L2: Polarity and regional identity: generating neuronal diversity
1/53
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
54 Terms
What this lecture is about, now that we know how the neurogenic region emerges during embryonic development
examine how this region is further polarized
and regionalized
leading to the generation of an great diversity of neuronal cell types for developed animals
What does the neuroectoderm anertior vs posterior half develop into?
Anterior→ brain and relative strucutres (eyes)
Posterior→ spinal cord (vertebreates) or nerve cord (insects and invertebrates) and complemtns of motor neurons for coordinating muscles groups in locomotion
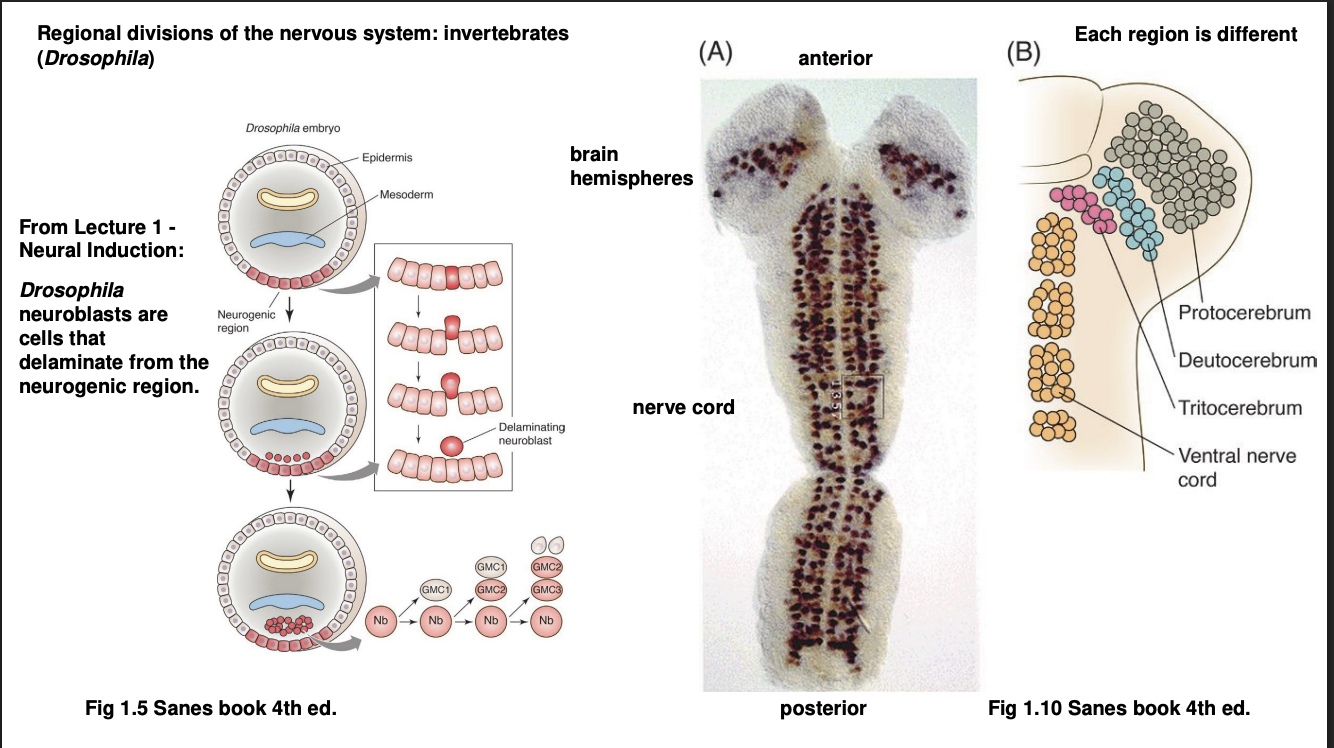
How are these regional divisions made in vertebrates vs invertebrates: Drosophila
Neuroectoderm delaminate (separate away?) into neurolast cells
Stereotyped sequence of asymmetric cell divisions
give rise to population of neurons but in subdivisions
Anterior→ brain, in subdivisions: protocerebrum, deuterocerebrum, tritocerebrum
Posterior→ sub-oesophageal ganglion (homolgous to vertebrate brain stem) and (more posteriorly)→ thoracic and abdominal segments of the nerve cord (homoglous to spinal cord)
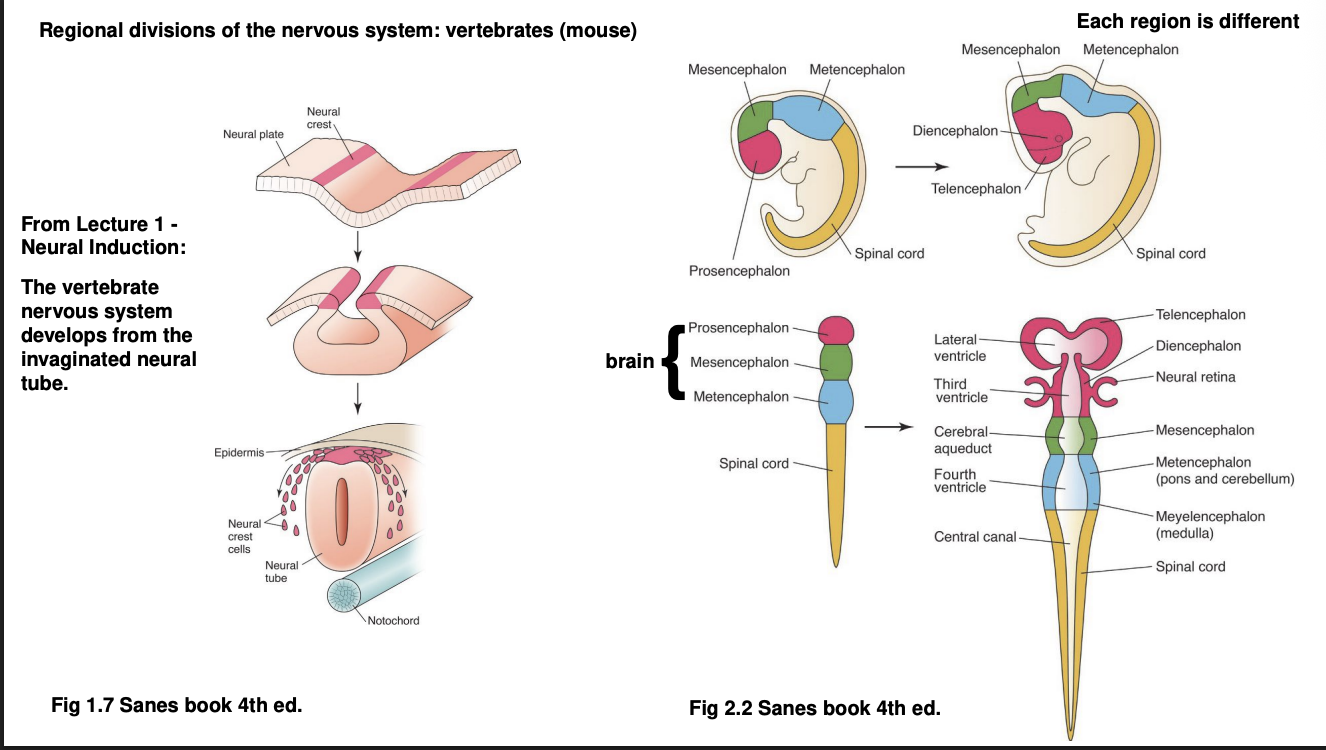
How are these regional divisions made in vertebrates vs invertebrates: Mouse
Neuroectoderm invaginates
forms neural tube alongside the notochord (a mesodermal strucutre)
Anterior neural tube→ brain, divided into
Prosencephalon (telecephalon and diencephalon)
mesencephalon
metencephalon
Posterior neural tube→ spinal cord→ further subdivided
Main difference between vert and invert
Inverts→ neuroblasts→ specialised neurons
Verts→ invagination first and then neural tube→ specialised neurons
From both of these sequences, the next question to ask is
How does each neuroblast/progenitor of the neural tube know which neuronal cell tpye and neural strucutures to develop?
how do they adapot a parituclar neural identiy and start expressing genes needed for developing a parituclar neural strucutre?
We will look at
Anterior-Posterior axis
Dorsal-Ventral
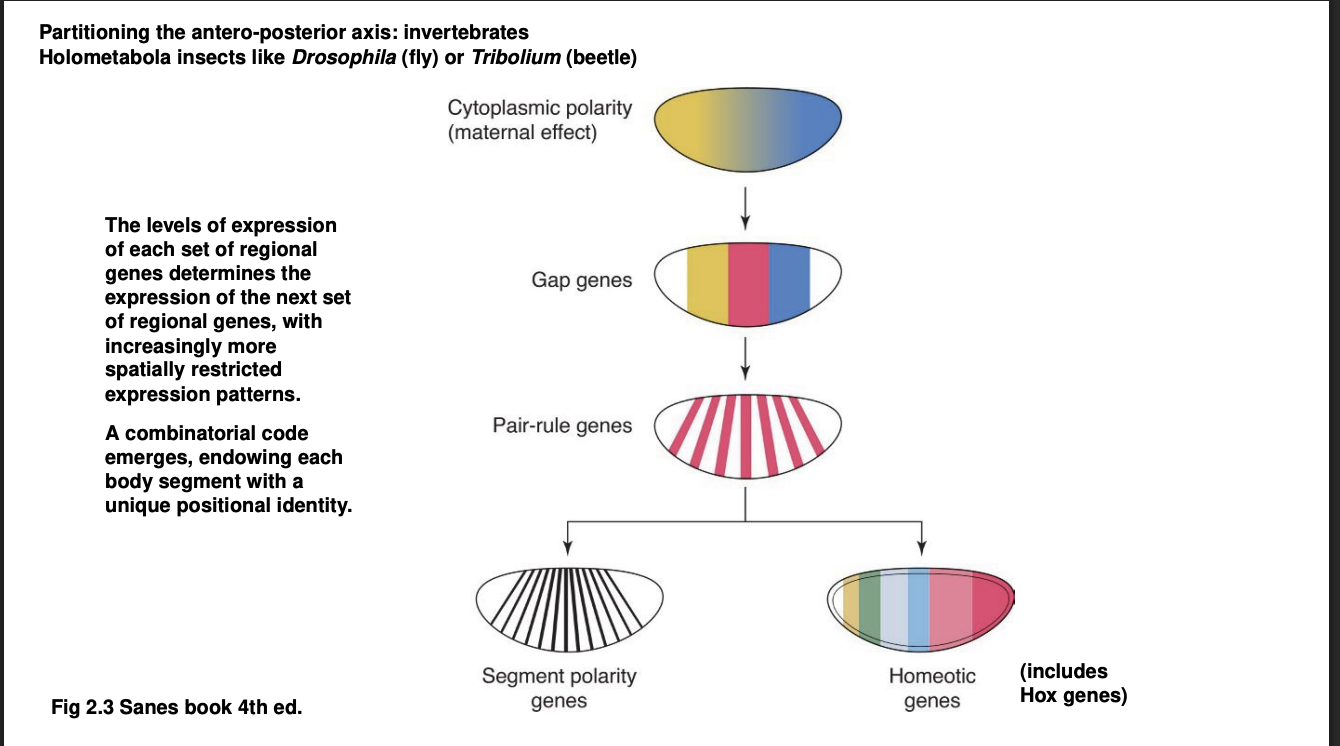
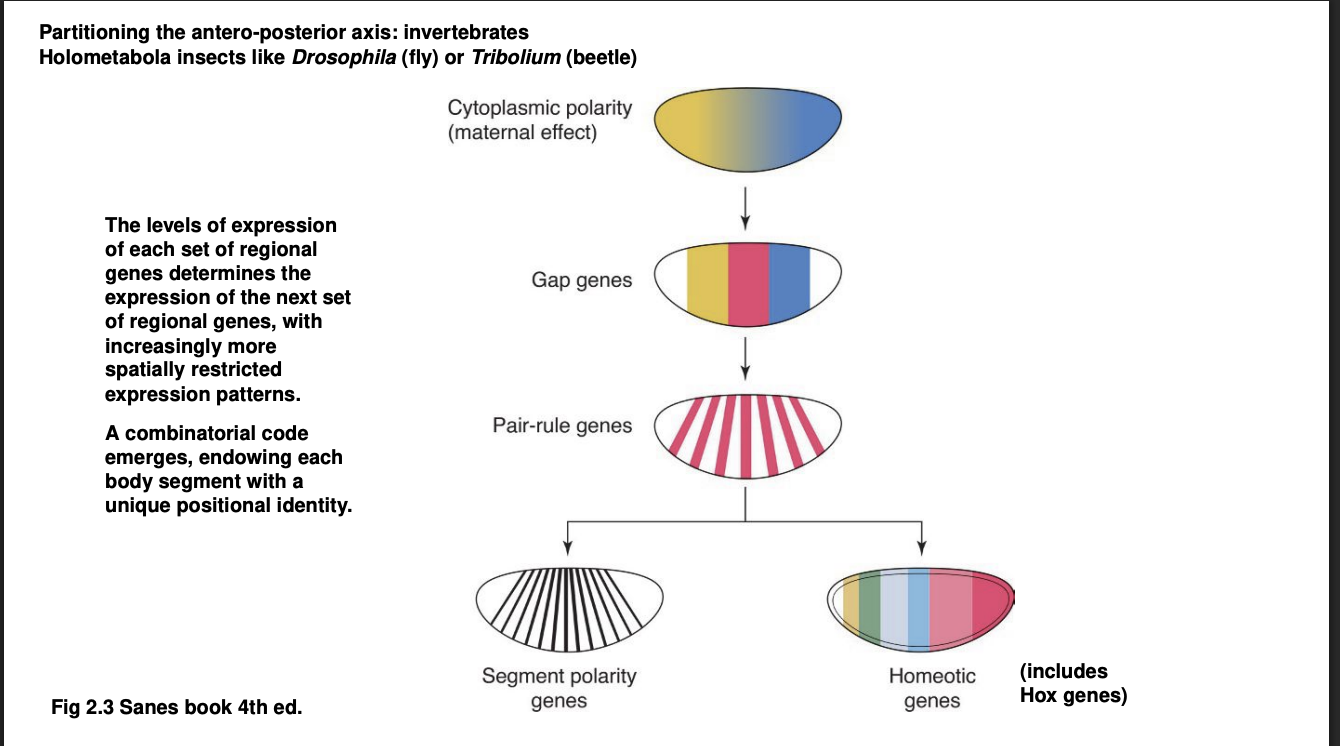
A-P axis→ Forms a kind of cascade of TF differential distributions: First Wave
Maternal mRNA
The egg itself is already polarised
differential distribution of maternal mRNA
these are translated into transciption factors
Why are TFs often referred to as morphogens
when applied experimentally
they can morph a part of the embryo into a particular tissue shape or type
A-P axis→ Forms a kind of cascade of TF differential distributions: First Wave→ How does the mRNA gradient activate different TFs
Promoter of each TF gene is differentially sensitive to the concentration of activating TFs
so the differential concentration of mRNA along the A-P axis will transcribe different TFs along when enough amount of activating TF protein is present
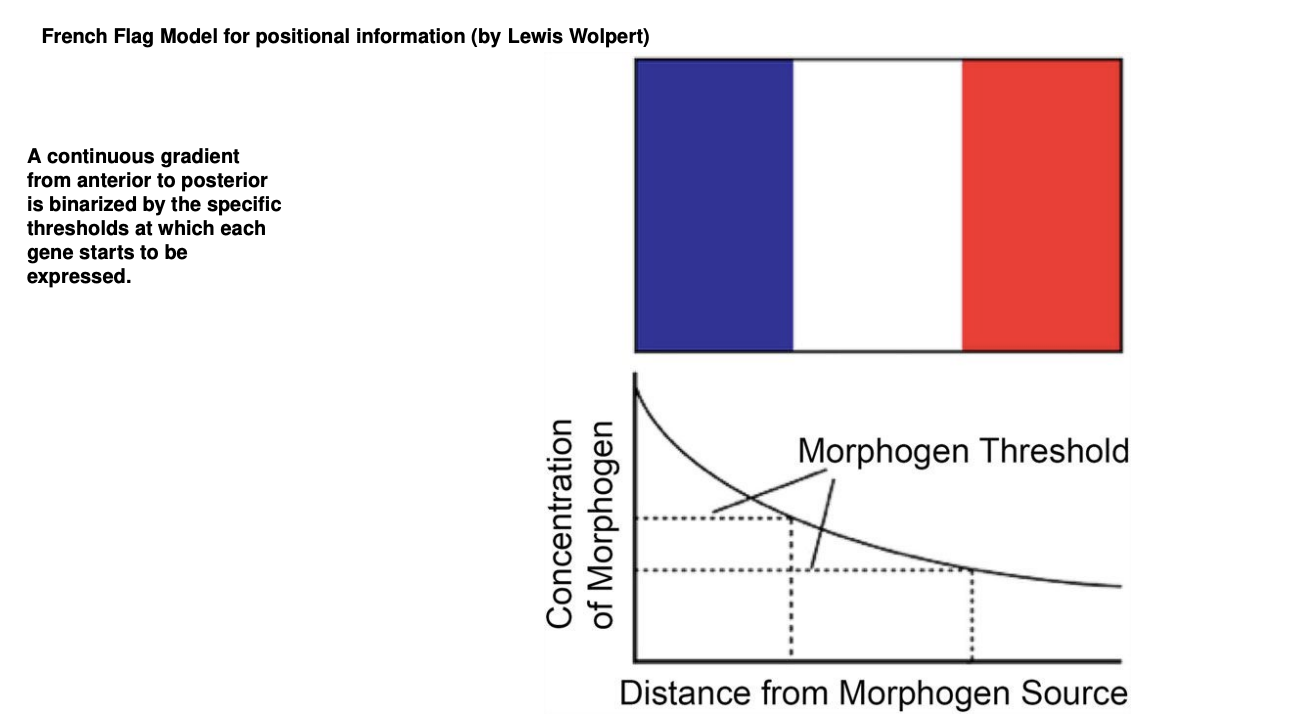
But→ this ia a continuous gradient→ it needs to be binarissez so that we have switching on and off of the TF genes
How is the continuous gradient binarized
French Flag Model
continuous gradient binarized into regions
boundaries are defined by concentration threshold of each subsequent TF gene promoter to be activated
A-P axis→ Forms a kind of cascade of TF differential distributions: Second Wave
Gap genes
Expression defines of large chunks of the A-P axis of the embryo
Experimental knock out→ removes substational and continuous portion of body regions
visible morphologically towards the end of embryonic development
hence the name ‘gap’
They are TFs themselves→ control the activation of the next wave…
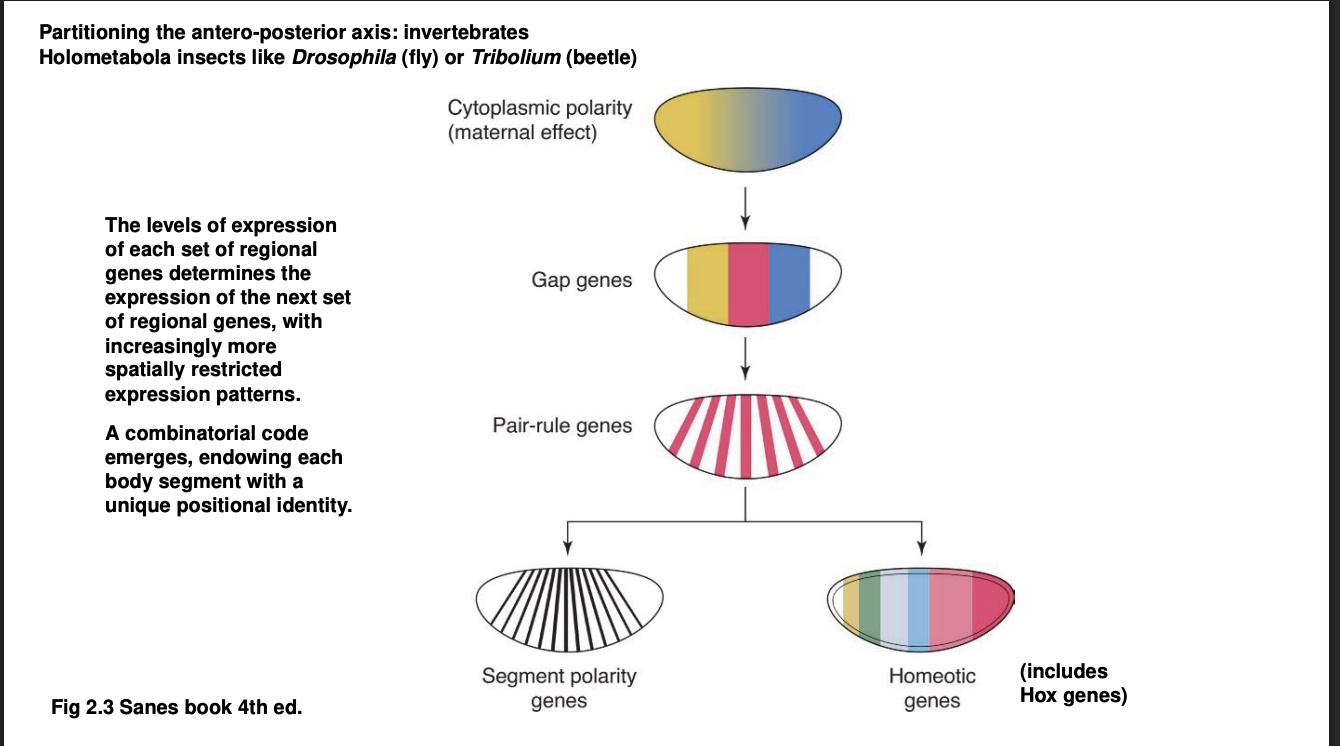
A-P axis→ Forms a kind of cascade of TF differential distributions: Third Wave
Pair-rule genes
expressed in alternaing pairs of body segments
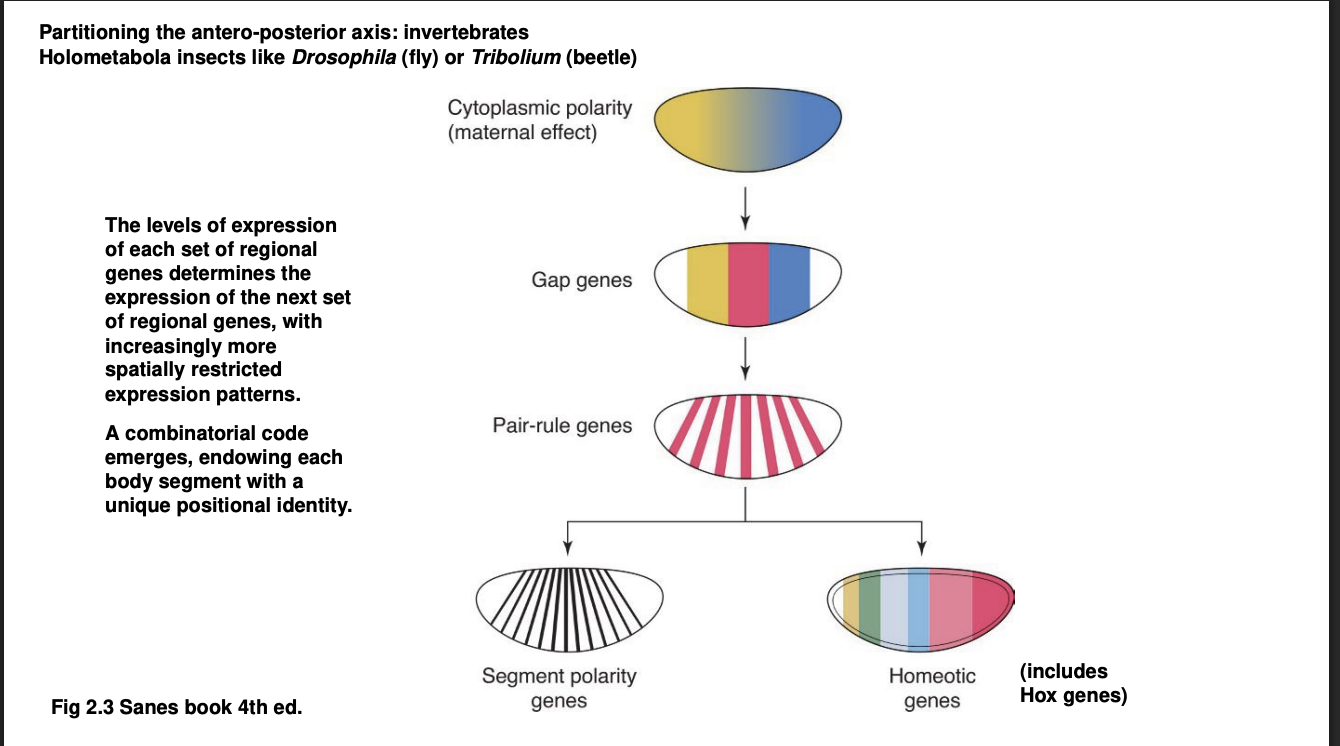
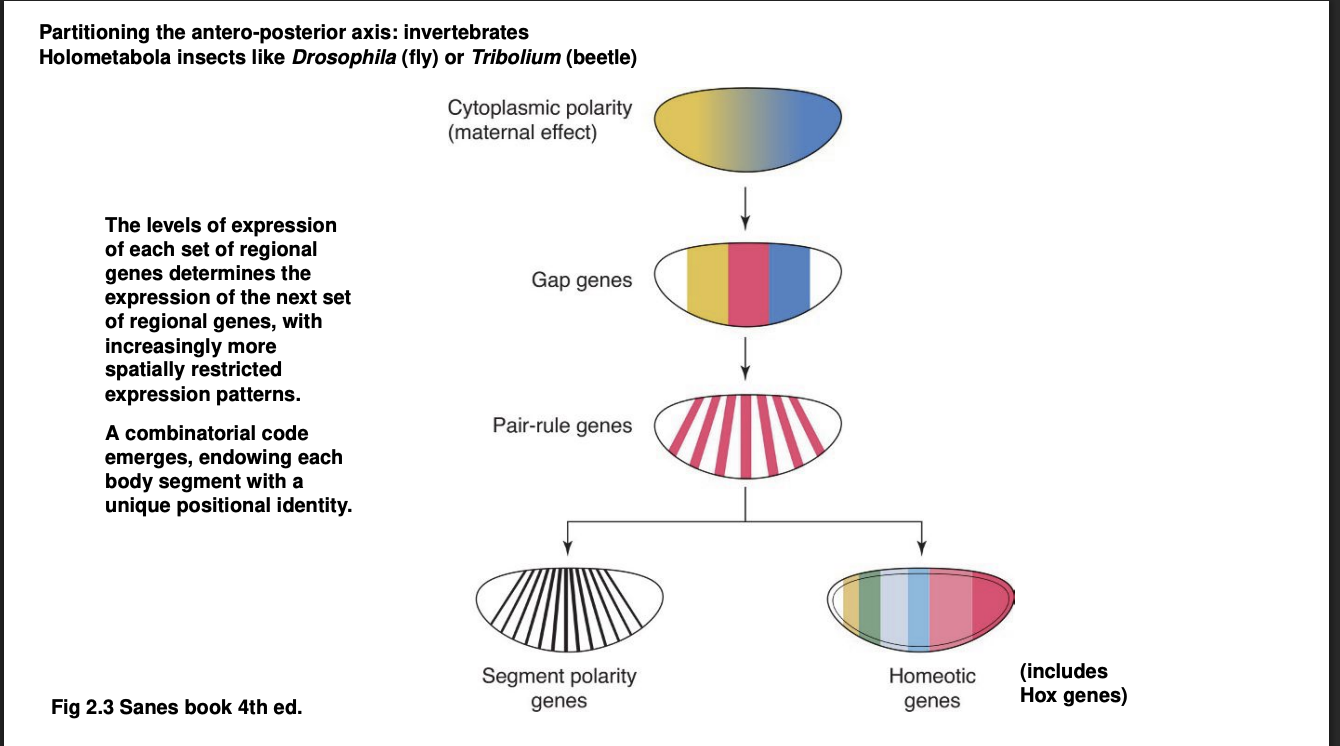
A-P axis→ Forms a kind of cascade of TF differential distributions: Fourth Wave
the combination of all or many of the TFs above control the expression of
Segment polarity genes→ expressed in a narrow band of cells of every body segment
Homeotic genes→ Hox genes are the most famous among the homeostic genes
What are Intermezzo Homeotic genes
genes that drive homeosis
→ the transformation of one organ into another
Famous examples from the history of homeotic genes
Ultrabithorax (Ubx)→ mutant in Drosophila= 2 sets of wings instead of 1
eyeless (ey) or vert homolog Pax6→ head and eye development and its induction ectopically
e.g tips of a fly leg or antennae
Hox genes features
key actos in patterning of A-P body axis
exist in all-freeliving animals
Sequential order of Hox genes corresponds to the region of expression along A-P axis
3’ to 5’ A→P
Individual Hox genes are homolgous across animals and play similar toles in development
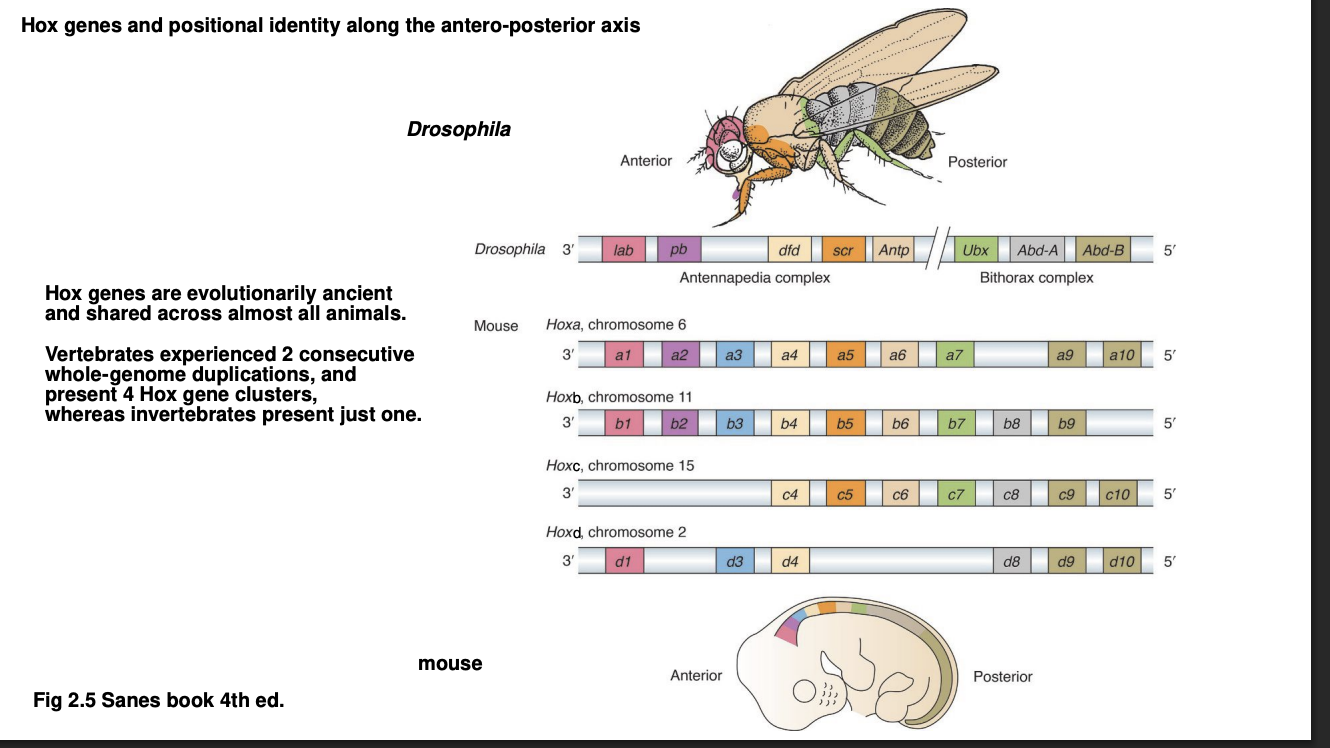
Experiments showing Hox genes conservation
DNA sequence for a specific Hox gene in a mammal can rescue the function of its homologous gene in Drosophila when knocked into its place
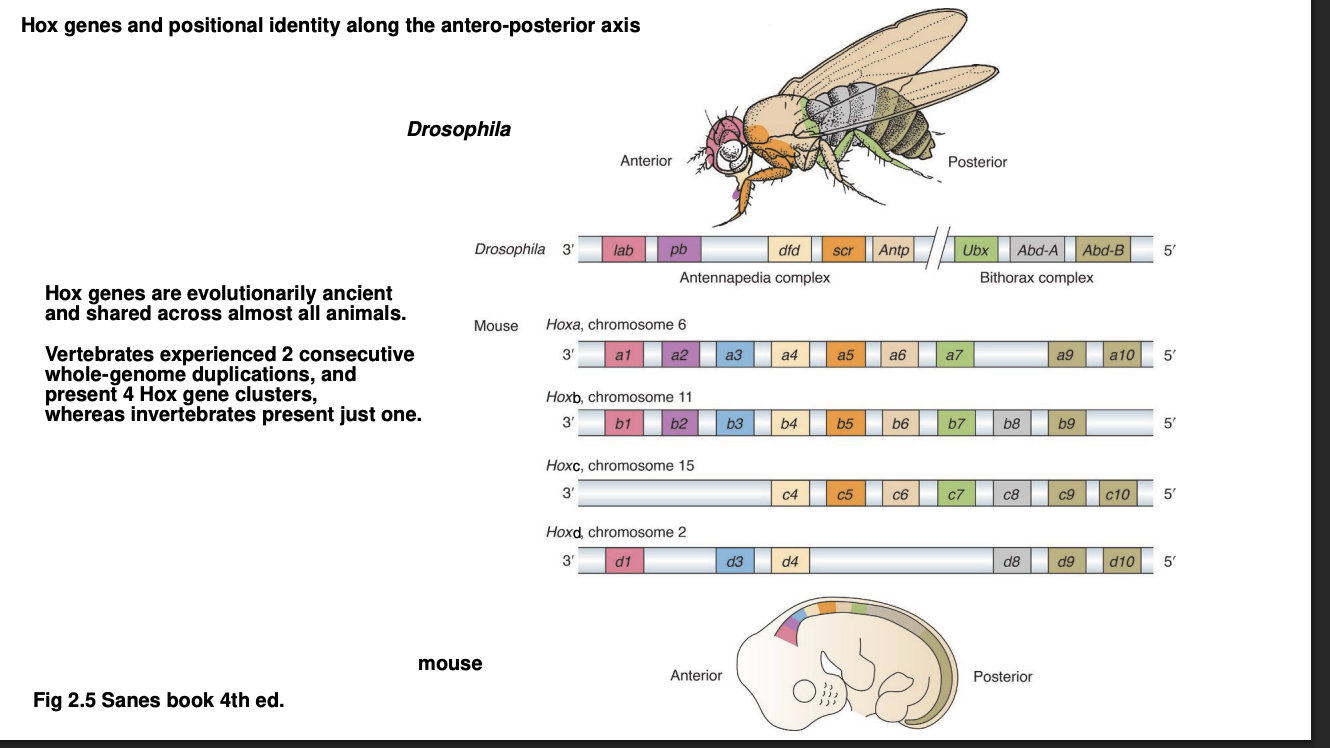
Why are there four clusters of Hox genes in vertebrate genome
experienced two round of whole-genome duplication
duplication easrly in the evolution of the chordates
But why have we mainainted the 4 clusters?
Clusters become specialised
now need them for limbs→ patterning along limb axis
What are the various Hox gene clusters called in vertebrates
Hoxa
Hoxb
Hoxc
Hoxd
→ Each gene in the cluster gets a numeric suffix
Hox gene clusters in invertebrates?→ Drosophila
Its only Hox gene cluster split into two throughout evolution:
Antennapedia complex
Bithorax complex
names reflect each cluster contains the homonious Hox gene
name itself derives from description of mutant phenotype
Role of Hox genes in patterning the A-P axis of the neural tube: (focusing on the mammalian hind brain)→ how does it work
Hox gene knockout shows
single gene can ultimately control the positional identiity and peculiarities of an organ or body region
Quick summary of the regions of the hindbrain
Subdivided into 8 Rhombomeres
each is a repeated subdivision→ a segment
distinct cranial nerves emerge from
each rhombomere
or
a combination of one or two adjacent rhombomeres
How do Hox genes help form these specific rhomobmeres with their specific cranial nerves
Genes from clusters Hoxa, Hoxb and Hoxd are expressed in particular pattern across rhombomeres
Each rhombomere has a unique combination of Hox genes expressed
each code can be shown using Hox gene knockouts to observe which rhombomeres have changed
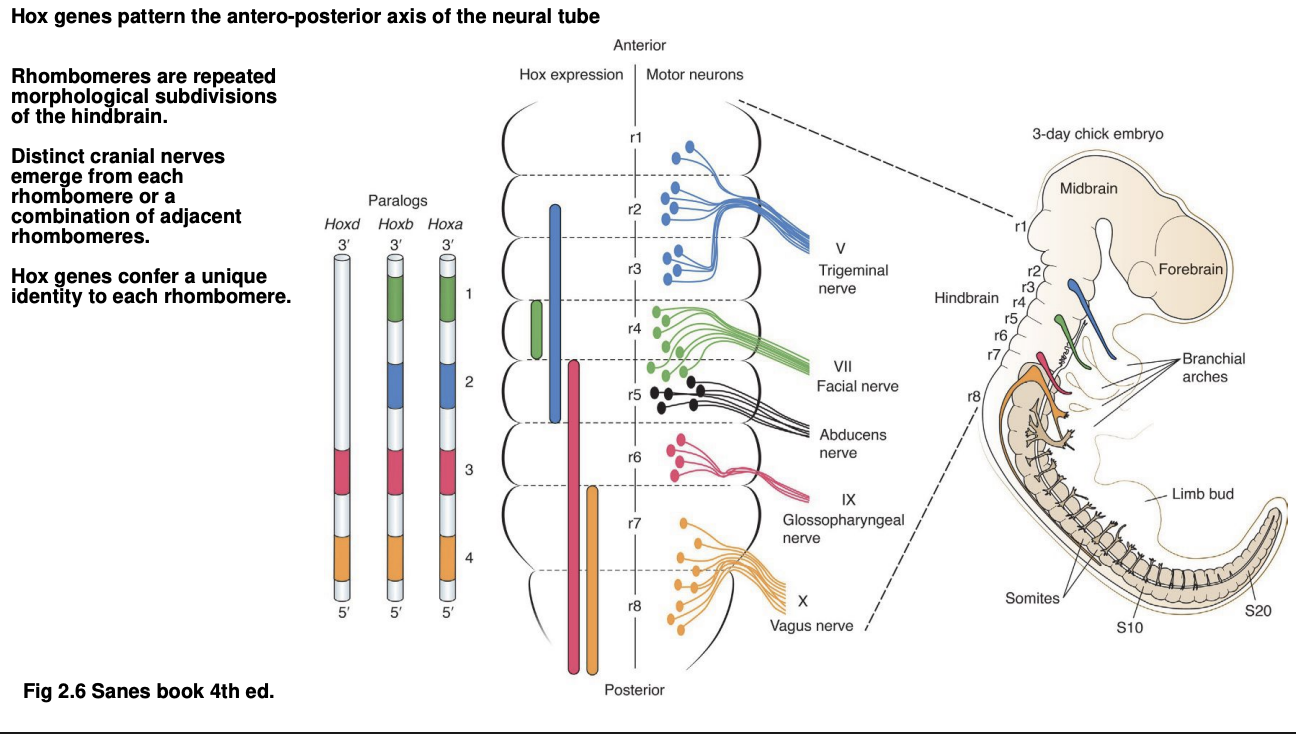
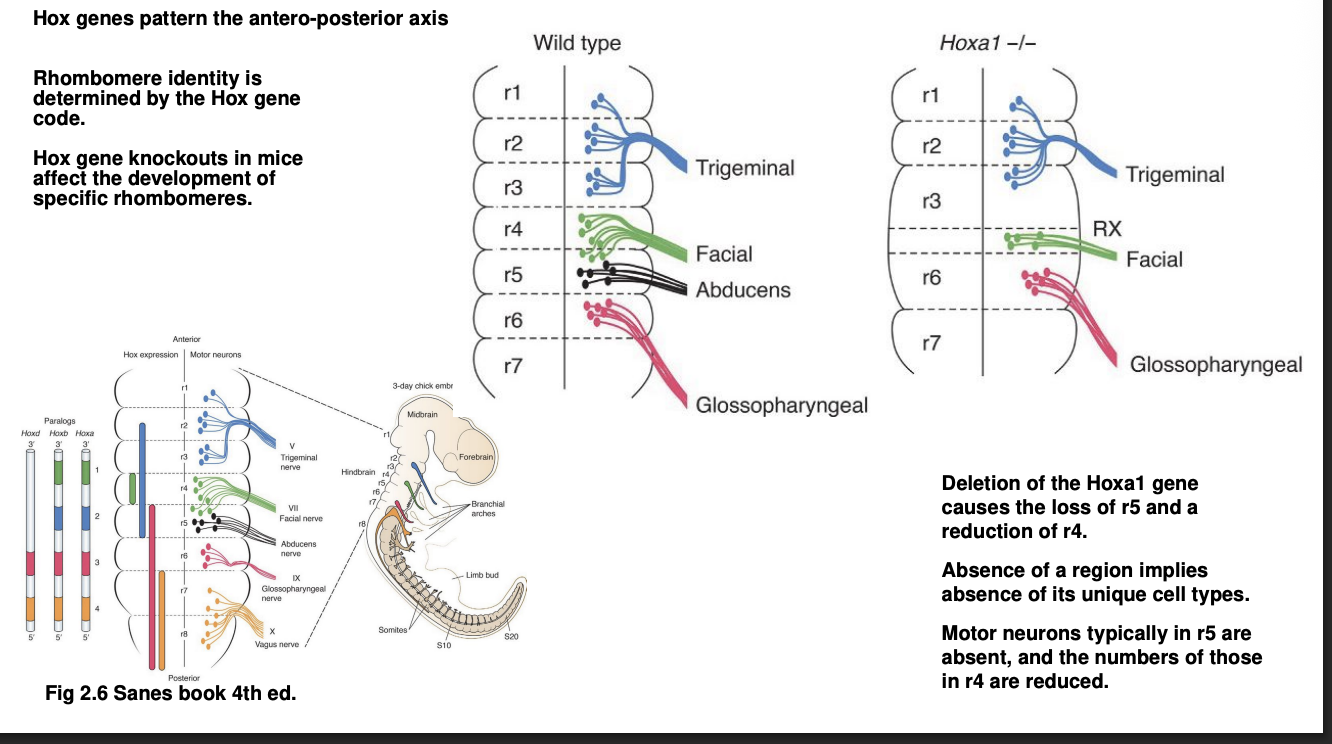
Examples of these codes
Hoxa→ expressed in r4
knowck out= loss of r4 and r5
abducens cranial nerve which normally emerges from r5 is lost entirely
facial nerve, from r4 and r5 neurons→ stronly affected
Cranial nerves from the other intact rhombomeres are themselves also intact
Why do we know Hox genes caudalize
Hox gene knockouts of whole Hox gene clusters show that all posterior segments resemble most anterior segments of the body
Example of experiment of this 1
Beetle→ Tribolium
removal of entire Hox gene cluster
→ all larval segments resemble the first, most anterior segment
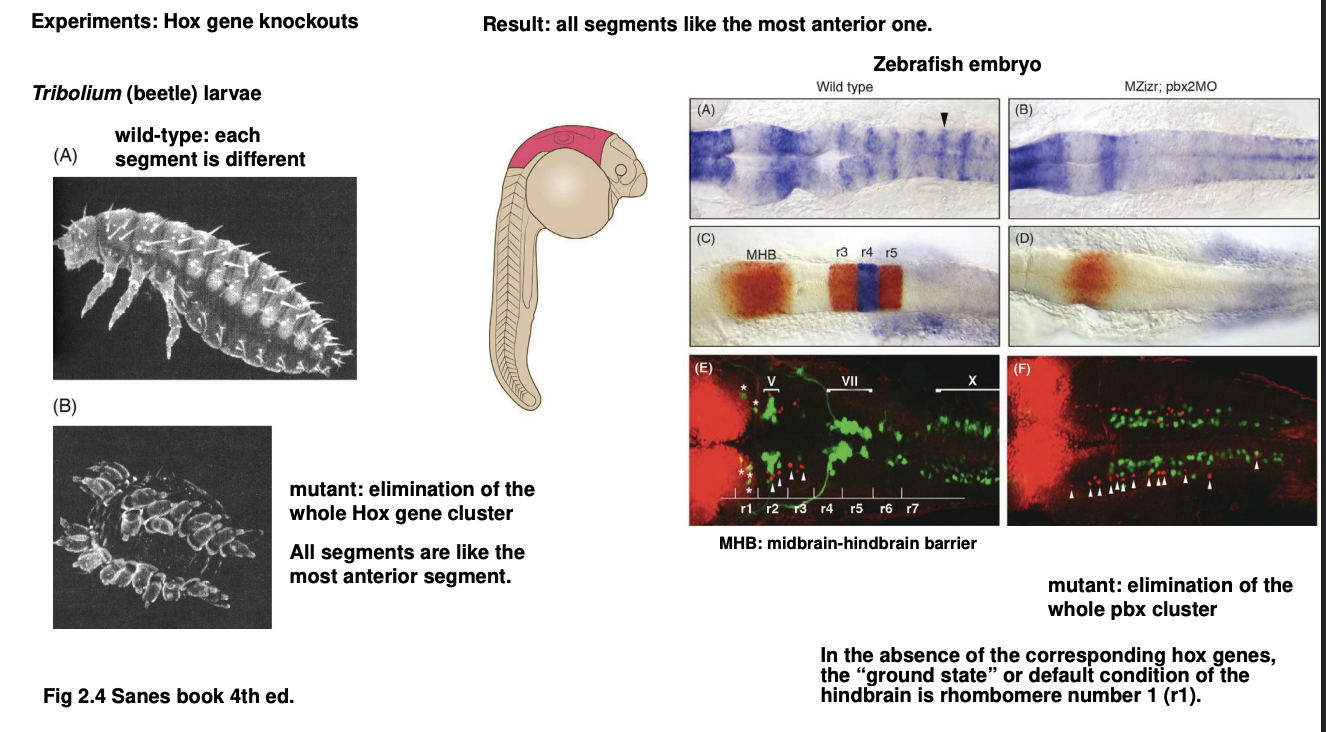
Example of this 2
Zebrafish
removal of whole pbx cluster (a group of Hox genes expressed i nthe hindbrain and its rhombomeres)
result→ entire region normally showing the rhombomeres→ now acquires the characteristics of the first rhombomere
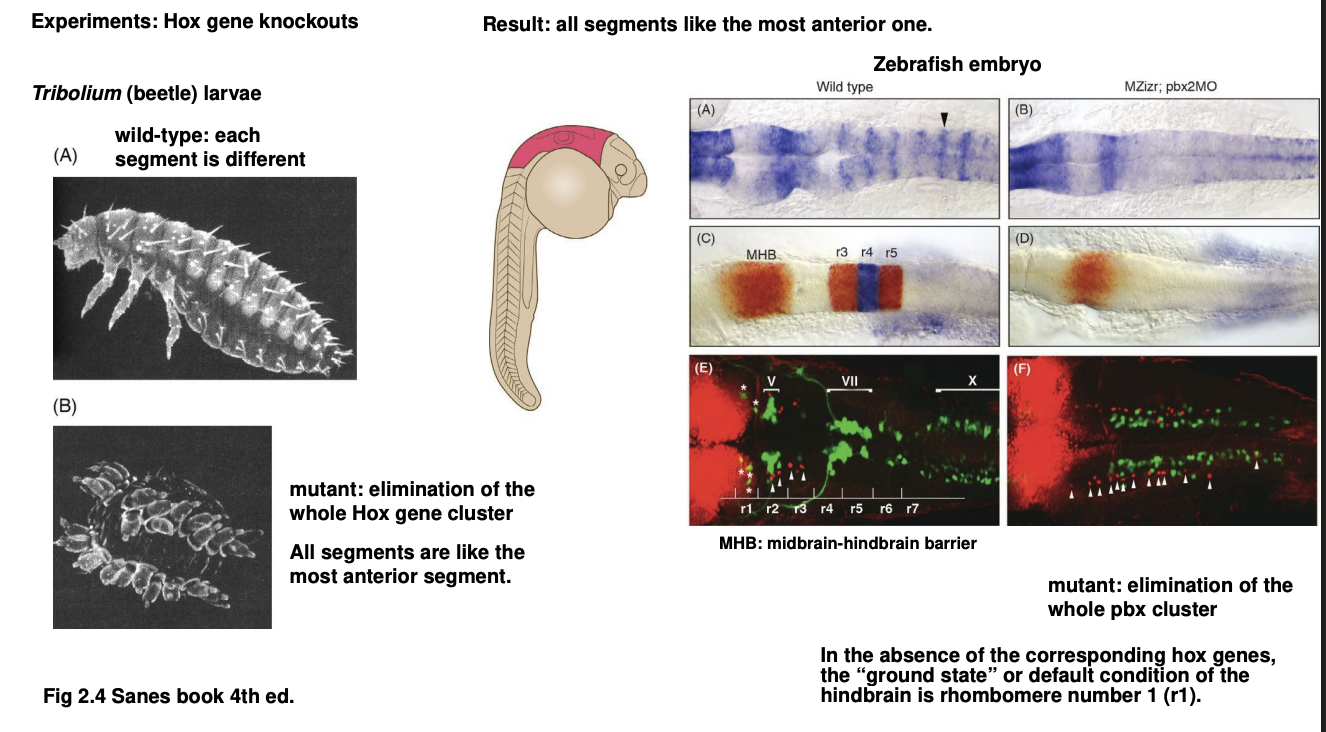
Therefore these 2 experiments show what about Hox genes
they causalize (posteriorize) the animal
the absence results in an otherwise posterior body region
presenting the identity of more anterior region
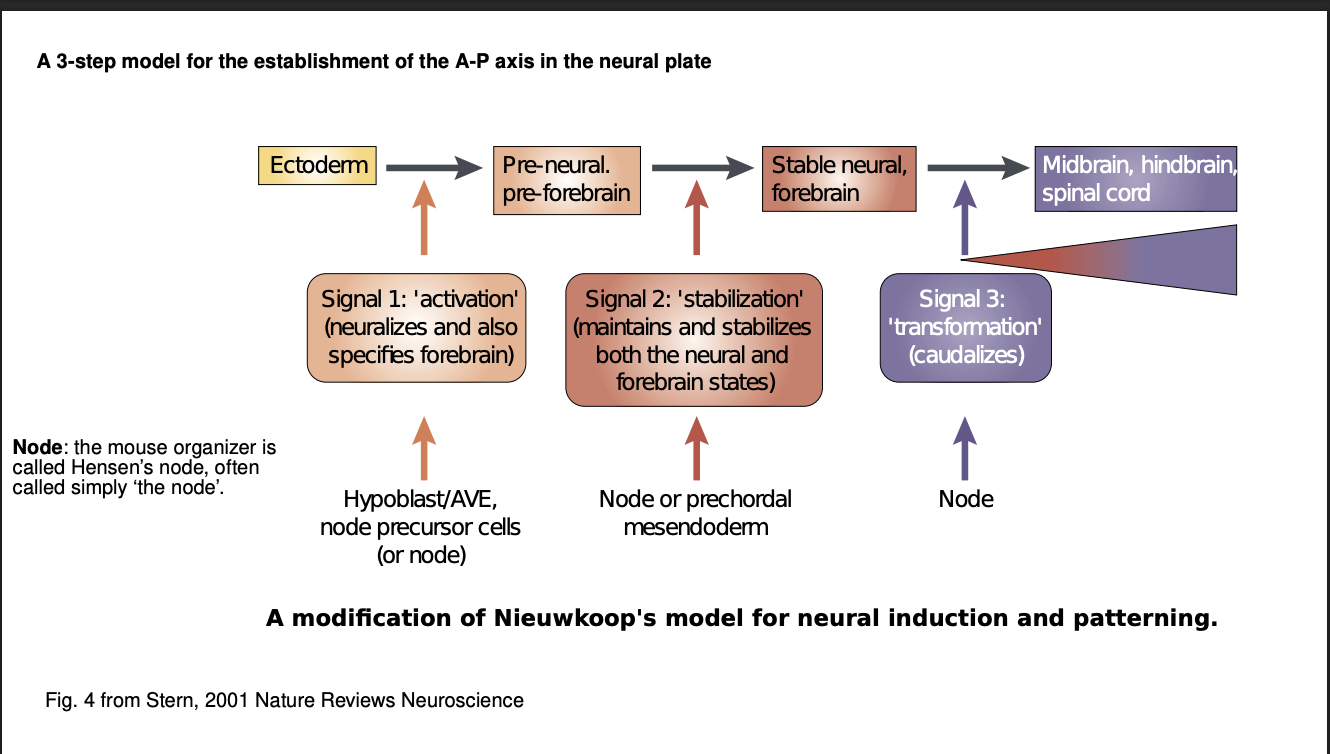
What is the 3-step model of nervous system induction and patterning
First signal→ activates (neuralizes) the gastrula’s ectoderm→ neruoectoderm
Second signal→ stabilizes neural fate of a region of neuroectoderm
defines anterior (forebrain) and posterior (the rest)
Third signal→Hox genes→ transforms (caudalizes) a region of the neural tube
details the subregions→ midbrain, hindbrain and spinal cord
But how is the Hox gene expression itself regulated along the A-P axis? (2 main facotrs)
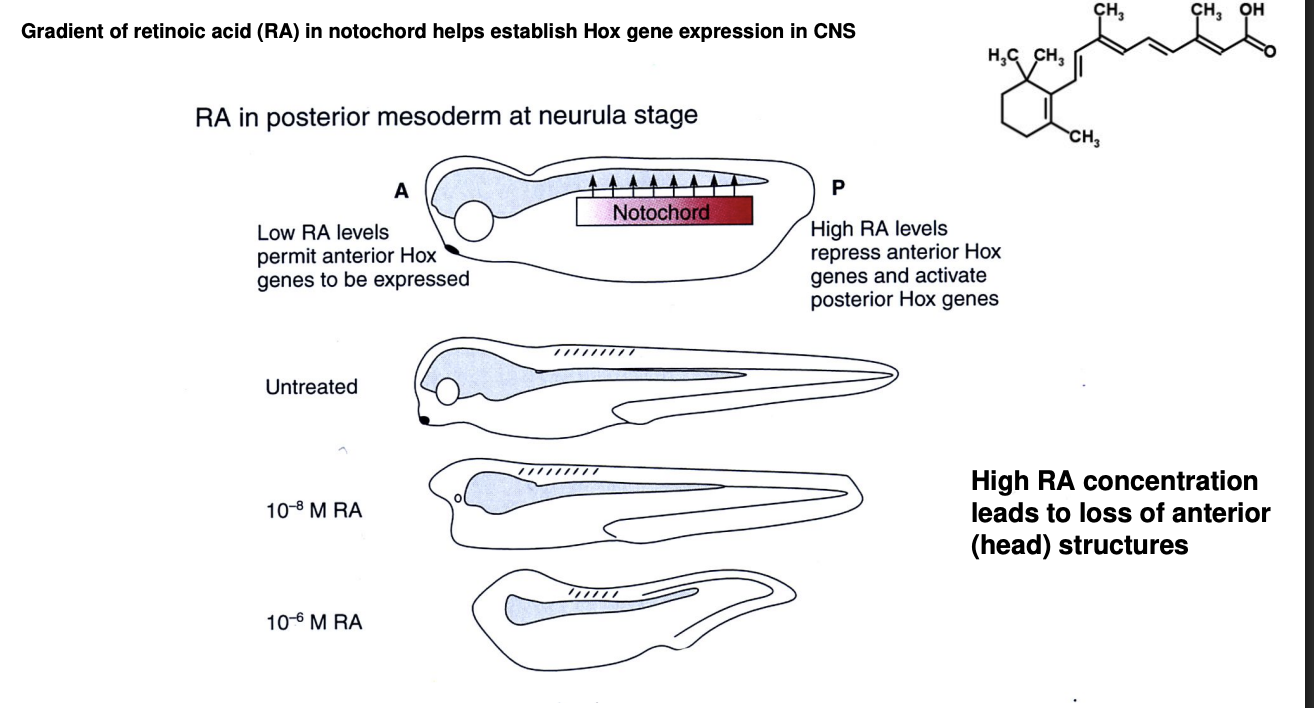
Retinoic acid (RA) gradient
FGF gradient (Fibroblast growth factor)
Where is RA released from
the notochord→ a mesodermal structure
Distribution of RA release along the notochord
Anterior→ little
Increases more and more…
Posterior→ highest level
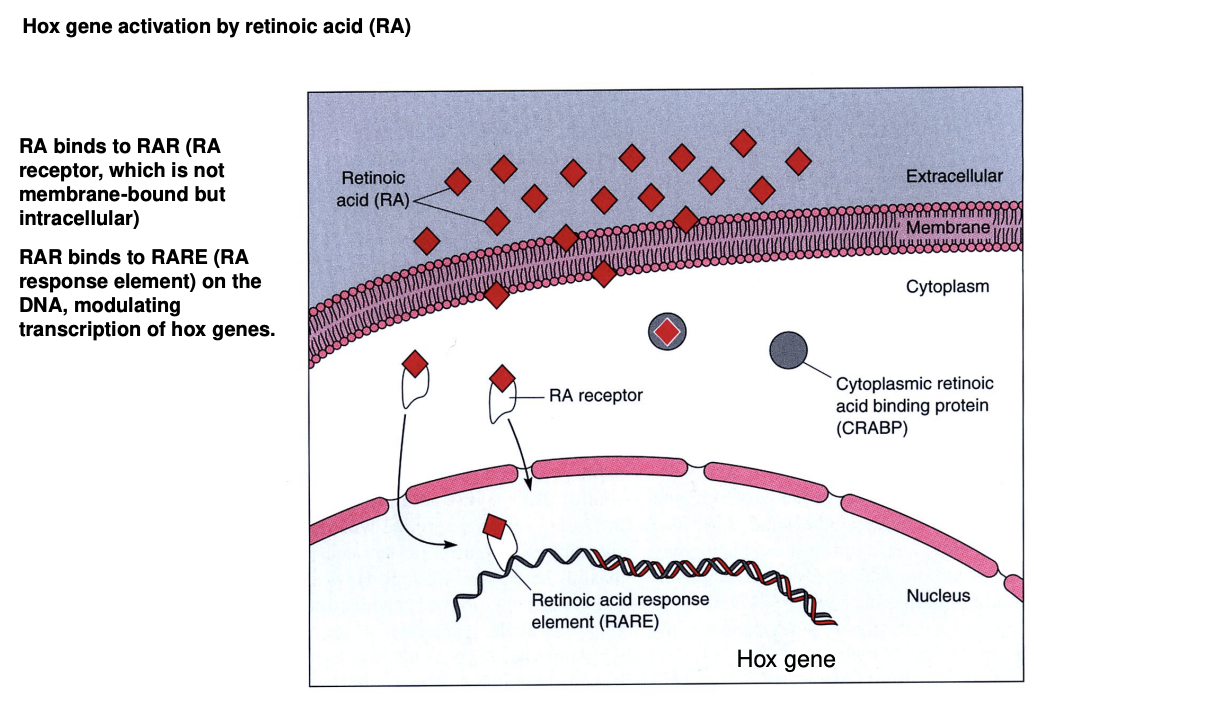
What does RA do
small molecule→ diffuses into the developing neural tube
crosses cytoplasmic membranes
binds to an intracellular receptor (RAR)
Binds to RA Response Element (RARE) on the DNA
activates Hox gene expression
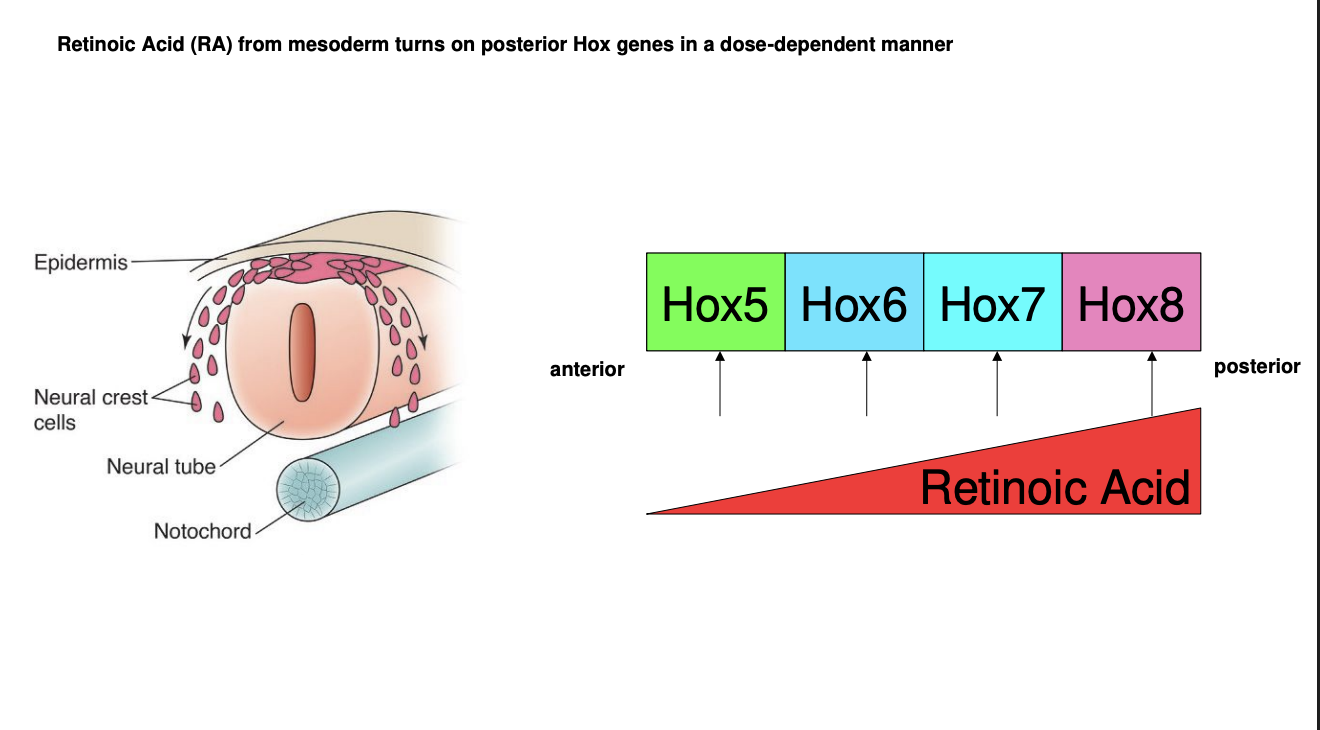
How does the gradient cause different Hox genes to be expressed
Different Hox gene promotoers are activated by different concnetrations of RA
Experimental evidence:
High RA→ loss of anterior strucutres
shows→ high levels drive expression of Hox genes for posterior identity
Low RA-. permits expression of anterior identity
Each Hox gene is activate at a different level of RA concentration→ therefore each region of the neural tube along the A-P axis will express one or more Hox genes
The combinatorial code further assists in narrowing down the identity of each neural tube region in the A-P axis
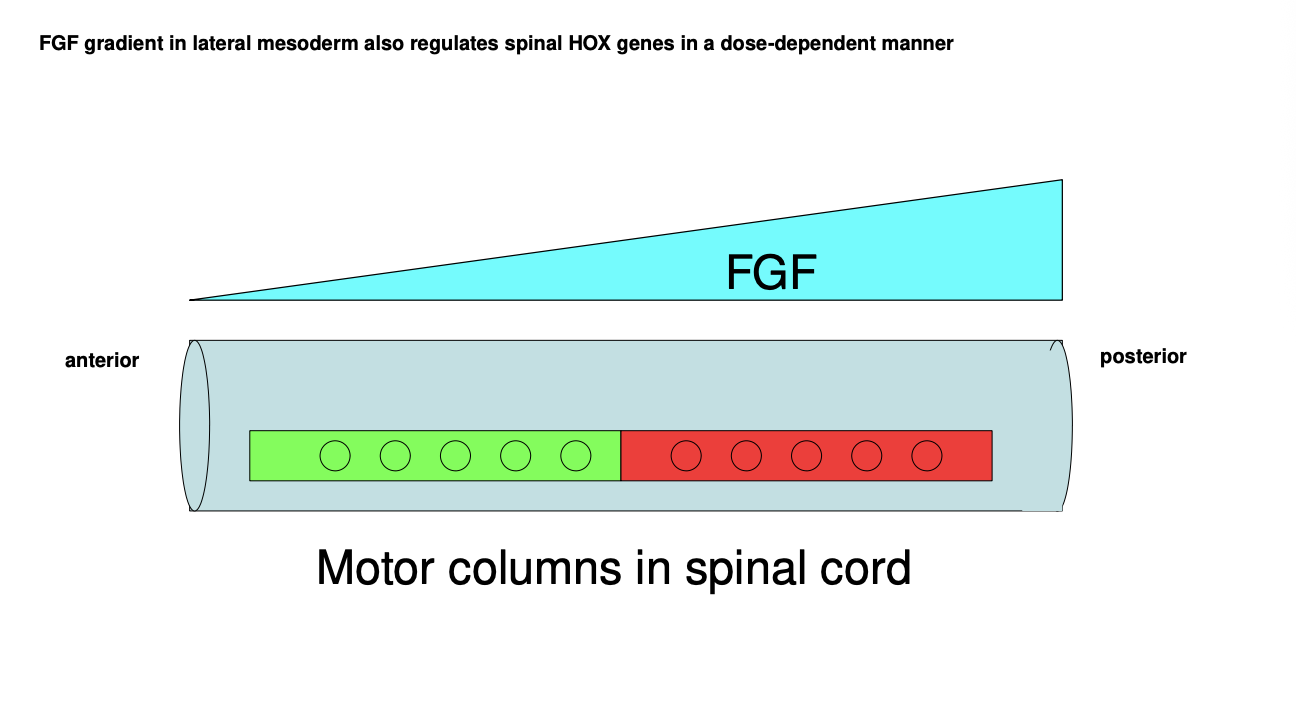
What is FGF where where expressed
secreted by the lateral mesoderm
(compared to RA emitted by the notochord , ventral to the neural tube)
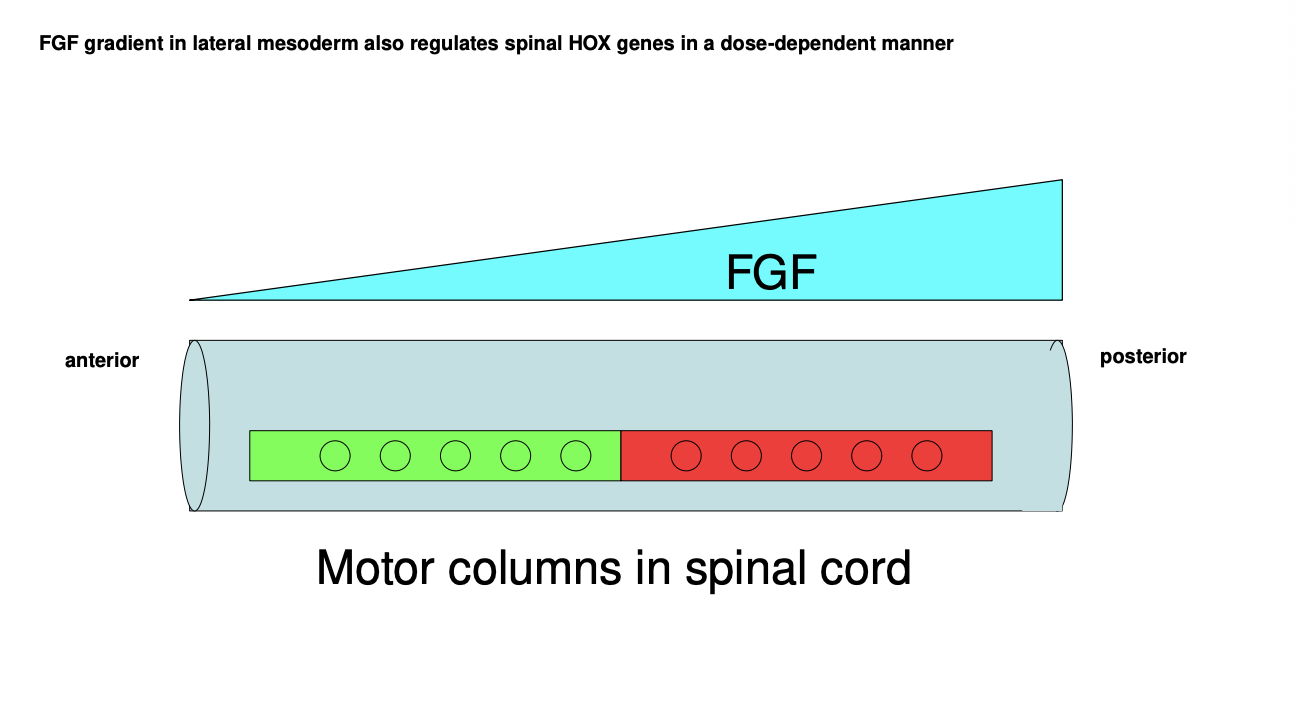
FGF gradient what it does
along the A-P axis
role in regulating Hox gene expression in a dose-dependent manner
combination further refines the posistional identity of every A-P region of the nerual tube
by differentially driving gene expression
Dorsal-Ventral axis is also patterned by→ protein gradients: Early development
Early in nerual tube development , when it invaginates
notochord already expresses Shh (sonic hedgehog)
what does Shh do
induces the formation of the floorplate
What is the floorplate
the most ventral region of the neural tube
it subsequentally releases Shh itself
So with the initial release and the induced release from floor plate of Shh
Shh forms a gradiet from ventral upwards
Further in development→ once the neural tube closes what happens
Most dorsal part forms the roof plate
What does the roofplate do
secrets BMP and Wnt
in a D-V gradient→ Dorsal down
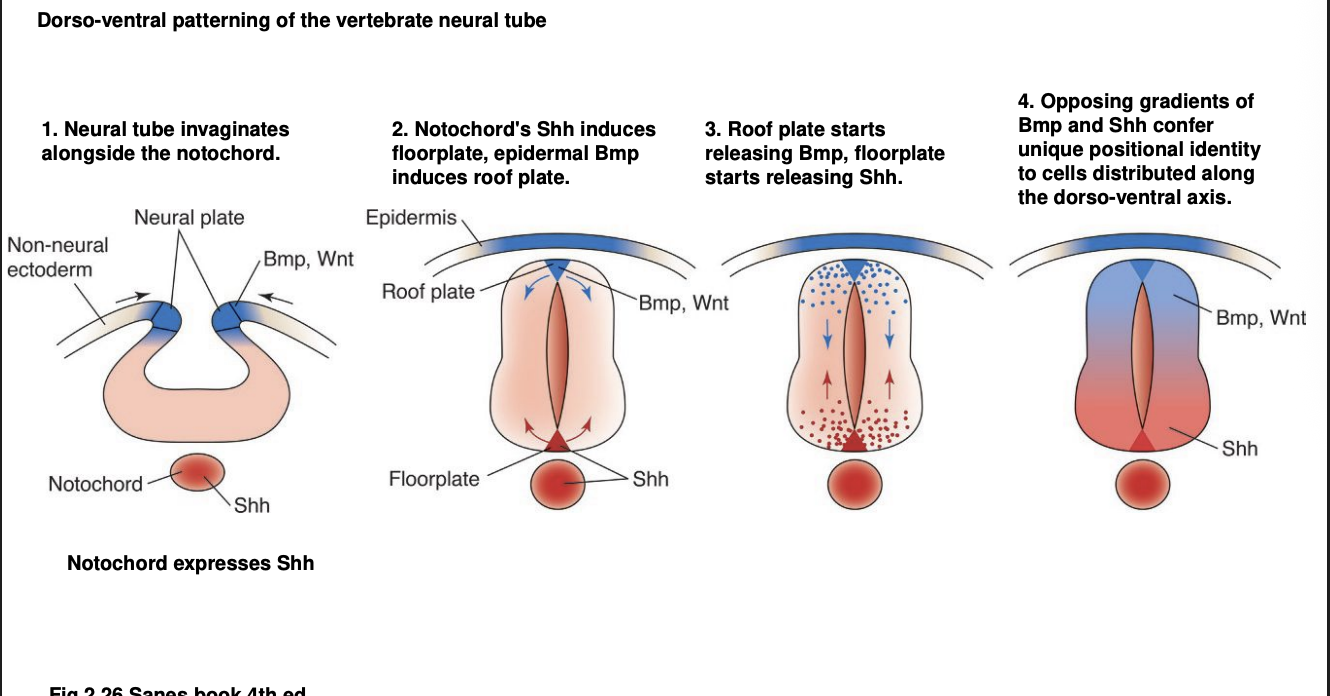
Overall summary of D-V patterning in vertebrates neural tube
Neural tube invaginates alongside the notochrod
Notochord Shh induces floorplate
epidermal Bmp induces roof plate
Roofplate starts releasing Bmp 9autonomously
floorplate starts releasing Shh autonomsouly
Opposing gradient of Bmp and Shh confer unqiues positional identity to cells distriuted along the D-V axis
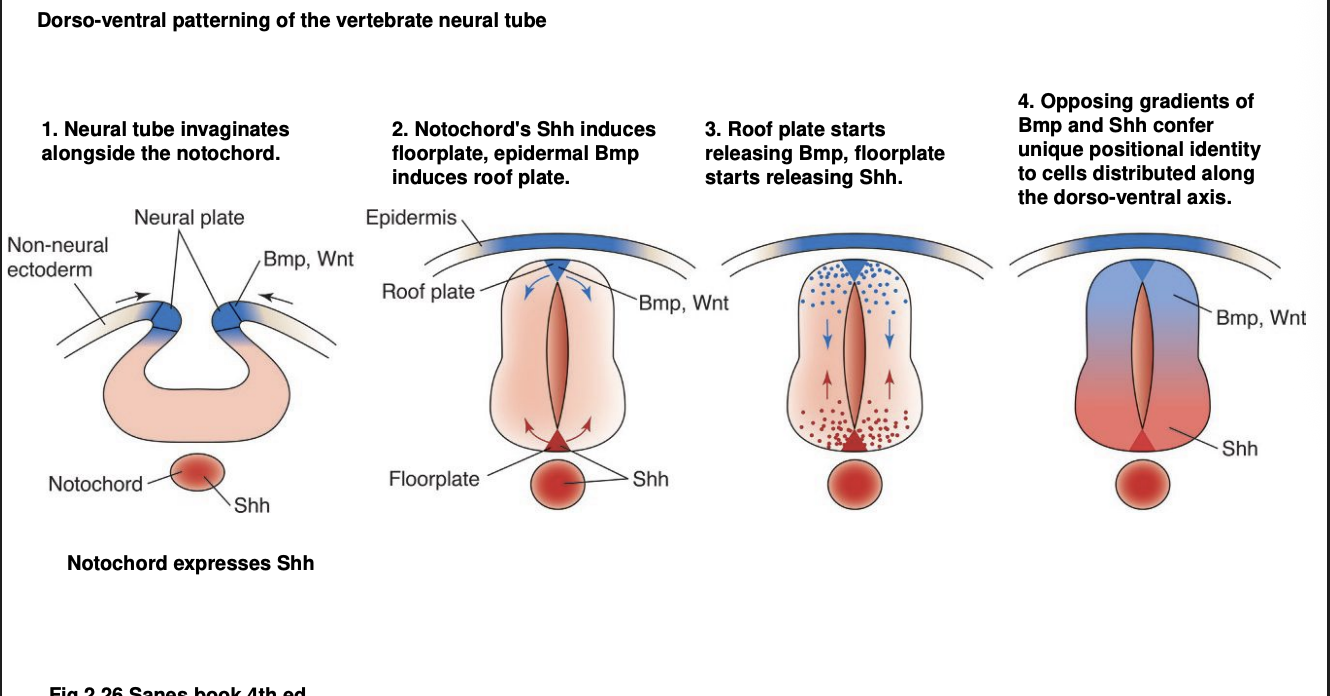
Overall we have a combination of D-V gradients
Shh→ Ventral up
BMP and Wnt→ Dorsal down
What this does:
gradient specifies a uniques D-V positional identity for differentiating neurons of the neural tube
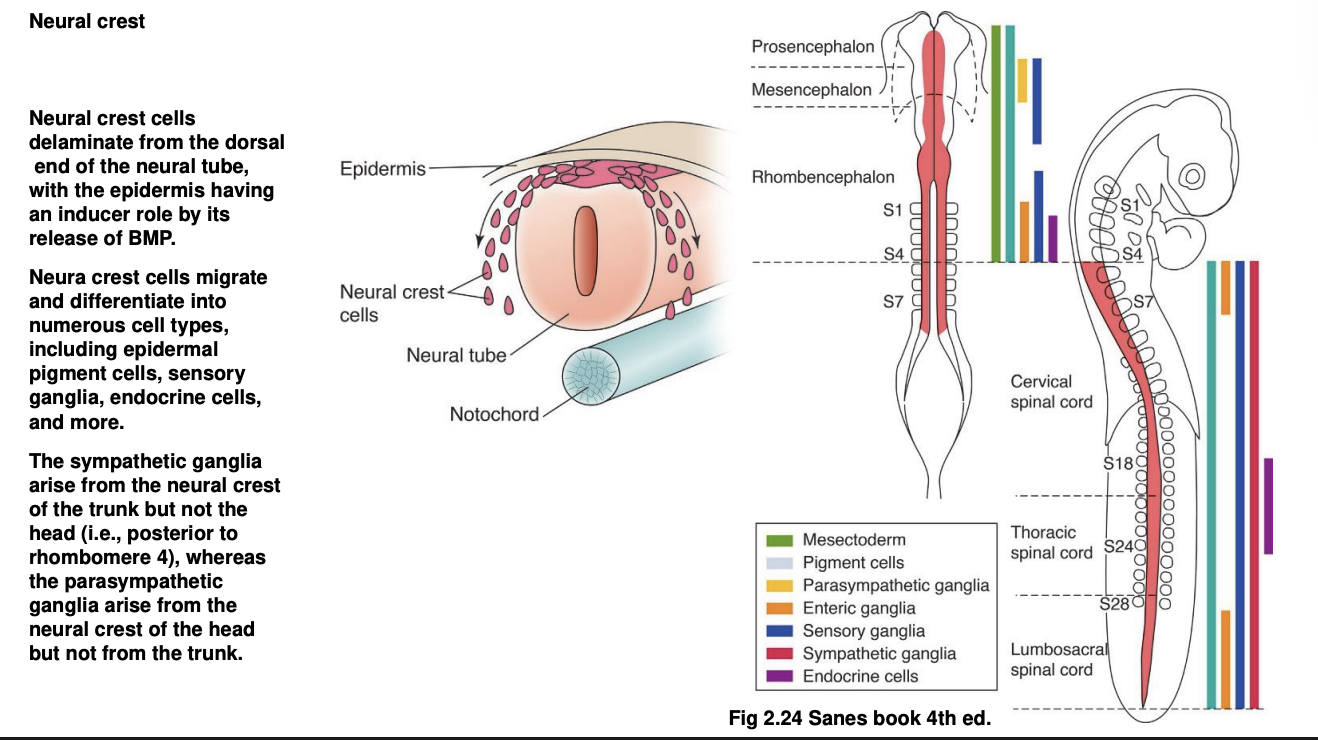
In addition, what does the interaction between the dorsal part of the nerual tube and epidermis cause (using the BMPs)
induces differentiation and subsequent migration of the neural crest cells
What do these neural crest cells do
Differentiate into numerous cell tpyes:
epidermal pigment cells
sensory ganglia
endocrine cells
etc
How do sympathetic vs parasympathetic ganglia arise from the neural crest
Sympathetic→ from the neural crest of the trunk but not the head
e.g posterior to rhombomere 4
Parasympathetic→ arise from neural crest of the head (but not the trunk)
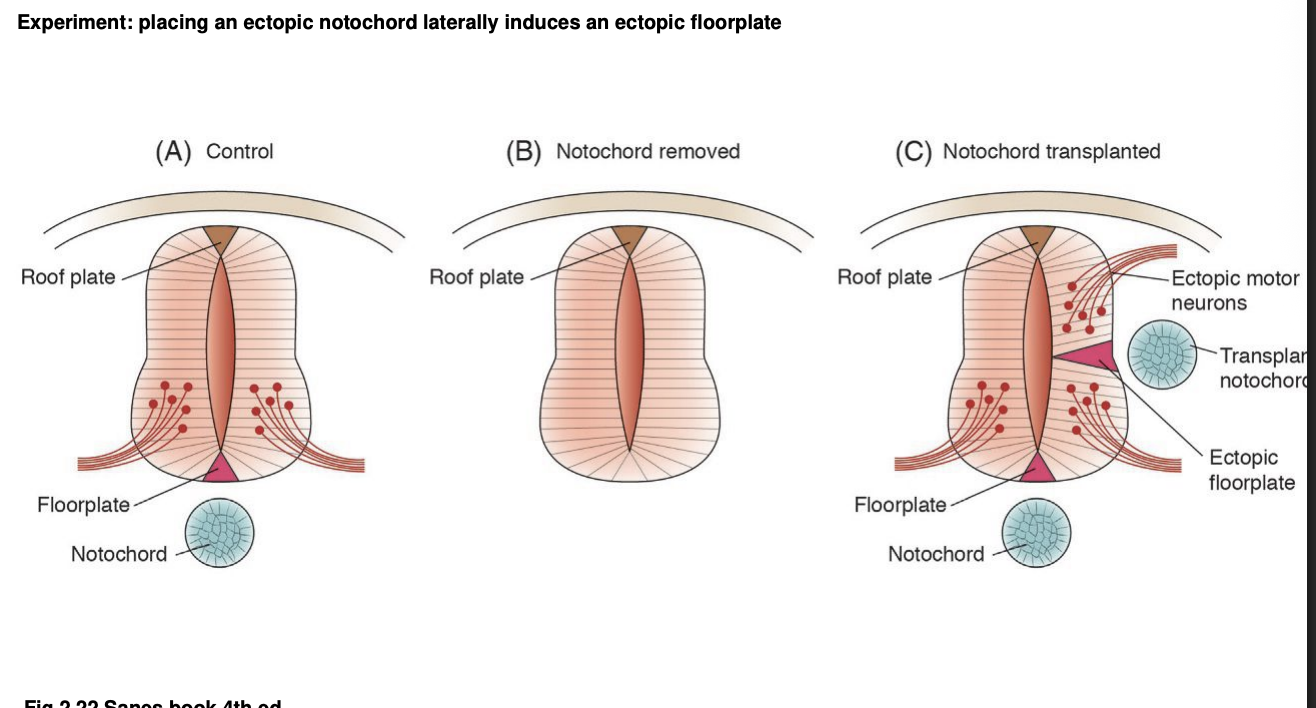
Experimental embryology and the D-V axis: role of the notochrod in inducing the floorplate formation has been determined
Procedure: ectopic tissue grafts
results:
Notochord removal→ prevents formation of floorplate →necessity
Notochord addition on the lateral side→ ectoptic floorplate→ sufficienty
complete with correspndinng complement of ventral motor neurons
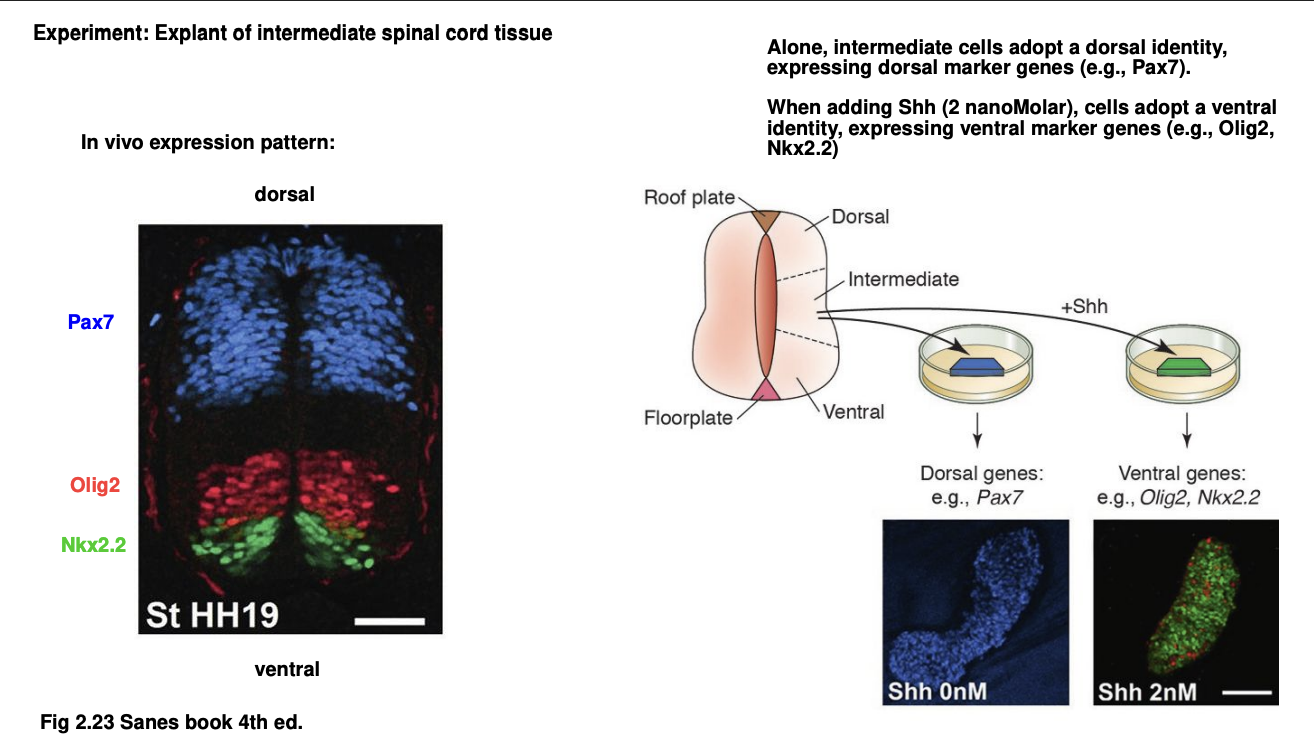
Experient 2: deafult D-V identity of spinal cord studied with tissue explants
Procedure: tissue explant alone or with added Shh, compared to in vivo
Results:
In vivo→ dorsal half of spinal cord expresses Pax7 gene
bottom third expressed Olig2 and Nkx2.2 genes
Cultivated intermediate part of spinal chord alone→ neurons express dorsal marker genes→ e.g Pax7
Addition of Ash→ shift identity to ventral→ observing then the expression of Olig2 and Nkx2.2 genes
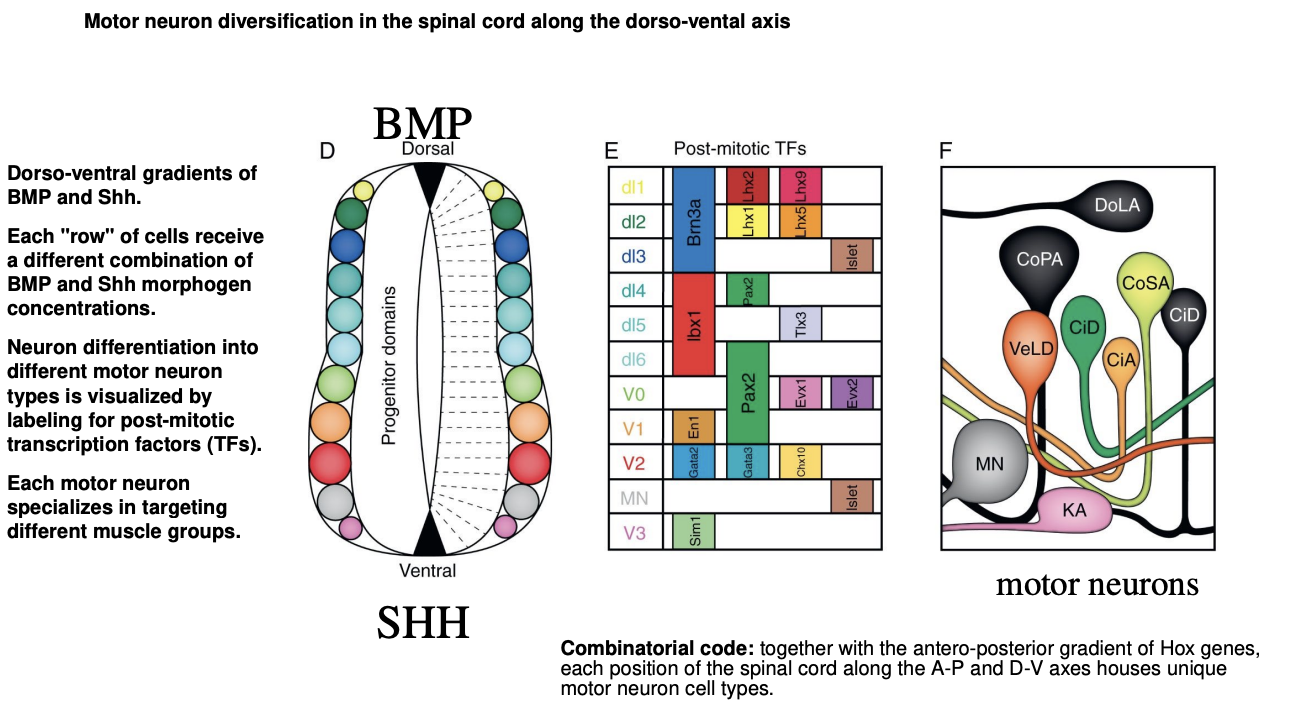
Moto neuron diversification in the spial cord along the D-V axis
DV BMP ad Shh gradients
each row of cells receive a different combination of BMP and Shh morphogen concentraions
neuron differentiation into different morot neuron types is visualized by labeling for post-mitotic transciption factors TFs
each moto neuron specializes in targeting different muscle groups
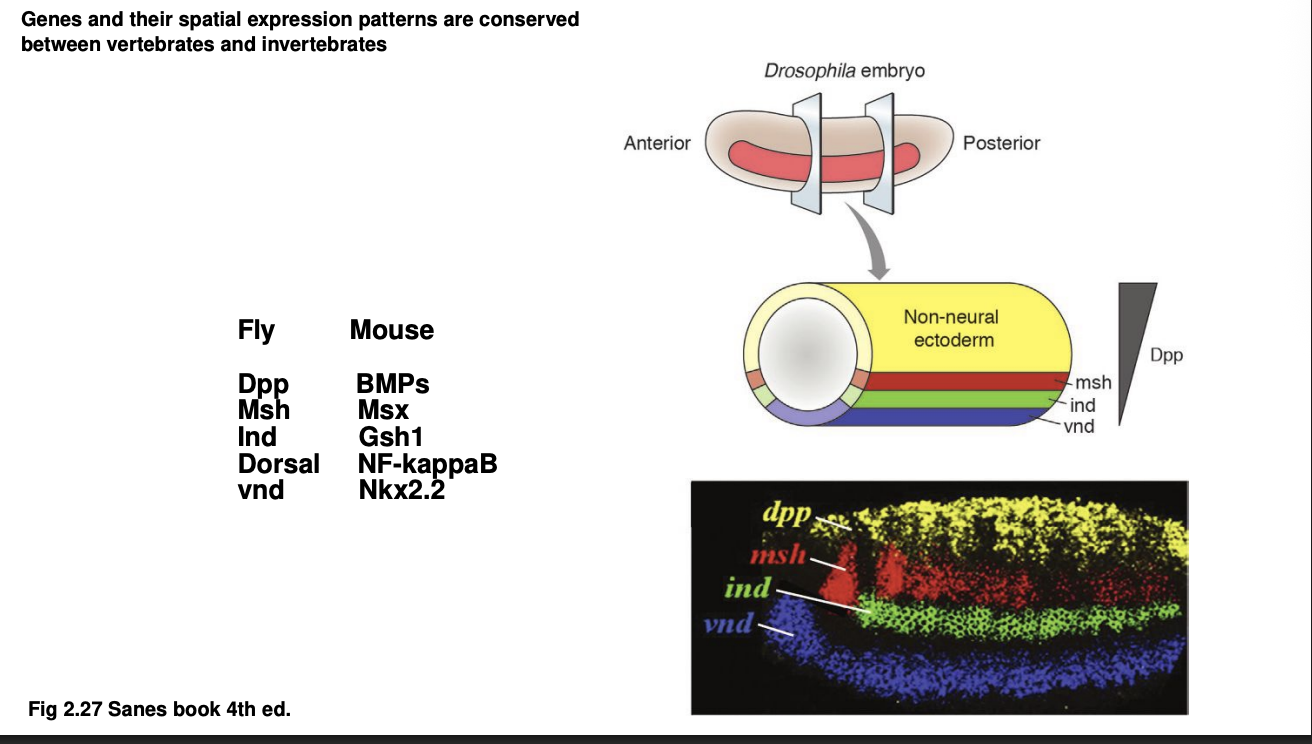
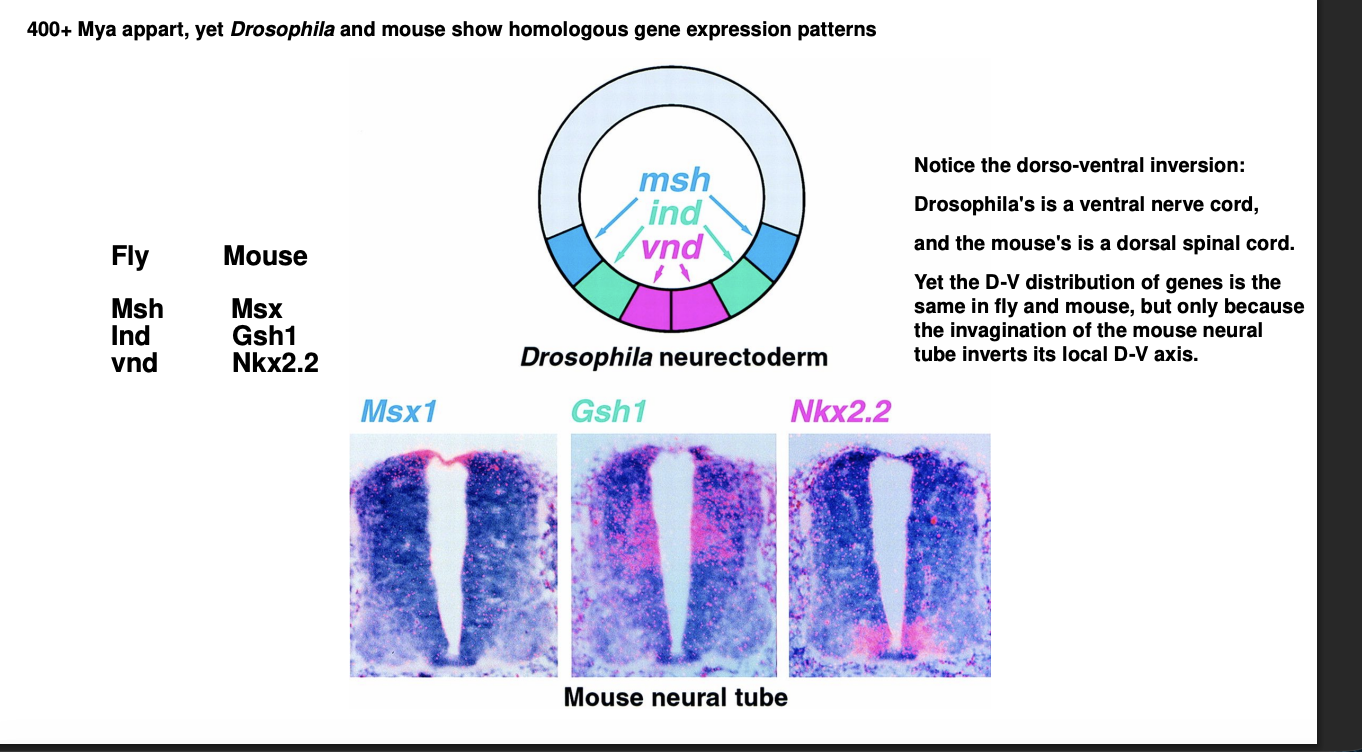
D-V patterning genes are evolutionarily ancient→ how
Similar how Hox patterns work in mouse vs Drosophila (sp both A-P axis and D-V axis are conserved)
Examples of homolgous:
Drosophila→ Msh, Ind, Vnd
Expressed in the neuroectoderm from droal to ventral
Mouse→ Msx, Gsh1, Nkx2.2
expressed in neurla tube dorsal to ventral
(How these genes are really homolgous even though look like they are being expressed in the oppsoite direction)
Must consider the Dorsal-ventral inversion in vertebrates
Neural tube is invaginated
therefore→ bottom of the neural tube is embryonically the most dorsal part of the neuroectoderm
the invagination of the neural tube reverses the relative position of the regions expressing these genes
therefore→ match with the absolute position of their homologues i nthe fly
recall: invertebrates are D-V inverted relative to vertebrates
Therefore: genes and their spatial expression patterns are conserved between vertebrates and invertebrates
Overall
D-V gradient along the spinal cord generate great diversity of motor neurons
as detected by the unique expression of myriad additional genes specific of each neuronal cell type
Together with A-P axis partiationing
each spatial location of the neural tube acquired a unique identity needed for development of the complete organism