RNA surveillance and Translation I (1)
1/31
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
32 Terms
What is mRNA surveillance?
The recognition of improperly processed pre-mRNAs and their earmarking for degradation
Nonsense-mediated decay (NMD)
Degradation of mRNAs that stop translating (stop codon) before the last splice junction
Errors in mRNA (2)
Error in splicing
Mutations (deamination, etc)
Exon-exon junction proteins
EJC or exon joining complex
How is a premature stop codon identified?
By ribosomes being unable to displace UPF3
What happens when UPF3 remains associated?
Formation of a complex that targets the mRNA for degradation: P-bodies
3 RNAs of translation
The rRNA
The tRNA
The mRNA
Amount of rRNA
80% of all cellular RNA
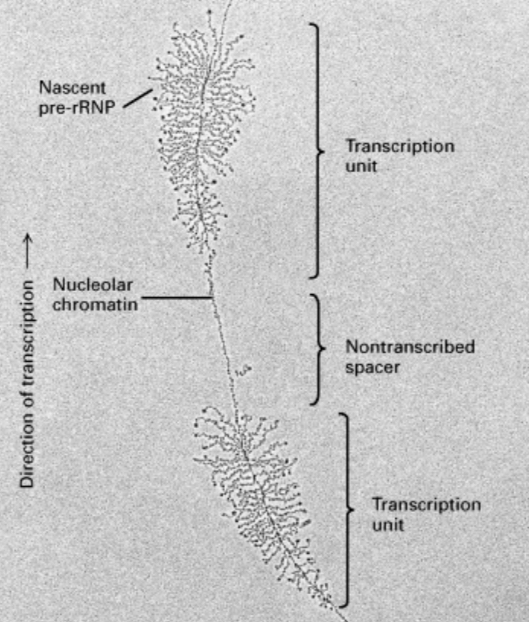
Arrangement of pre-rRNA
Repetitive clusters
What RNA Pol makes rRNA?
Pol III
Bacterial ribosome composition
50S+30S=70S
Eukaryotic ribosome composition
60S+40S=80S
Highly conserved secondary structure of rRNA
Stem-loop structures = SIMILAR 16S and 18S small subunits
Position of the anticodon
Bottom of the tRNA
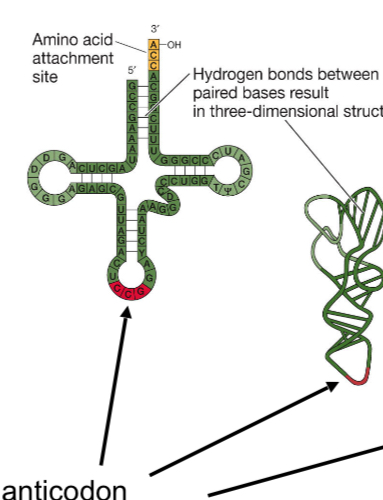
Amicoacyl-tRNA synthetases
Carry the AA
Recognize and bind to cognate tRNA
Rule ab aminoacyl tRNA synthetases
Can bind to more than 1 tRNA
Degenerate code
Different codons can code for the same AA
The tRNAi Met
+methionine = INITIATION
The tRNAi Met in bacteria
Needs to be modified by formyl (CHO) group before +methionine = initiation
The tRNA Met
+methionine = ELONGATION
Pre-initiation complex
40S subunit+ elF2 ternary complex+ elF5 (GTPase)
elF2 ternary complex
= elF2-GTP + tRNAi Met
How can protein synthesis be negatively regulated?
By phosphorylation of elF2 bc it won’t be able to bind ATP= energetically taxing
Why is the 5’ cap required for efficient translational initiation?
Bc the dimeric guanylyltransferase binds to P-CTD
2nd step of pre-initiation
elF4E binds to 5’ cap of mRNA
How is elF4 activity a limiting factor?
It’s under tight regulation bc its overexpression leads to tumour formation
3rd step (2)
elF4G…
Bind elF3 from preinitiation complex
Bind PABPC (3’ end)
How do 5’-3’ interactions favour re-intiation?
Protein:Protein interactions (elF3-elF4)
4th step of pre-initiation
elF4A-RNA helicase complex removes secondary structures in 5’ region→3’
What happens to elF4E?
It stays bound to the 5’ cap resulting in the formation of a loop
5th step of pre-initiation
AUG codon is recognized=
elF2-GTP hydrolysis
Large subunit joining
Once translation is initiated…
The ribosomal complex is irreversibly bound until termination of translation