MCB*2050 Quiz 2: RNA Modifications and Quantification
1/29
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
30 Terms
noncoding RNAs (ncRNAs)
Accomplish a remarkable variety of biological functions.
Regulate gene expression at the levels of transcription, RNA processing, and translation.
Protect genomes from foreign nucleic acids.
Can guide DNA synthesis or genome rearrangement.
For ribozymes and riboswitches:
RNA structure itself provides the biological function.
Most operate as RNA-protein complexes (including ribosomes, snRNPs, snoRNPs, telomerase, microRNAs, and long ncRNAs).
Many, though not all, exploit the power of base pairing to selectively bind and act on other nucleic acids.
small RNAs (miRNA, siRNA)
Small (21-29 bp_ RNA molecules that regulate the stability or the translation of the mature mRNAs. They regulate nearly every biological process! Abnormalities in their levels play a major role in the development of many diseases, including cancer.
They are generated from larger double-stranded RNA molecules that must be processed to produce a mature miRNA and siRNA.
More than 3000 different types in humans, each capable of binding many different mRNA transcripts.
Each mRNA transcript can potentially bind many different ones.
mRNA
Messenger RNA with 5’ cap, introns removed removed by RNA splicing, and a poly(A) tail.
pre-mRNA
An mRNA precursor containing introns and not cleaved at the poly(A) site.
hnRNA
Heterogeneous nuclear RNAs. These RNAs include pre-mRNAs and RNA-processing intermediates containing one or more introns.
snRNA
Five small nuclear RNAs that function in the removal of introns from pre-mRNAs by RNA splicing, plus two small nuclear RNAs that substitute for the first two at rare introns.
pre-tRNA
A tRNA precursor containing additional transcribed bases at the 5’ and 3’ ends compared with the mature tRNA. Some also contain an intron on the anticodon loop.
pre-rRNA
The precursor to mature 18S, 5.8S, and 28S ribosomal RNAs. The mature RNAs are processed from this long precursor RNA molecule by cleavage, removal of bases from the ends of the cleaved products, and modification of specific bases.
snoRNA
Small nucleolar RNAs. These RNAs base-pair with complementary regions of the pre-rRNA molecule, directing cleavage of the RNA chain and modification of bases during maturation of the rRNAs.
siRNA
Short interfering RNAs, ~22 bases long, that are each perfectly complementary to a sequence in an mRNA. Together with associated proteins, cause cleavage of the target RNA, leading to its rapid degradation.
Degrade viral RNA and RNA produced by transposons.
Produced from cleavage of double-stranded RNA. They bind PERFECTLY (no mismatches) to the target RNA and cause its rapid degradation by a mechanism called RNA interference, which controls multiple processes.
miRNA
MicroRNAs, ~22 bases long, that base-pair extensively, but not completely, with mRNAs, especially over bases 2 to 7 at the 5’ end of the microRNA (the “seed” sequence). This pairing inhibits translation of the target mRNA and targets it for degradation.
IMPERFECTLY (there are mismatches) bind to the target mRNA; binding usually at the 3’-UTR → leads to repression of translation.
“seed” sequence
Bases 2 to 7 at the 5’ end of miRNA. Tend to base-pair extensively but not completely with mRNAs.
alternative splicing of mRNA
Along with alternative cleavage at different poly(A) sites!
Yields a significant variety of different mRNAs from the same gene (resulting in different phenotypes) in different cell types or at different developmental stages.
Some of the proteins generated this way have drastically different activities.
In neurons: perception of sound, neuron connectivity
Sex determination in Drosophila
RNA-binding proteins (RBPs)
Recognize specific sequences near splice sites and regulate alternative splicing.
e.g. Sxl, Tra
Sxl
An RNA-binding protein that acts as a suppressor of splicing / an intron splicing silencer (inhibitor).
Controls its own splicing to produce exons 2,4 mRNA.
Tra
An RNA-binding protein that acts as an activator of splicing / an intron splicing activator.
Dsx
A transcription activator/repressor.
Exons 1-3 encode a protein domain that binds to the same DNA element in gene promoters in both male and female organisms.
The female isoform (Exon 4) contains a transcriptional activation domain.
The male isoform (Exons 5-8) contains a transcriptional repression domain.
Lin-4 and Let-7
Genes that are involved in the development of the worm C. elegans. They do NOT encode any proteins. Instead, they produce small RNAs that are complementary to other mRNAs.
The imperfect binding of the microRNAs (miRNAs) to the 3’-UTR (untranslated region) of the mRNAs dramatically reduce the translation of the mRNAs → protein is made at a low level or is not made at all.
In other species, the perfect binding of siRNA to mRNAs interferes with the stability of the mRNA, leading to their quick degradation.
RNA-induced silencing complex
RISC
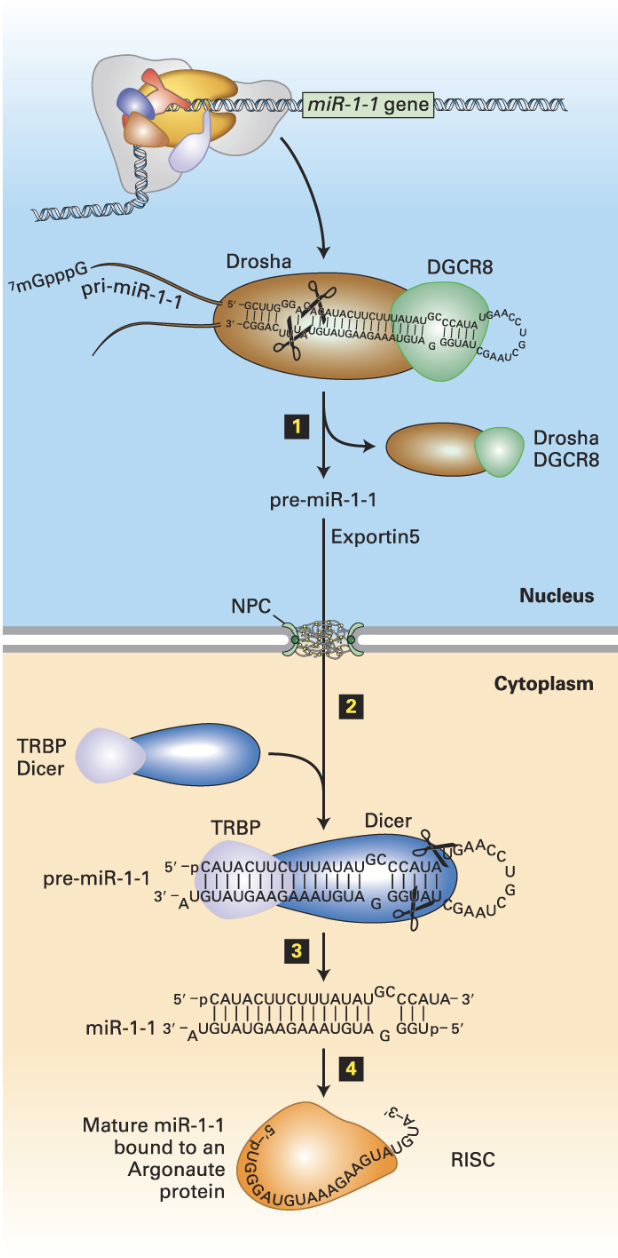
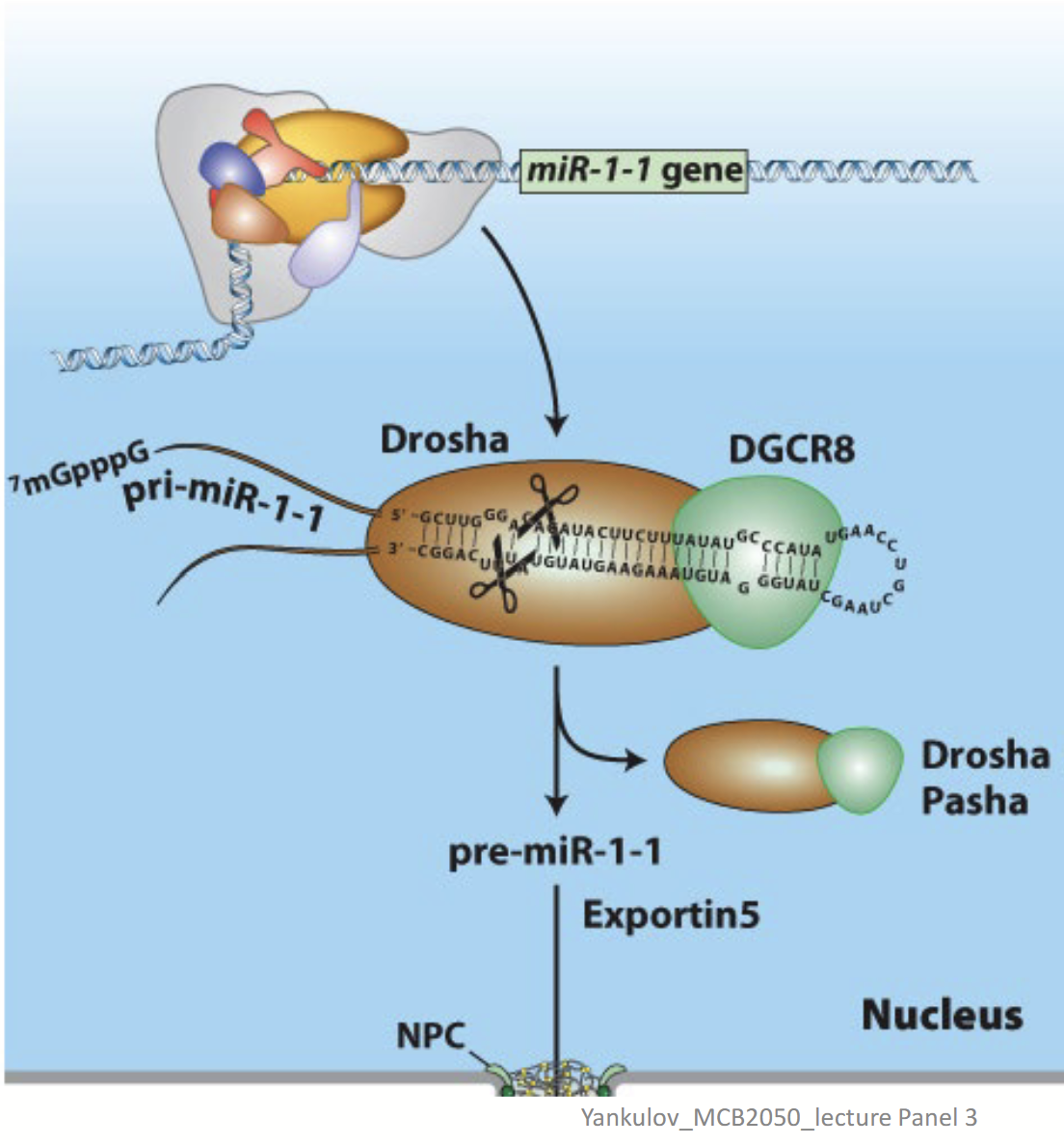
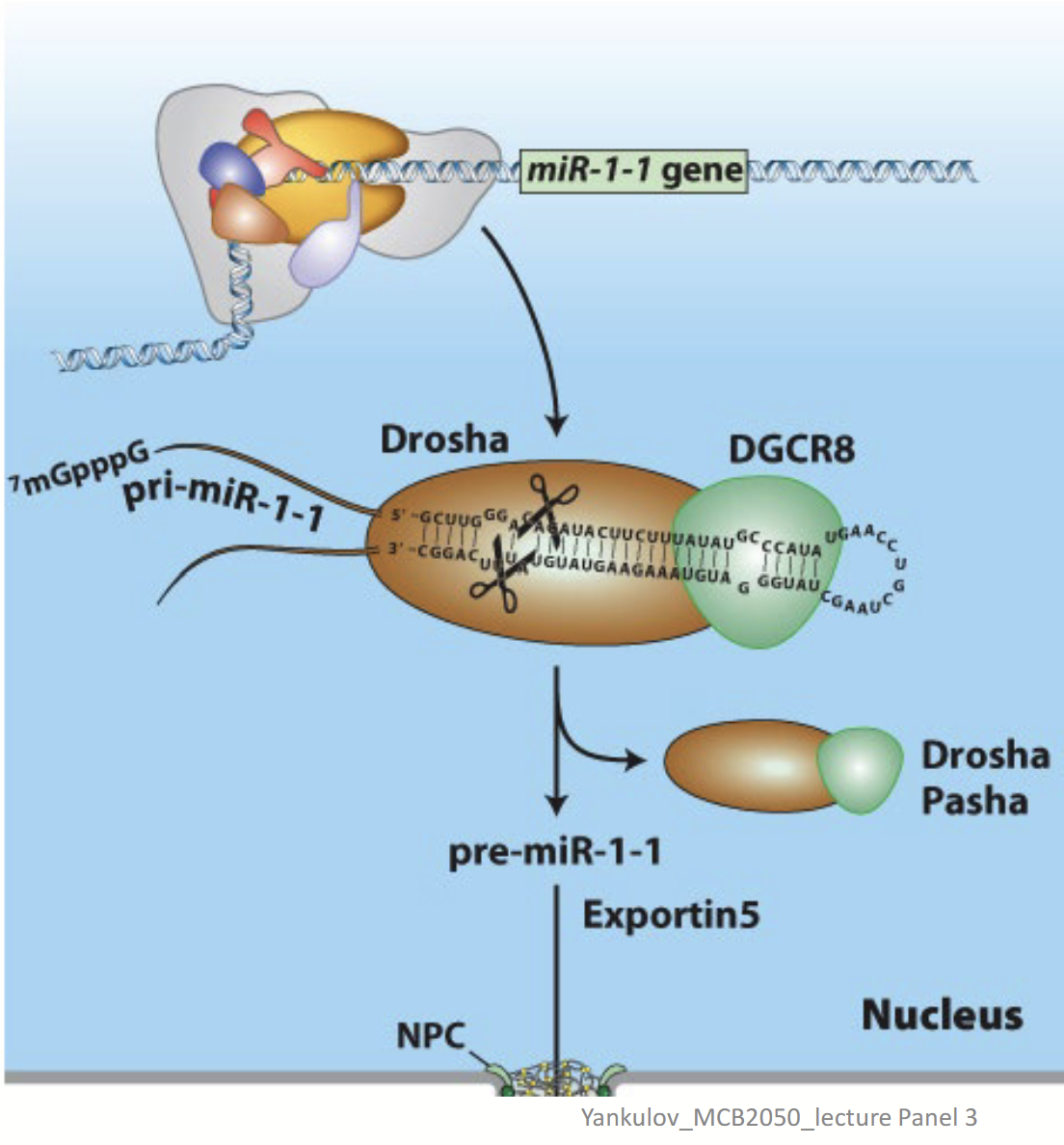
Drosha/DGRC8
In miRNA (siRNA) biogenesis, is responsible for the removal of hairpins in double-stranded RNA. Binds pri-miRNA double strand regions.
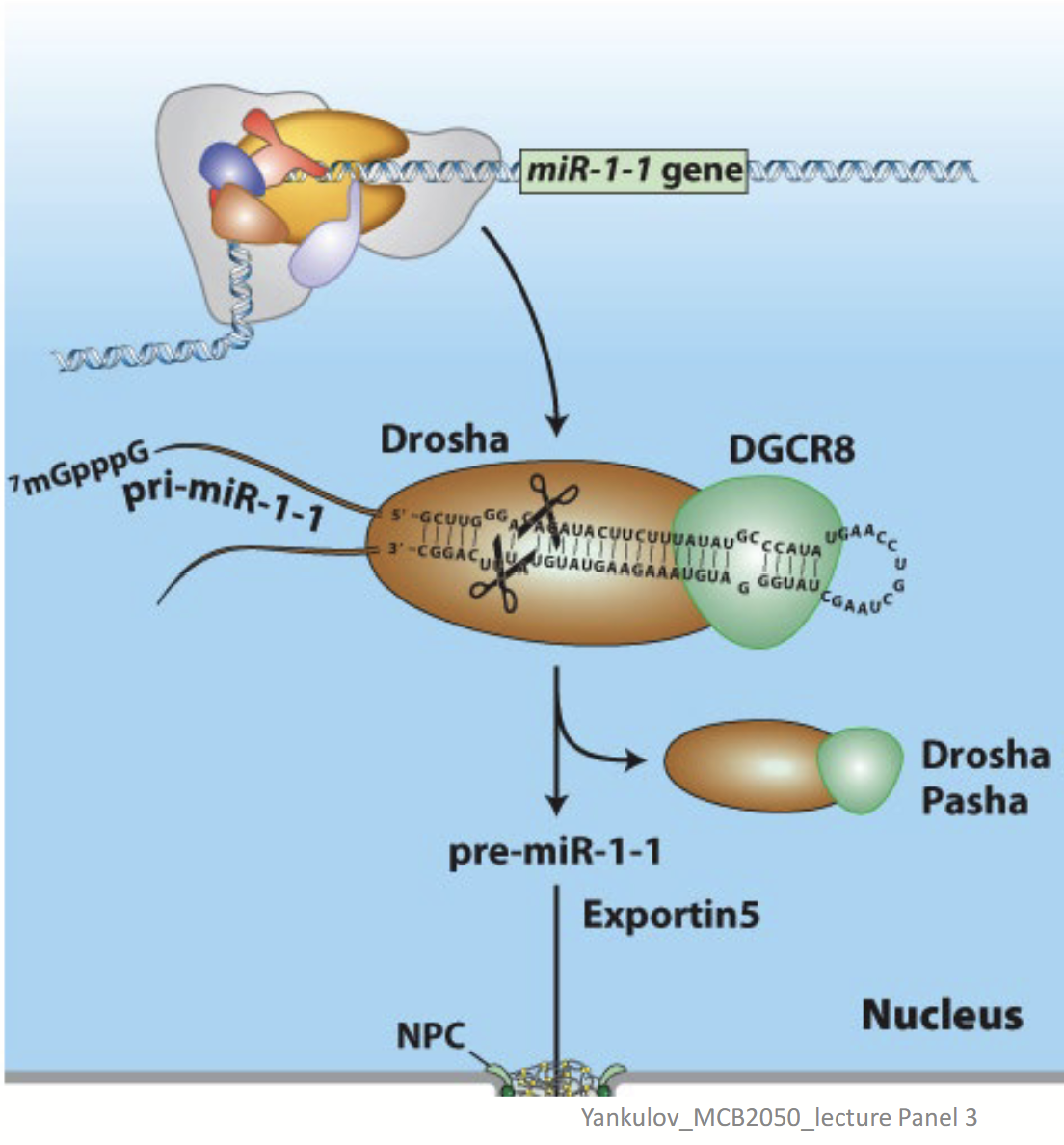
Exportin 5s
In miRNA (siRNA) biogenesis, the nuclear transporter transports the processed miRNA to the cytoplasm.
miRNA (siRNA) biogenesis
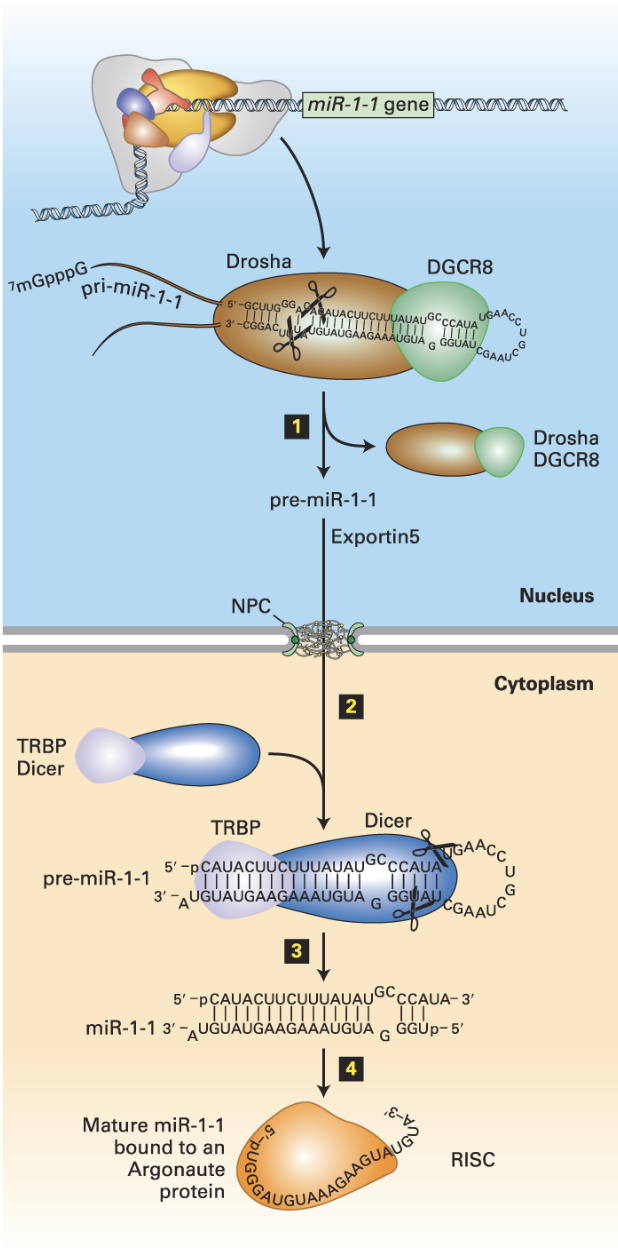
Drosha
Nuclear double-strand RNA–specific endoribonuclease that works with DGRC8 to bind pri-miRNA double strand regions. Also cleaves the pri-miRNA – generates a ~70-nucleotide pre-miRNA.

DGCR8
Double-strand RNA–binding protein that works with Drosha to bind pri-miRNA double strand regions.
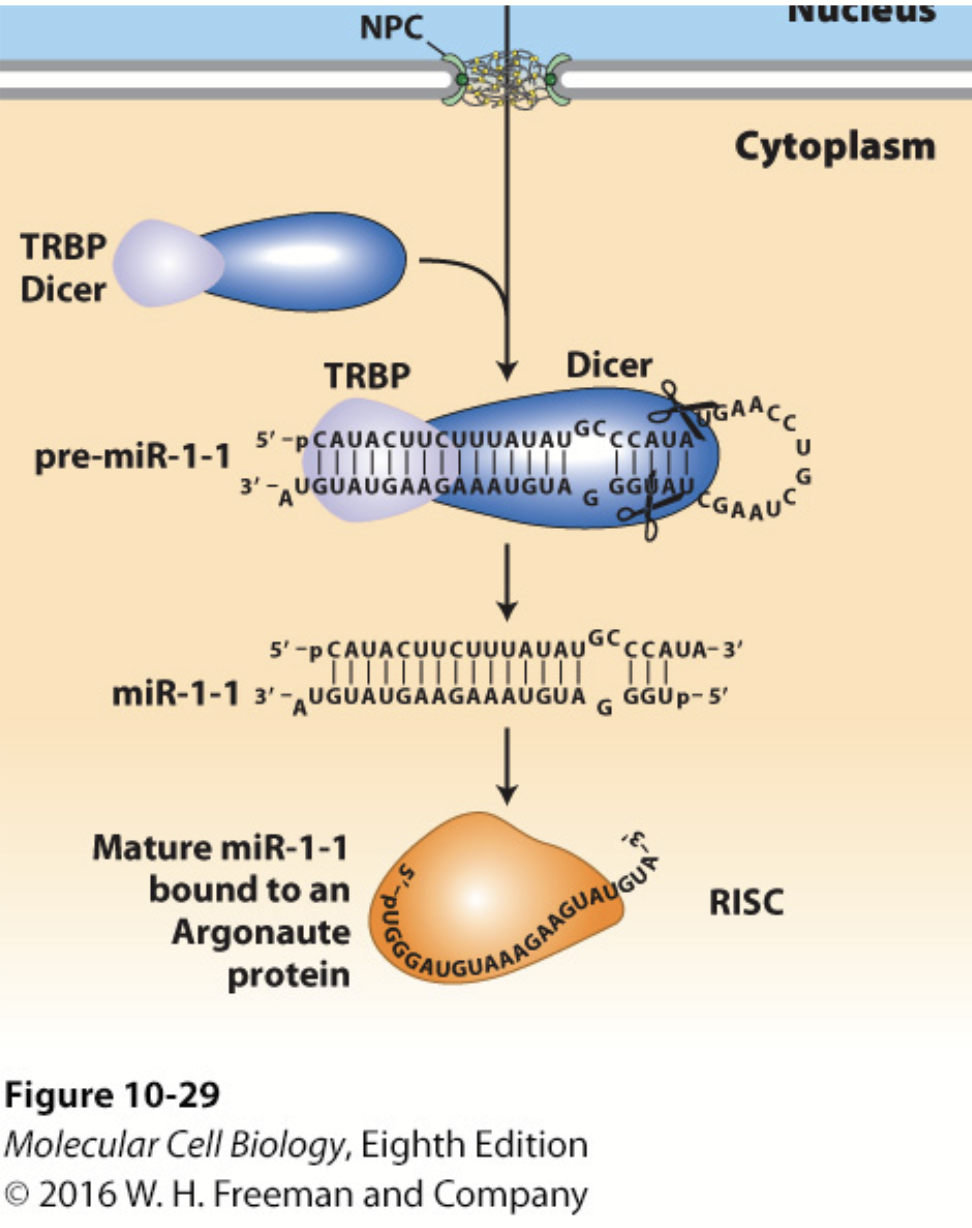
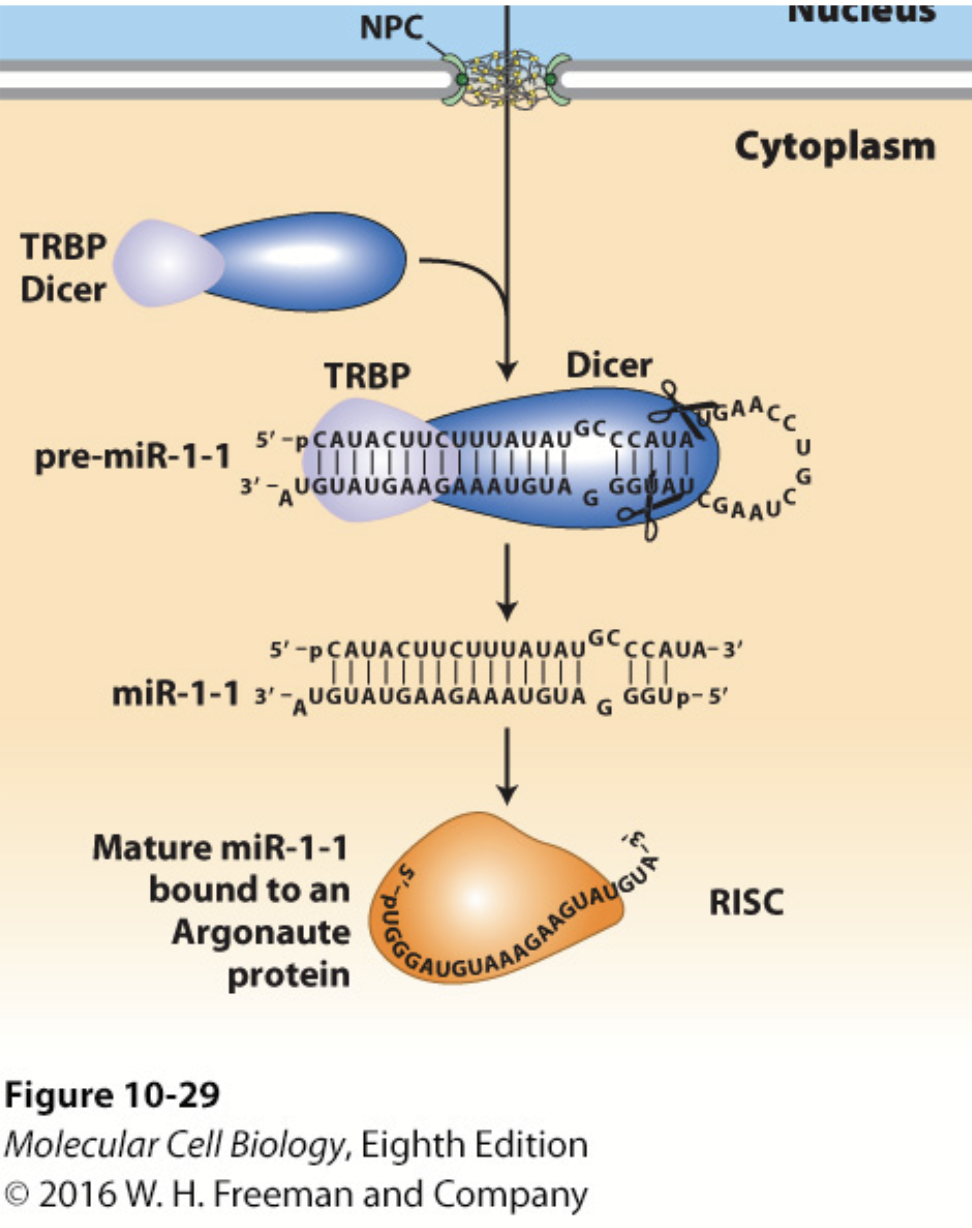
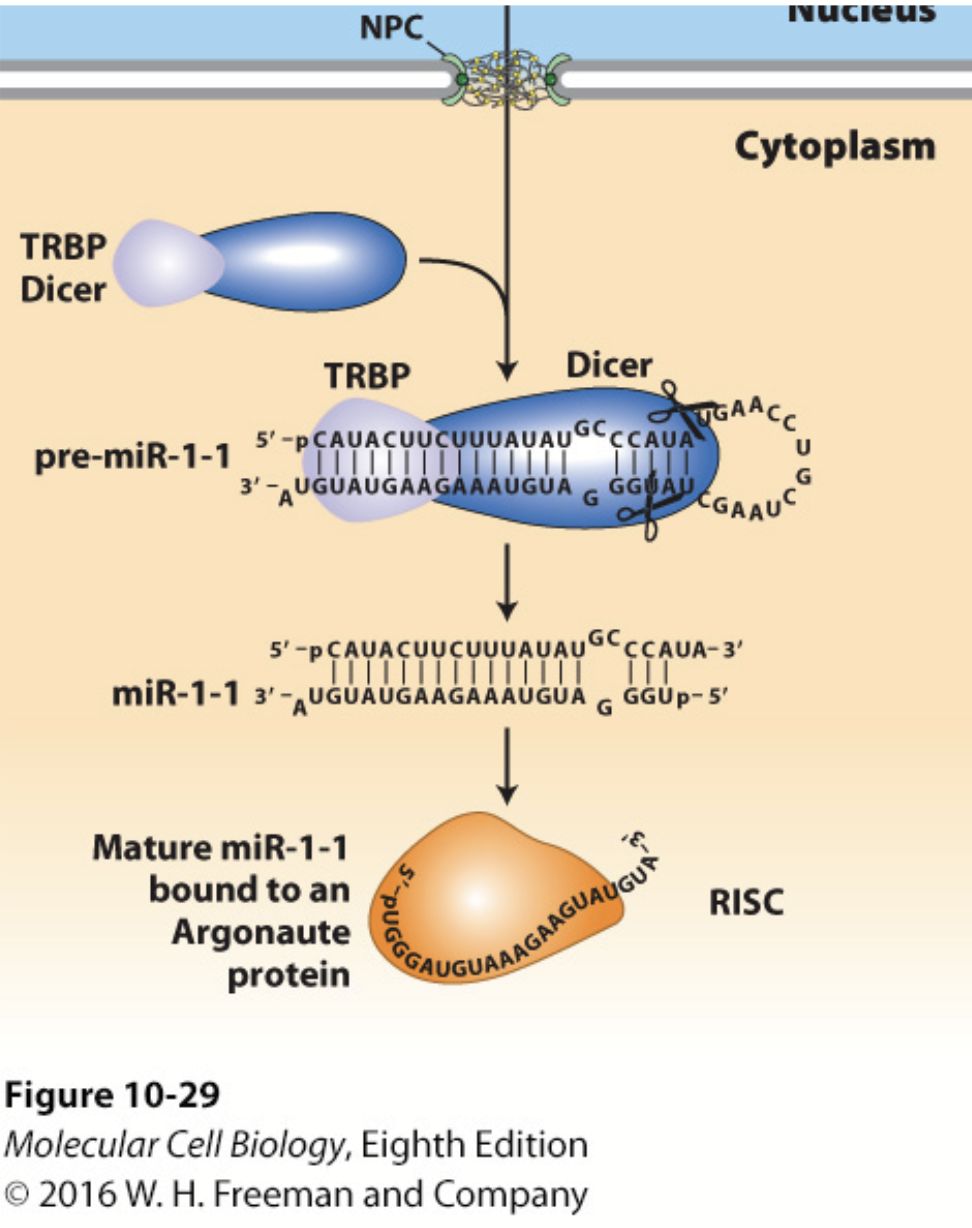
DICER
In conjunction with the double-stranded RNA–binding protein TRBP (Loquacious in Drosophila), processes pre-miRNA into a double-stranded miRNA with a two-base single-stranded 3′ end.
Cuts the double-stranded RNA into small pieces (21-28 bp).
Argonaut
An RNA helicase that removes one of the strands of the double-stranded RNA (unwinds double-stranded RNA). Leads the trimmed siRNA to the target mRNA. Active RISC is formed.
RNA-Induced Silencing Complex (RISC)
Binds one of the two strands. Incorporates mature miRNA into complex with Argonaut proteins.
RISC-siRNA complex
Complex that can target the siRNAs to the mRNAs complementary to the guide siRNA in it. This process is called RNA interference.
RNA interference (RNAi)
The targeting of the siRNAs to the mRNAs complementary to the guide siRNA in the RISC-siRNA complex.
Double-stranded RNA is cleaved to small (21-28 bp) RNA fragments by a double-stranded RNA-endonuclease called DICER.
Small double-stranded RNAs bind to RISC (RNA-Induced Silencing Complex) and form an inactive RISC complex.
Argonaut in RISC is an RNA helicase. It unwinds double-stranded RNA. Active RISC is formed!
Active RISC, guided by single-stranded siRNA, cleaves target mRNA (“SLICER” activity by Argonaut).
Amplification of RNAi: the cleaved target RNA and RISC produce more RISC-siRNA to target more long RNA molecules.
PAX6
A developmentally regulated gene. Contains:
Three promoters and three transcription initiation sites
More than three enhancers
It is expressed at different times and in different organs during embryonal development; not expressed at all times.
It is expressed in certain parts of the brain in adult organisms.
It encodes a transcriptional activator.
Alternative splicing of the mRNA produces proteins with different DNA binding specificity; the translation of these mRNAs is regulated by several miRNAs.