished (DNA pol. Can remove itself for repair mechanism)
Only has modest proofreading capabilities
What is RNA splicing?
introns (and exons) are removed from primary transcript using protein complex (spliceosome)
Occurs within the nucleus
Can change the meaning of an RNA molecule
Splicing:
Is flexible (alternative splicing) depending on alternative proteins necessity
Depends on:
The affinity of splicing machinery (3 mRNA signals (the 2 splice junctions and branch point)
Spliceosome assembly
Exon definition based on enhancer/activator
What is a rRNA gene
genes that code for rRNA
Found in the nucleolus
What is rRNA
non-coding RNAs that catylize protein synthesis and have ribosomal function
Transcribed by RNA pol. I
Doesn't have a cap or Poly A tail so can’t leave nucleus (because pol. I doesn’t have a tail to add this)
Needs to be combined with other proteins to work
4 types:
Small subunit: 18s
Large subunits: 5.8s, 28s, & 5s (5s made by pol. III)
What is snRNA
Small nuclear RNAs that join exons after splicing by recognizing splice sequences
What is a spliceosome
a large (60s) complex made of RNPs (6 snRNAs + their proteins)
snRNAs + 7 protein subunits = snRNP (ribonucleotide proteins)
snRNPs form spliceosome core
“complex of multiple snRNPs on mRNA”
What is a TATA box
A sequence of DNA Ts and As located ~30 nucleotides upstream from transcription start site
TFIID binds to it
NOT the only transcription start sequence
EX: BRE, INR, & DPE (all bind to different general TFII)
What is a terminator?
Signal in bacterial DNA that halts transcription; in eukaryotes, transcription terminates after cleavage and polyadenylation of the newly synthesized RNA
where the polymerase halts and releases both the newly made RNA molecule and the DNA template
What is a transcription unit
Each transcribed segment of DNA
Information in just one gene for just one RNA molecule or single protein (or group of mRNAs if there is splicing)
Typically carries the information of just one gene, and therefore codes for either a single RNA molecule or a single protein (or group of related proteins if the initial RNA transcript is spliced in more than one way to produce different mRNAs).
What is a mediator?
allows activators to communicate (make contact) with transcription complexes
Important in regulation of gene expression
Chromatin and histone remodeling proteins & enzymes are recruited via a mediator
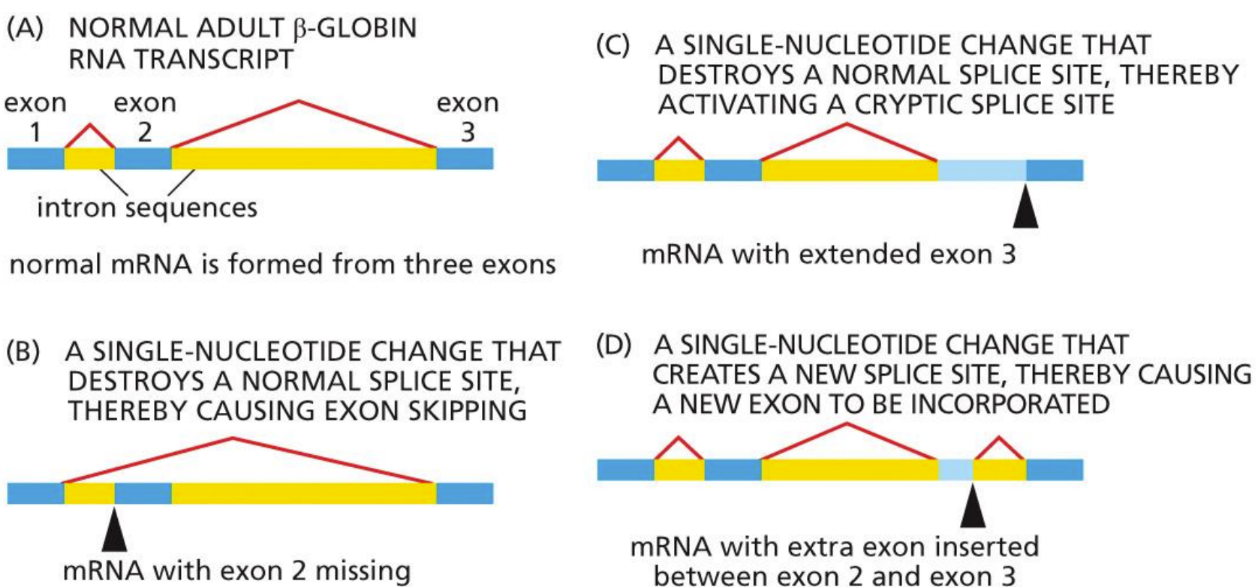
What is alternative splicing
Different splicing patterns leading to different protein production
What is the nucleolus
site of rRNA processing and incorporation of rRNAs into ribosome subunits (ribosome factory)
Not membrane bound (allows chromosomal DNA to move around)
Contains rRNA genes, precursor rRNAs, mature rRNAs, processing enzymes, and partly built ribosomes (ribosomes assembled outside nucleus)
Telomerase assembled & tRNA processed here
Merge RNA + protein to make complete structures
How is mRNA exported from the nucleus
Mature mRNA is selectively transported from the nucleus into the cytoplasm
Proteins help export (particular proteins indicate mRNA processing is incomplete and NOT ready to leave the nucleus)
Requires a nuclear transport receptor (the “key”) to leave the nucleus through the nuclear pore
What is the function of subnuclear structures
Thought to be involved in processing and storage of RNA-processing components (forms “staging area” for assembly of spliceosome parts)
Cajal bodies
GEMs
Interchromatin granule clusters (“speckles”)
* all of these (subnuclear aggregates) are needed to get appropriate splicing
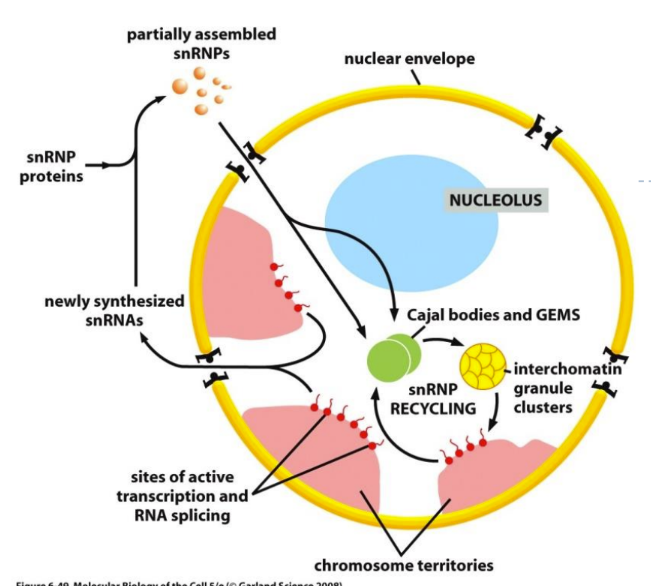
What is disorder that relates to subnuclear structures
Spinal muscular atrophy (SMA) = incorrect splicing because of mutated ribonuclear proteins
Mutated SMN1 gene