Lecture 36-Mutations
1/44
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
45 Terms

What is a mutation?
heritable change in a DNA sequence that can lead to a change in phenotype; can occur spontaneously (SNPs, recombination events) or via horizontal gene transfer
What is a wild type strain?
•Typically refers to a strain isolated from nature
“Wild-type” can also refer to just one gene
What is a mutant?
a cell or virus derived from the wild type strain that carries a nucleotide sequence (genotype) change
Observable properties (phenotype) may also be altered
Can also be obtained from a parental strain which was derived from a wild-type strain
Genotype is designated by what?
•italics (three italicized lowercase letters followed by a capital letter) for a gene (e.g., the hisC gene encodes the HisC protein)
Mutations are designated with what?
numbers referring to the order of isolation (e.g., hisC1, hisC2)
Phenotype is designated by what?
a capital letter followed by two lowercase letters and +/– to indicate presence/absence (e.g., His+ or His– = able/unable to synthesize histidine)
Virtually what characteristic of an organism can be changed by mutation?
any
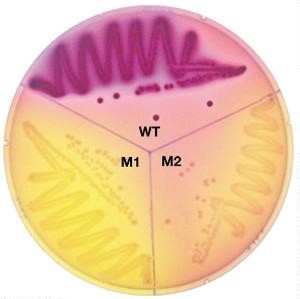
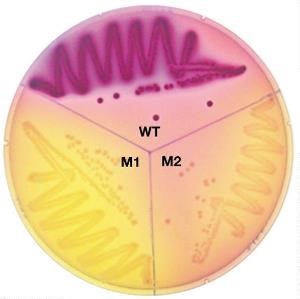
•Selectable mutations confer an advantage. How?
•Under certain environmental conditions, progeny outgrow and replace the parent (e.g. antibiotic resistance)
•The isolation of a single mutant from a population of millions/billions of cells
Ables to visually see because only ones able to grow!

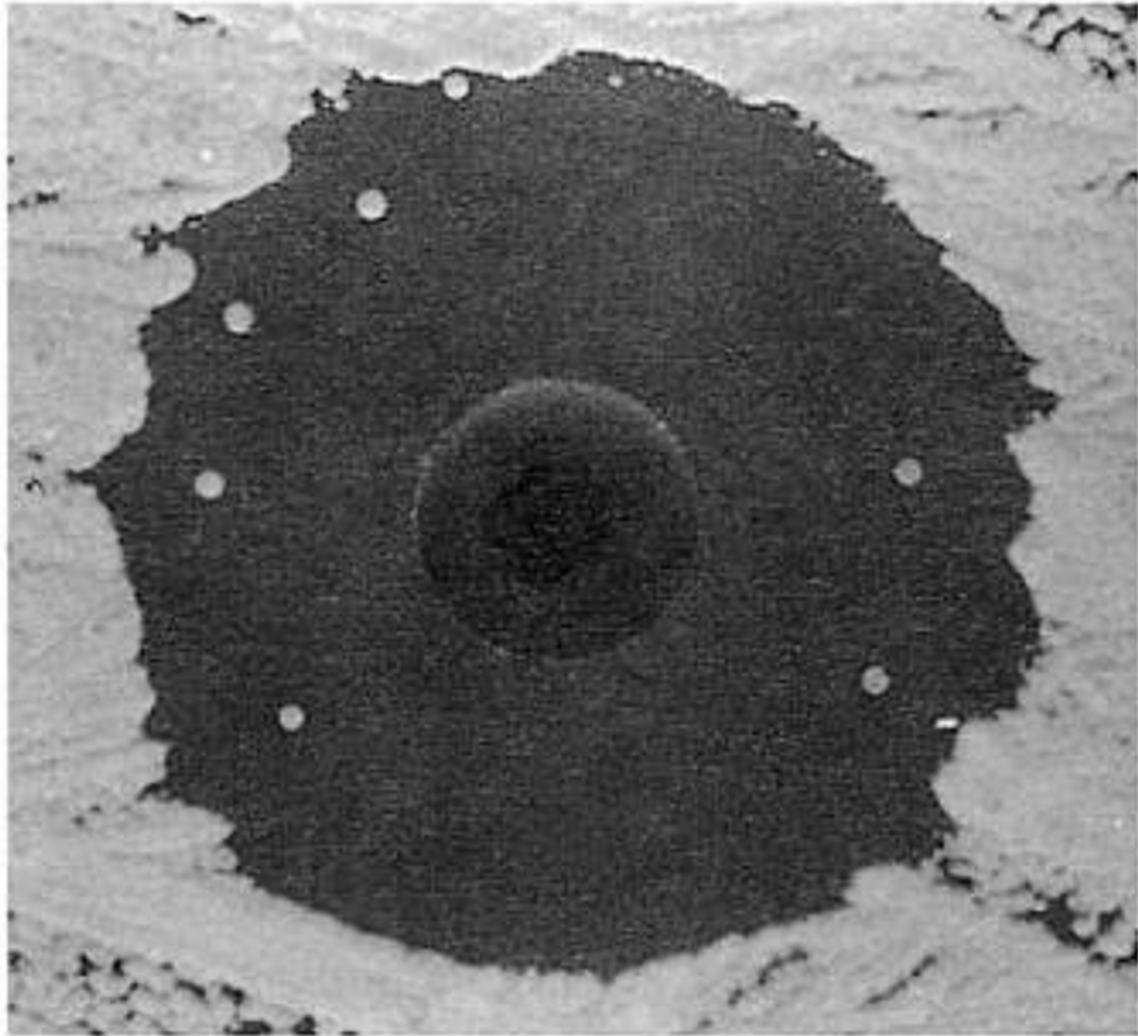
What happens with non selectable mutations?
•Nonselectable mutations do not confer an advantage even though they may lead to a phenotypic change (e.g. color loss in a pigmented organism) (can’t see!)
•Isolation of nonselectable mutants requires laborious, time-consuming screening (= examining large numbers and looking for differences)
Although screening is more tedious than selection, useful methods, such as ________, have been developed for screening large numbers of certain types of mutants
replica plating
What is an auxotroph? What is its parent called?
Mutants that require a particular nutrient
prototroph
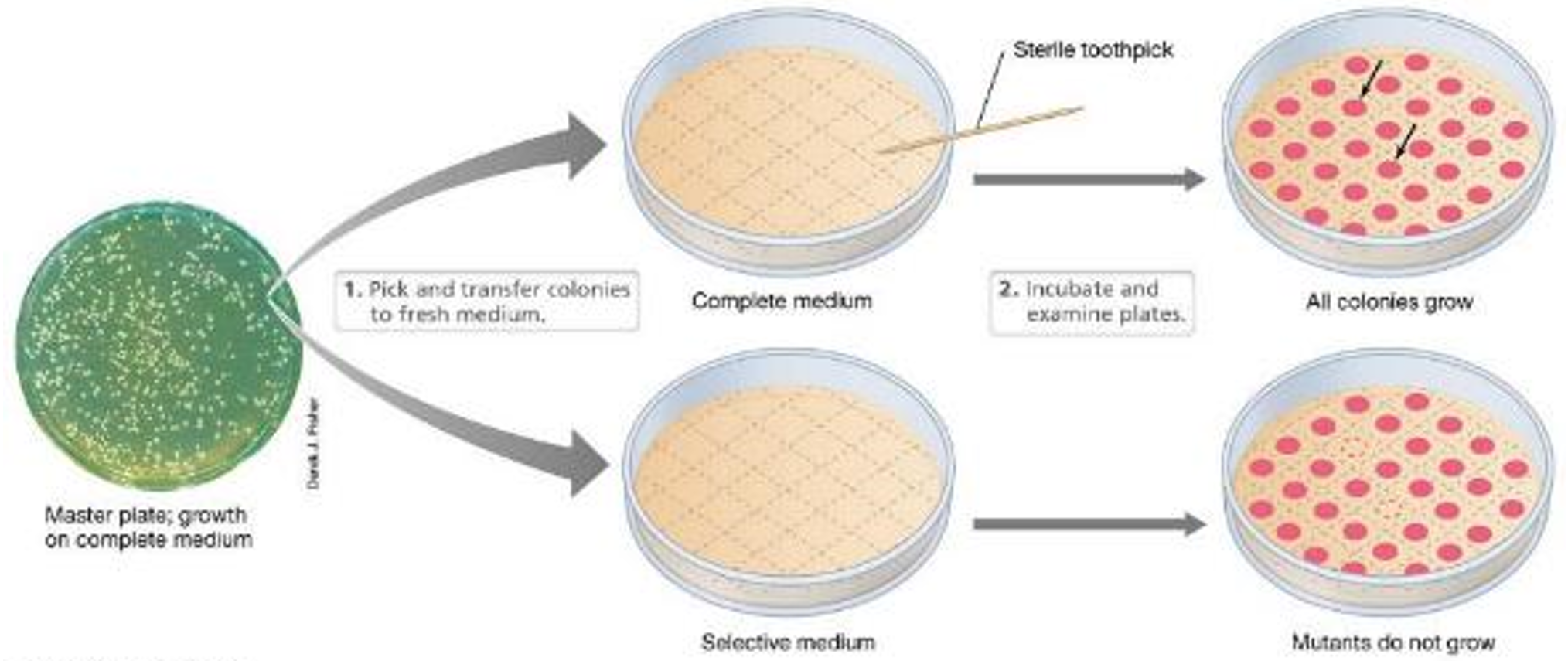
Describe the isolation of nutritional auxotrophs.
•Transfer colonies from master plate to agar lacking the nutrient —> inability of colony to grow on the medium lacking the nutrient indicates a mutation
•Colony on master plate is picked, purified, characterized
Mutations can be _____________.
spontaneous or induced
What are spontaneous mutations?
Occur without external intervention
Most result from occasional errors by DNA polymerase during replication
What are induced mutations?
Caused by environmental agents (e.g. carcinogens or radiation —> alter DNA structure) or made deliberately in the lab
What are point mutations?
Mutations that change only one base pair
Can lead to single amino acid change in a protein, an incomplete protein (stop codon), or no change at all
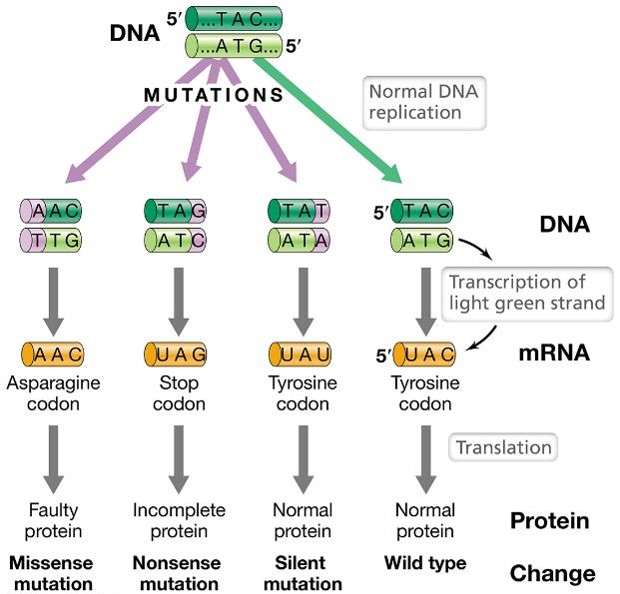
What are examples of point mutations (substitutions)?
Missense, nonsense, and silent mutations
Due to _______in the genetic code, not all mutations change the polypeptide amino acid sequence
degeneracy
What are silent mutations?
Silent mutations change the nucleotide sequence, but do not affect the amino acid sequence of the encoded polypeptide so do not change the phenotype
Almost always change the third nucleotide of a codon
What are missense mutations?
Missense mutations change the sequence of amino acids in a polypeptide as a result of nucleotide changes; can affect protein structure —> change the phenotype
What are nonsense mutations?
Nonsense mutations change the nucleotide sequence to a stop codon; typically result in truncated (incomplete) proteins that lacks normal activity
What are terms used to describe the precise change that occurred in a point mutation?
Transitions/transversions
What are transitions?
point mutations in which a purine is substituted for another purine or a pyrimidine is substituted for another pyrimidine (e.g. A—>G, G—>A, C—>T, T—>C). More common than transversions
What are transversions?
point mutations in which a purine is substituted for a pyrimidine or vice versa (e.g. A—>C, A—>T, G—>C, G—>T, C—>A, C—>G, T—>A, T—>G)
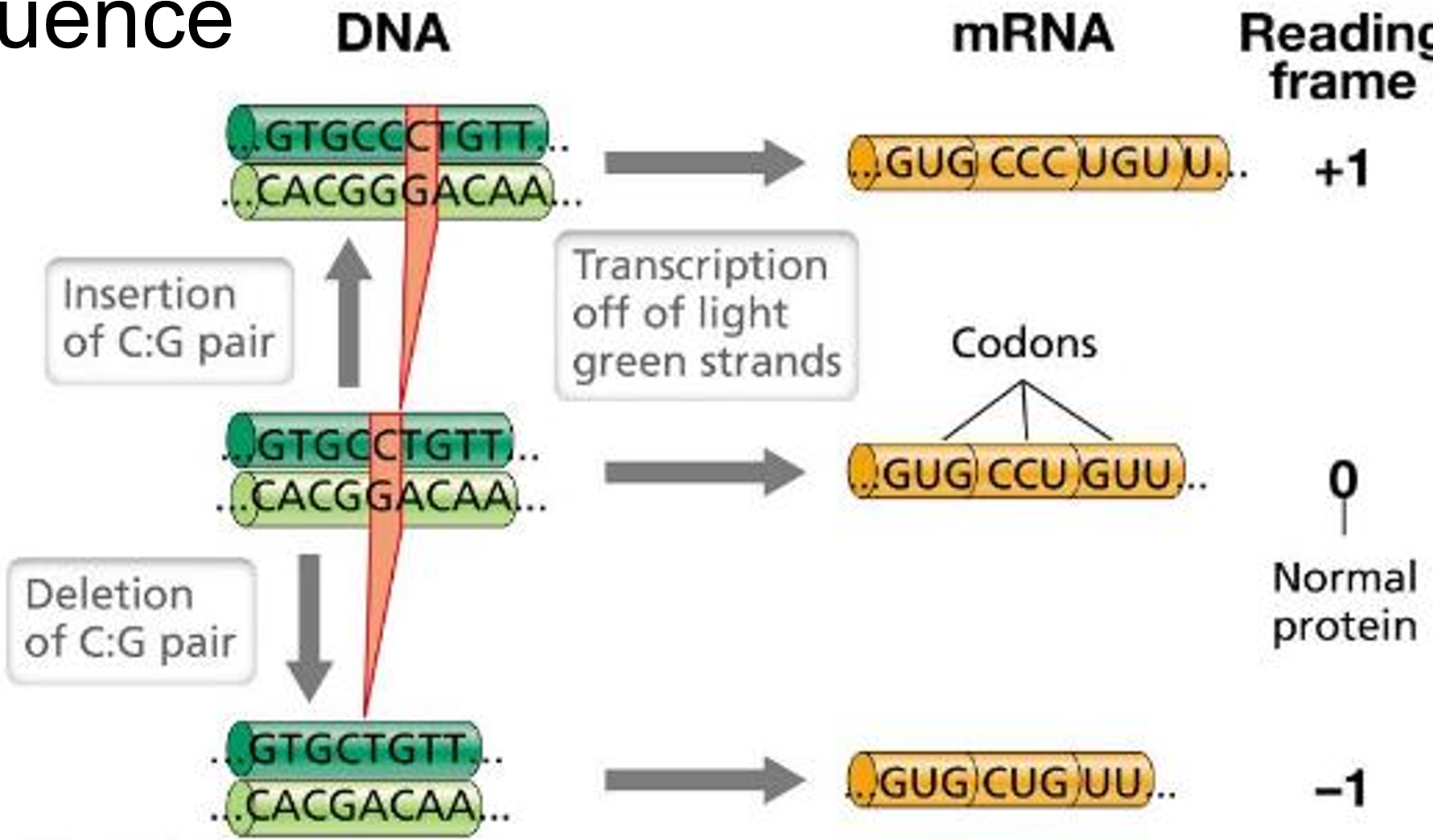
What is a frameshift mutation?
deletion or insertion that results in a shift in the reading frame
Insertion/deletion of three base pairs adds/deletes an amino acid, which is usually not as detrimental to the encoded protein structure/function as a frameshift
Frameshift mutations scramble the entire downstream polypeptide sequence
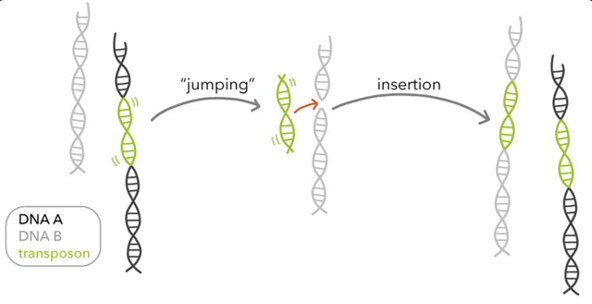
Large insertions or deletions can result in the gain or loss of hundreds to thousands of base pairs. Often result in complete loss of gene function
May arise from errors during genetic recombination. What else?
transposable elements
The rates at which different types of mutations occur vary widely and depend on the ____ of DNA changes and the efficiency of DNA repair
frequency
What are reversions?
Point mutations are typically reversible; process is known as reversion
What is a revertant?
strain in which the original phenotype, which was changed in a mutant, is restored by a second mutation
What is a same-site revertant?
phenotype/activity-restoring mutation occurs at the same site as the original mutation; true revertants restore the original sequence as well as the original phenotype/activity
What are second-site revertants?
phenotype/activity-restoring mutation occurs at a different site in the DNA than the original mutation
These mutations restore the original phenotype by compensating for the mutation
Also known as suppressor mutations and include:
Mutation elsewhere in the same gene that restores function to the encoded protein
Mutation in another gene that restores function to the encoded protein (e.g. protein-protein interactions, tRNA genes)
Mutation in another gene that results in the production of an enzyme to replace the nonfunctional one
What are some common mutation rates?
For most microorganisms, errors in DNA replication occur at a frequency of 10-6 to10-7 per thousand nucleotides during a single round of replication
Because a typical gene is ~1 kb, the frequency of a mutation occurring in a single gene is in the same range
Eukaryotes have replication error rates ~10-fold lower than typical bacteria
DNA viruses have error rates 100-1000x greater than cellular organisms
RNA viruses have even higher error rates due to less proofreading and the lack of RNA repair mechanisms
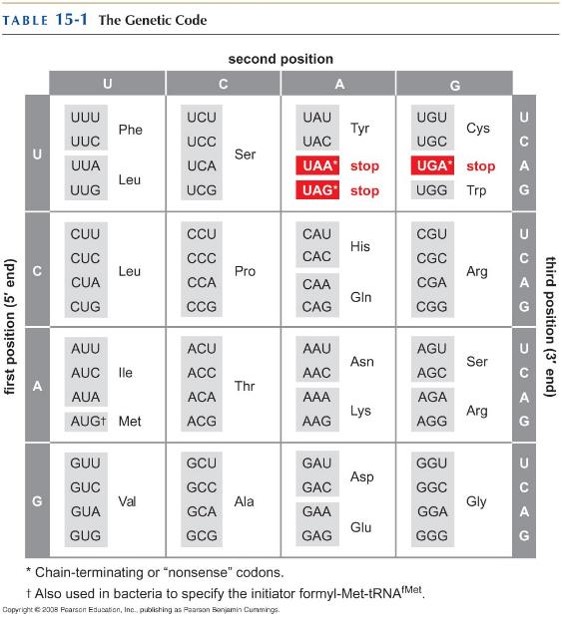
The genetic code evolved in a way that minimizes the effects of mutations: how?
Mutation in the 1st position: usually gives a similar (if not the same) amino acid
Transitions are the most common type of mutation and have minimal effect:
Codons with a pyrimidine in the 2nd position mostly specify hydrophobic amino acids; codons with a purine in the 2nd position mostly specify polar amino acids
So a transition in the 2nd position will substitute a similar amino acid
Transition in the 3rd position rarely changes the amino acid
What are mutagens?
chemical, physical, or biological agents that increase mutation rates
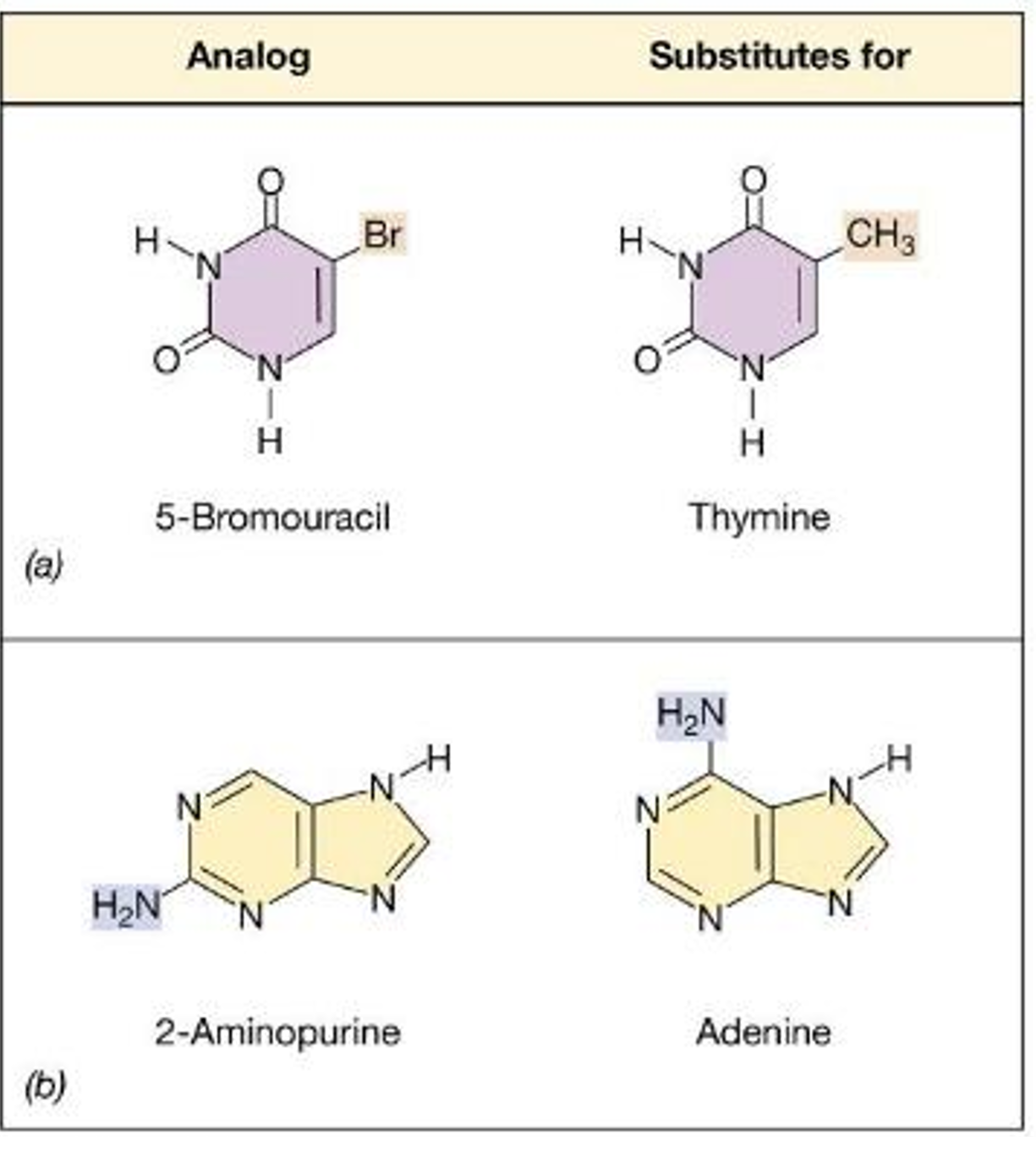
What are some chemical mutagens?
Nucleotide base analogs = molecules that resemble nucleotides but have faulty base-pairing —> lead to DNA sequence changes
Mutagenesis occurs during DNA replication
5-Bromouracil can base-pair with guanine —> causes AT to GC changes
2-Aminopurine can base-pair with cytosine —> causes AT to GC changes
What are the effects of chemical mutagens?
Chemical mutagens that induce chemical modifications in the base of a nucleotide —> lead to faulty base-pairing —> leads to permanent changes in the nucleotide sequence
what are alkylating agents?
(chemicals that substitute an alkyl group for an amino, carboxyl, or hydroxyl group) such as nitrosoguanidine induce mutations at a higher frequency than base analogs; can introduce changes in both replicating or nonreplicating DNA
What are intercalating agents?
agents such as acridines push two base pairs apart —> trigger single base-pair insertions or deletions; typically induce frame-shift mutations rather than point mutations
![<p>Describe <span><em>Nonionizing radiation</em> (<em>e.g., </em>Ultraviolet [UV] radiation)</span></p>](https://knowt-user-attachments.s3.amazonaws.com/fcfbaa23-9bc0-48bd-afd7-f3d65d24b9f9.png)
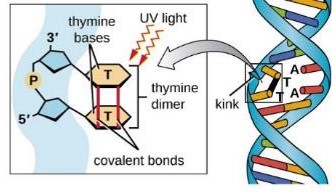
Describe Nonionizing radiation (e.g., Ultraviolet [UV] radiation)
Purines and pyrimidines strongly absorb UV (quantify nucleic acids by measuring absorbance at 260 nm)
UV can lead to pyrimidine dimers (two adjacent Cs or Ts on the same strand become covalently bonded) —> impedes DNAP and/or increases likelihood of DNAP misreading the template
Killing cells by UV is primarily due to the effect of UV on DNA
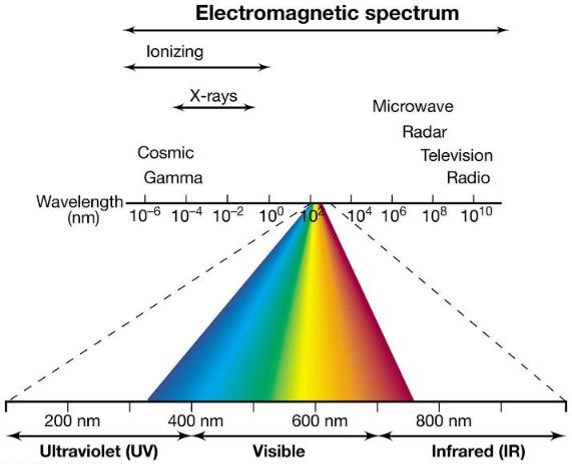
UV radiation consists of wavelengths just shorter than visible light; the shorter the wavelength, the higher the _____.
energy.
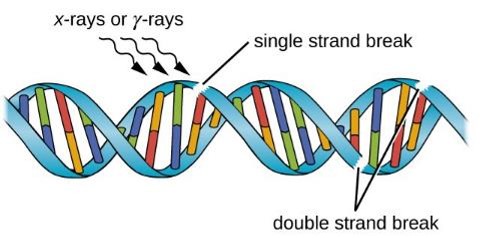
Describe Ionizing radiation (e.g., X-rays, cosmic rays, and
gamma rays).
More powerful than UV (greater penetration)
Causes water and other substances to ionize
Results in the formation of free radicals such as hydroxyl radicals (OH·) that damage cellular macromolecules, including DNA
Cause double-stranded and single-stranded breaks that can lead to rearrangements or large deletions
By definition, a mutation is a ______; therefore, if damaged DNA can be corrected before the cell divides, no mutation will occur
heritable change
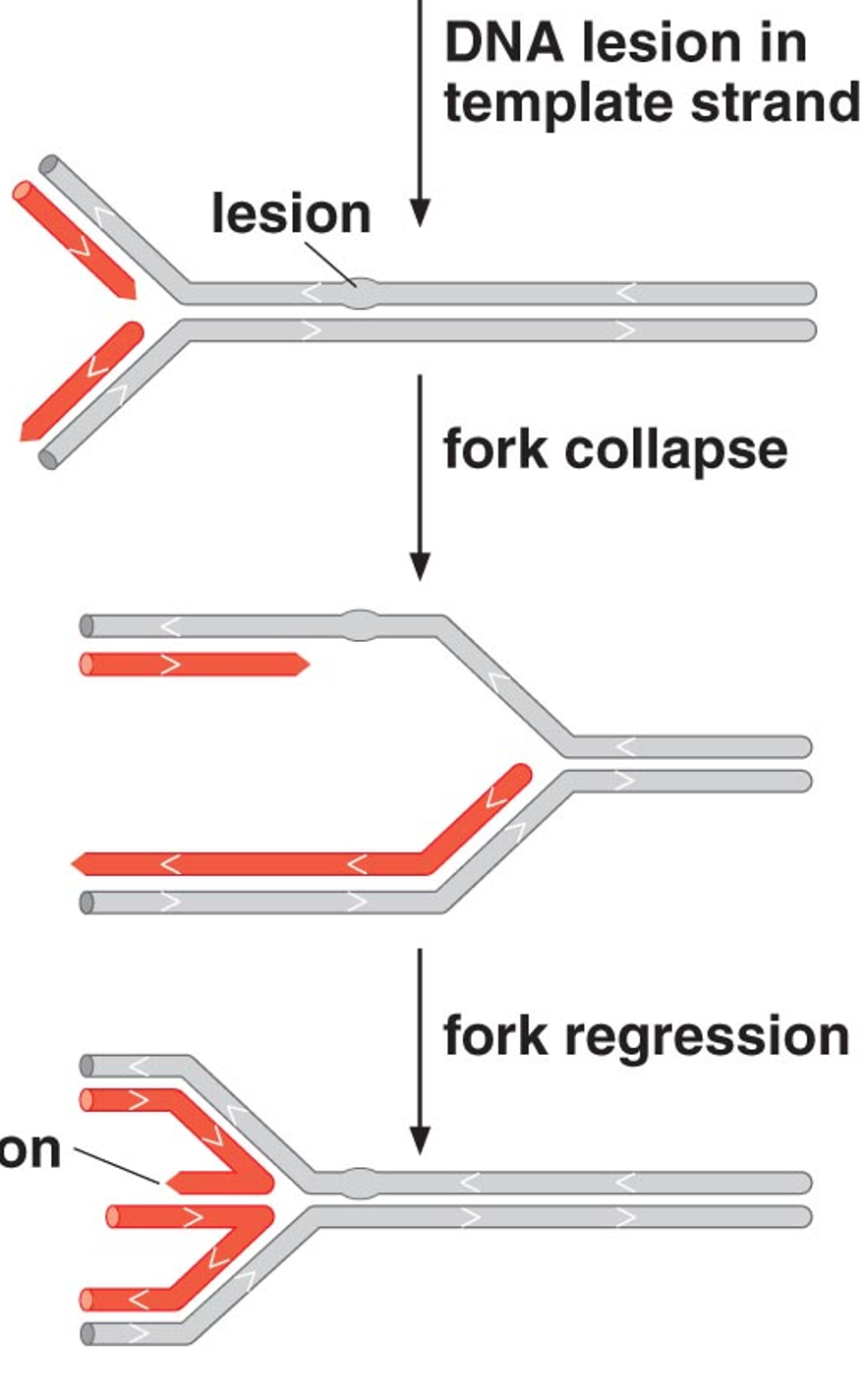
Cells have a variety of different DNA repair processes to correct mistakes or repair damage; some are error-prone and the process itself introduces mutations. What’s an example of this?
Some types of DNA damage can cause lesions that interfere with replication; if these lesions are not removed prior to replication, DNAP will stall and lethal “breaks” in the chromosome will result.
What is the DNA repair and SOS system? What is translesion synthesis?
In Bacteria, stalled replication or major DNA damage activates the SOS repair system which initiates many DNA repair processes, some of which are error-free
However, this system also allows DNA repair to occur without a template (with random incorporation of dNTPs); this process is called translesion synthesis and results in many errors and mutations
In E. coli, the SOS repair system does what?
controls the expression of ~40 genes (SOS regulon) that encode proteins that participate in DNA repair and DNA damage tolerance (translesion synthesis; lesion is not actually repaired, just bypassed)