BIOL 207 - Genetics & Heredity
1/306
Earn XP
Description and Tags
everything for the midterm ig.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
307 Terms
Genetics
Uses manipulation of genes/genomes to study how genes function in fundamental biological processes.
Alter gene sequence and/or gene function (i.e. mutations) and see how it affects a process.
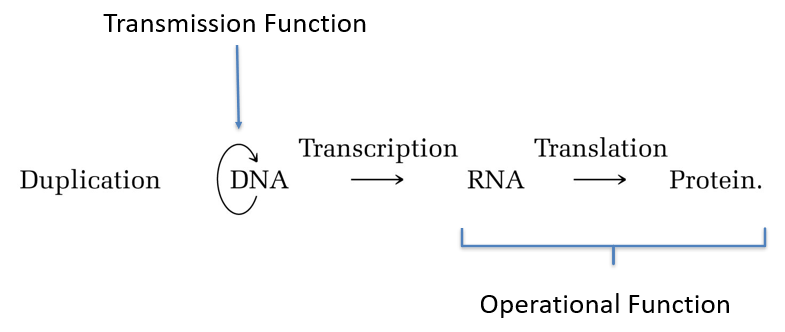
Gene
Has two main functions:
Operational.
Transmission.
Transcribed region + all sequences necessary for correct expression.
Gene - Operational Function
A gene is a (usually continuous) stretch of DNA (or in some organisms RNA) that contains information encoding a gene produced (usually an RNA and a protein).
Codes a gene.
Gene - Transmission
A gene carries information from one generation to the next.
Or it carries information from parental to daughter cells.
Types of Genetic Material
DNA or RNA as genetic material (RNA only in viruses).
Linear (eukaryotes, many viruses) or circular (bacteria, mitochondria, some viruses).
Double- or single-stranded.
Segmented (some viruses) vs unsegmented (bacteria, eukaryotes, many viruses).
+ve vs. -ve Strands
Positive strand → can go right in to be translated.
Negative strand → must do reverse replication before translated.
DNA as Dynamic
Not static.
Can be heavily expressed, somewhat expressed, or not expressed at all.
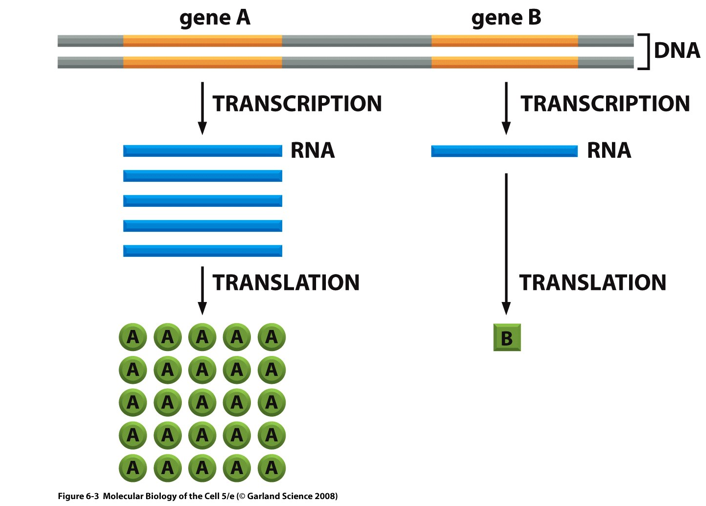
Gene Activity
Genes can produce different amounts of RNA.
Gene A → many RNA transcripts → many protein molecules.
Gene B → few RNA transcripts → few protein molecules.
Level of transcription controls how much protein is made.
Gene Components
Gene = transcribed region + all sequences necessary for correct expression.
RNA corresponds to a region of DNA, called the transcribed region (transcription unit).
Regulatory Sequences
Sequences in the gene tell the cell to turn “off” or “on.”
Allows the gene to be appropriately expressed.
Can be upstream, downstream, or within the gene.
Activator Protein
Can recognize the regulatory region.
Will recruit RNA polymerase which begins transcribing.
Repressor Protein
Binds regulatory region.
Blocks RNA polymerase.
No transcription occurs.
Gene inactive = not expressed.
Gene Expression (Active)
RNA polymerase transcribes DNA.
RNA is produced.
Gene is expressed.
Gene Expression (Inactive)
Transcription blocked.
No RNA produced.
Gene is not expressed.
“…omics”
The characterization of the genome and what it means.
High throughput sequencing.
i.e. to study the expression of all genes at once.
Many types of these disciplines exist (metabolomics, lipidomics, glycomics, etc).
Genomics
The study/cataloging of entire genomes.
Transcriptomics
Studies genome-wide responses to different conditions.
The quantification/cataloging of all RNAs in a sample.
Powerful when comparing a controlled variable with a dependent variable.
Proteonomics
The quantification/cataloging of all proteins in a sample.
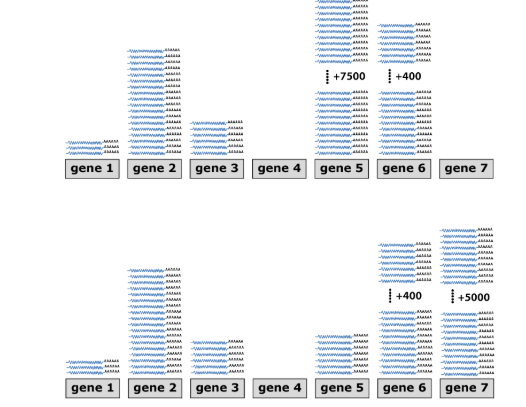
Transcriptomics - Example
Sample 1 (Control): Gene 5 = highly expressed. Gene 7 = not expressed.
Sample 2 (Drug-treated): Gene 5 = repressed. Gene 7 = induced.
Other genes → unaffected by drug.
The drug treatment represses gene 5, while it induces gene 7 (all other genes remain unaffected).
Proteomics & Transcriptomics vs Genetics (Insect Example)
Proteomics/Transcriptomics: find mRNAs or proteins that respond to a stimulus (e.g., light exposure changes expression).
Genetics: find genes that are required for a process (e.g., mutants can’t fly to the light).
Transcriptomics or Proteonomics (Insect Example)
Will identify mRNA/proteins that respond to a stimulus or upregulated during a biological process (light perception).
Done by comparing tissues from flies receiving a light stimulus vs. flies who did not receive a light stimulus.
Genetics (Insect Example)
Identifies genes that are required for the biological process.
Mutanizes flies, breaks, checks results, analyzes, and finds the gene.
Tests thousands of mutant fly lines and selecting those with a defect (flying to the light source).
Forward Genetics
Genes first identified by their mutant phenotype.
Mutations mapped to the corresponding gene.
Gene characterized using molecular, cellular, or biochemical tools.
Reveals genes with previously unknown functions in known processes.
Forward Genetics Advantage
Unbiased and powerful.
Identification of genes that nobody has ever linked to the biological process.
Finds the unknown unknowns (the “surprises”).
Forward Genetics Disadvantage
Slow.
Identifying mutated gene can be tricky.
Reverse Genetics
You mutate, remove, or modify a gene to test its function.
Studies a known gene in a known or unknown process.
Understanding the processes involved or understanding how modification affects the function of a gene product.
Reverse Genetics Advantage
Extremely specific and versatile.
Allows for the manipulation of genes with precision.
Reverse Genetics Disadvantage
Biased (tests the known unknowns).
The gene that you modified may not have a obvious phenotype.
One gene at a time.
Cell Biology
Study cells directly (i.e microscopy) to learn about fundamental cellular processes.
Biochemistry
Study biological phenomena on a molecular level to understand the mechanistic aspects of a process.
i.e. protein-protein interactions.
Genome
A complete set of genetic material found in the somatic cell.
Includes the coding and non-coding strands.
Preformationism
Sperm were homunculi.
Preformed humans.
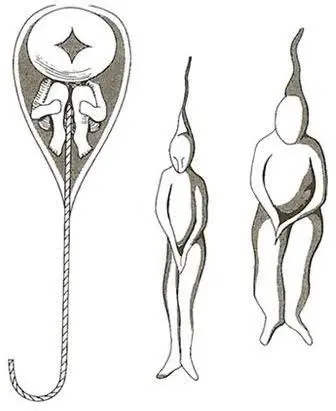
Inheriting Traits
Genetic traits are inherited by both parents.
A child can only inherit 50% of genetic information from their parents.
Information
First level of DNA structure.
Encodes the characteristics of an organism.
Replication
Second level of DNA structure.
Can be copied.
Transmission
Third level of DNA structure.
Is passed from parents to offspring during reproduction.
Variation
Fourth level of DNA structure.
Subject to occasional modifications that result in differences between individuals.
Forms of Genetic Information
Different forms of the same kind of information exist: alleles and polymorphisms.
Genotype
Entire genetic information inherited by an organism: refers to all of the specific information stored in the genome.
The genetic information with respect to a single gene or group of genes.
More used definition.
Phenotype
Can be genetic or environmental.
Detectable manifestation of a specific genotype.
Includes morphological, behavioral, physiological, or molecular traits.
Reaction Norm
Two genetically identical individuals can look very different.
This is usually due to environmental differences.
Limited nutrients or stress may restrict growth.
This reflects the “flexibility” of certain traits in response to different environments within an individual.
“Extended Phenotype”
Argues that phenotypes should not be limited to organismal traits.
Proposed by Richard Dawkins.
Would apply to gene-dependent, visible impact on habitats, driven by behaviours.
Central Dogma
Central dogma of molecular biology.
DNA → RNA → Protein.
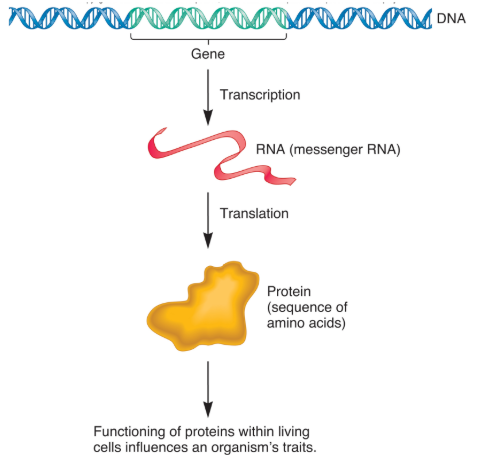
Gene Expression
Information stored in DNA is accessed through the process of transcription (“copying”) into (messenger) RNA molecules.
Many RNAs are translated into proteins, leading to observable traits.
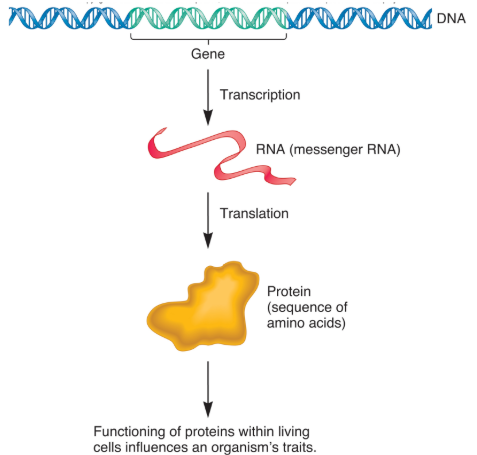
Genetic Variation
Describes differences among individuals within a population.
Morphs
Contrasting, but recurring forms or types within a single population of a species.
i.e. melanistic forms of cats.
A race/subspecies is different because they form a geographically isolated and distinct population.
Mutation
A process that introduces a lasting change into the genetic material (DNA & sometimes RNA).
If the change occurs in the germline, the mutation can be passed from parent to offspring.
In somatic cells, mutations are transferred to daughter cells.
Point Mutation
A type of mutation that affects only a single nucleotide.
This may cause changes in gene expression and/or affect the function of the corresponding gene product.
Chromosomal Alterations
Multiple genes are affected by the loss, rearrangement, reattachment of a chromosome (also considered mutations).
Continuous Variation
Refers to traits for which the phenotypes change gradually.
i.e adult height in humans.

Discontinuous Variation
Traits that fall into distinct groups (i.e. blood type).
Morphs and mutants would also fall into this group, such as albinos.

Polymorphisms
Generally considered relatively frequent discontinuous variants in a population.
Individual forms are called morphs.
It is frequency-based, and one has to make an arbitrary cutoff (often stated as 1%).
Mutants
Rare discontinuous variants in a population.
Polymorphisms and Mutants
Both result from occasional changes in the genetic material (mutations in DNA or RNA).
For some reason, changes in the morph are relatively common. However, mutations remain rare.
∴ No conceptual difference.
Alleles
Gene variants are different versions of the same gene within a population or cell.
They include sequence differences in coding, intron, or regulatory regions.
Many differences are inconsequential; from a geneticist’s view, such variants are functionally equivalent.
The term applies only to variation of a gene within a species.
Homologs
Gene inherited in two species from a common ancestor.
Pair of chromosomes, one from each parent.
Identical in size, banding pattern, and gene locations.
Human cells have 23 pairs of homologous chromosomes (not alleles).
BRCA1 Gene
Located on chromosome 17.
Involved in repairing damaged DNA.
Chromosome 17 contains ~1100–1200 genes.
A single mutant copy greatly increases the risk of breast cancer.
The mutant copy is an allele of the wild-type gene (the normal version).
Chromosome
DNA + associated proteins.
Linear in eukaryotes, usually circular in prokaryotes.
For viruses, the term “viral genome” is used.
For mitochondria, the term “mitochondrial DNA” (mtDNA) is used.
Historical Milestones for Chromosomes
1880s - Dyes reveal the thread-like chromosomes; number is constant in a species.
Cell division - Chromosomes condense and segregate equally to daughter cells.
1900s - Mendel’s work rediscovered.
1902 - Sutton & Boveri: chromosome movements mirror Mendel’s factors → genes are on chromosomes.
Chromosome Location in E&P
Gene Location
Found at specific locations.
The location does not change!
Gene - Locus (Loci)
Gene are usually represented with some sort of symbol (letter, number, symbols, etc).
Specific physical location of a gene or other DNA sequence on a chromosome, like a genetic street address.
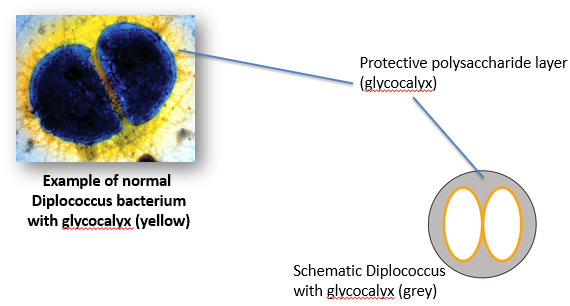
Fred Griffith (1928)
Studied Streptococcus pneumonia, which causes pneumonia in mice.
Forms a diplococcus, where a pair of cells remain attached due to incomplete cell division.
He cultured the infectious bacteria on plates and later saw a “rough-looking” strain.
Found that the new strain lacks the glycocalyx.
S. pneumonia “S” Strain
“S” forms smooth colonies, is virulent (causes pneumonia).
→ inject → mice dead.
“S” form is virulent because the glycocalyx protects bacteria from mouse immune system.
Colonies look smooth because they have the glycocalyx.
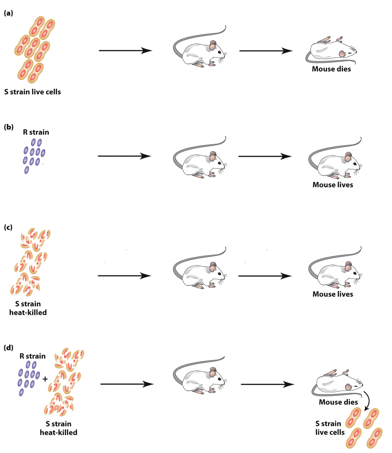
S. pneumonia “R” Strain
“R” forms rough colonies— not virulent (no pneumonia).
→ inject → mice live.
Not virulent because they lack the glycocalyx.
Colonies look rough because they lack the glycocalyx.
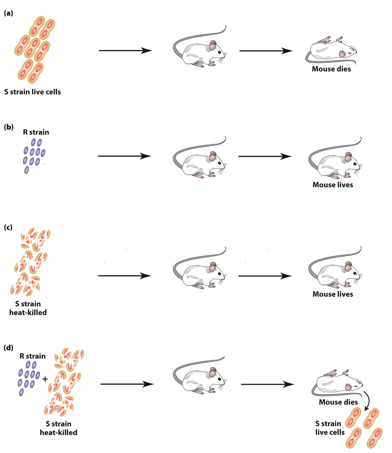
Inactivating S. pneumonia “S” Strain
Heat treatment inactivates the S strain.
Heat-killed “S” is not virulent.
→ inject → mice live.
No S strain can be obtained from the mice after injection.
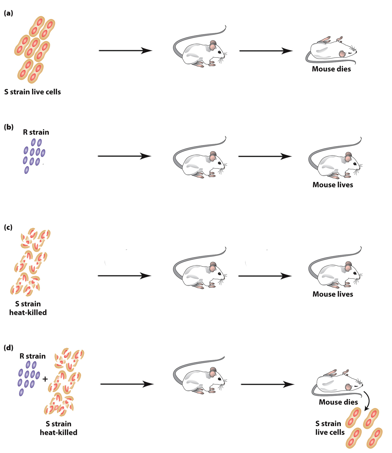
S. pneumonia Co-Injected S&R Strain
Heat-killed “S” nor “R” are virulent on their own.
Injected together → mice dead.
Both “S” and “R” strains can be obtained from the mice after injection.
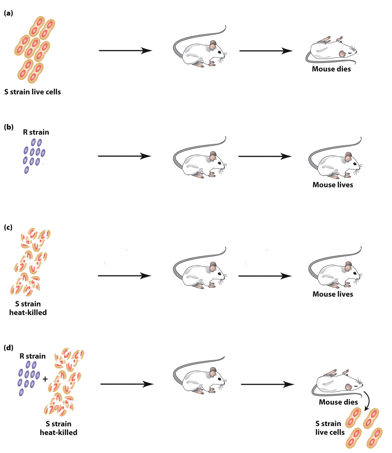
Fred Griffith’s Transformation Experiment
Live S. pneumonia cells could be recovered from these dead mice.
Streptococcus cells contained “S” strain (formed smooth colonies).
The “S” strain bacterial were virulent upon subsequent injections.
Transforming Principle
Living cells "(“R”) could be “transformed” by dead (“S”) cells.
Something in the S-cell debris could convert R cells into S cells.
Called transformation.
Competent
State in which bacteria can take up DNA from the environment.
Requires specific proteins called competence factors.
Enables incorporation of foreign DNA into the genome.
Used in molecular biology to introduce and amplify DNA.
Competence Factors
Proteins that bind single-stranded DNA.
Induce or maintain bacterial competence.
Help incorporate ingested DNA into the genome.
Support laboratory transformation techniques.
Homologous Recombination
Type of genetic recombination in which nucleotide sequences are exchanged between two similar or identical molecules of DNA.
Similar to meiosis.
Homologous Bacterial Transformation
Donor DNA from another strain aligns with a homologous site on the host chromosome.
Requires breaks in the host DNA.
Donor DNA acts as a “repair template,” replacing the host sequence at that locus.
Produces a transformed cell carrying the donor gene (e.g., S DNA for the glycocalyx locus).
Avery, MacLeod & McCarty Experiment (1944)
S-strain extracts treated with destroying agents.
Removing polysaccharide, lipids, RNA, or proteins → activity intact.
Removing DNA (with DNase) → activity lost.
∴, DNA is the transforming substance.
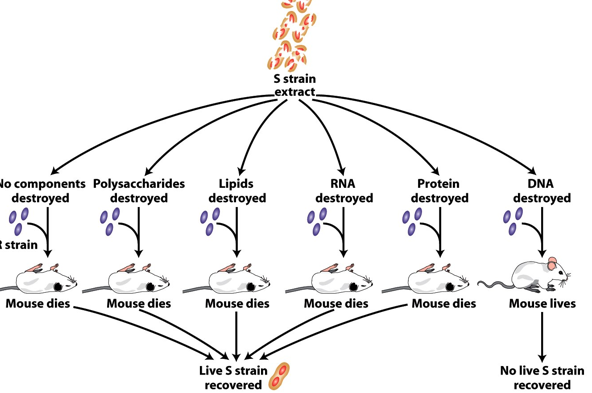
Avery’s + Co. Experiment Aftermath
The data from the experiment was conclusive, but there was reluctance to accept DNA as the genetic material.
DNA didn’t seem complex enough to be the base for genetic material.
Additional experiments added more evidence.
∴ acceptance that DNA is the carrier of all genetic material.
Hershey and Chase Experiment (1952)
Grew T2 phages in media containing either ³²P (to label DNA) or ³⁵S (to label protein).
Allowed labeled phages to infect E. coli.
Blended and centrifuged to separate bacterial cells from phage “ghosts.”
Found ³²P inside bacteria, while ³⁵S stayed with ghosts → DNA, not protein, entered cells.
Hershey & Chase - T2 Phage
Bacteriophage T2 is made of protein and DNA (no RNA or saccharides).
Genetic material had to be either protein or DNA.
Composition of the two components was unclear at the time.
Showing which part entered the cell would reveal the genetic material.
Hershey & Chase - Radioactive Isotopes
³²P (radioactive phosphorus) labels DNA; P not found in protein.
³⁵S (radioactive sulfur) labels protein; S not found in DNA.
Two cultures prepared: one with ³²P-labeled DNA, the other with ³⁵S-labeled protein.
Used to follow each component during phage infection of E. coli.
Hershey & Chase - Insertion
Phages infected E. coli, then cells were blended and centrifuged.
³⁵S (protein) radioactivity stayed with phage “ghosts,” not in bacteria.
³²P (DNA) radioactivity was found inside the bacteria.
DNA, not protein, entered the cells.
Hershey & Chase - Conclusion
DNA entered E. coli and could be recovered from new phage progeny.
Protein (³⁵S) remained outside in phage ghosts.
Therefore, for bacteriophage T2, genetic information is in DNA, not protein.
DNA is the hereditary material, at least for T2 phage.
Proteins Contain Sulfur
Some proteins are phosphorylated after translation by protein kinases.
Phosphorylation targets the non-carboxyl –OH groups in serine, threonine, and tyrosine.
Cysteine and methionine instead contain sulfur.
DNA Contains Phosphorus
Rich in P but lacks sulfur.
If DNA is the Hereditary Material…
Cells must be able to replicate DNA.
Carries hereditary information.
Transfer information to control a cell’s activity.
Must be able to change (mutate).
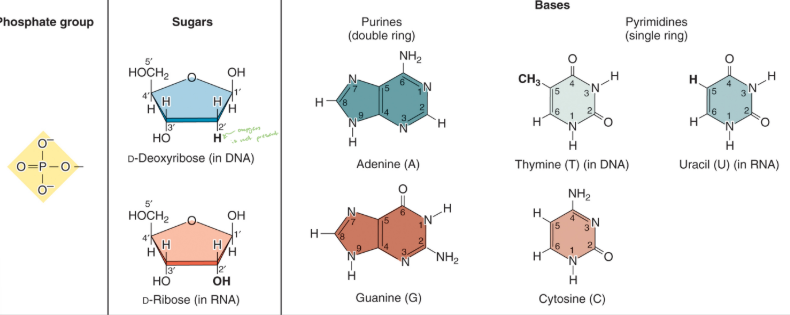
Nucleotides
Composed of a phosphate group, pentose sugar, and a nitrogenous base.
Base + sugar + phosphate = nucleotide.
Named after the base used.
Numbering system of atoms in sugars and nitrogenous bases.
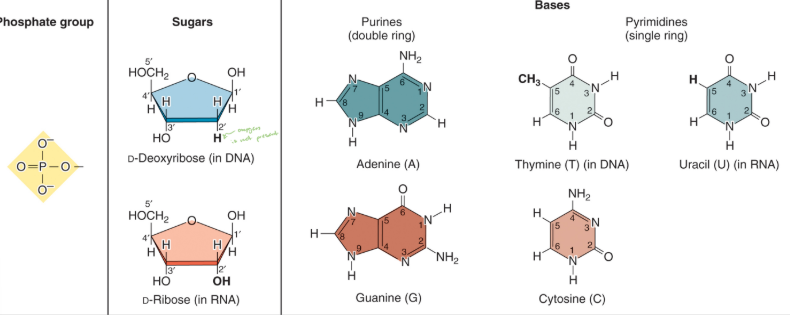
Pyrimidines & Purines
Pyrimidines: thymine, cytosine, uracil (RNA).
Purine: adenine, guanine.
Nucleotide Rotation & Flipping
Nitrogenous base is linked to the sugar by a single (glycosidic) bond.
This bond allows the base to rotate freely relative to the sugar.
Sugar-phosphate backbone bonds (3′ and 5′ carbons) also rotate, giving flexibility.
Bases can “flip out” of the helix for repair or binding events.
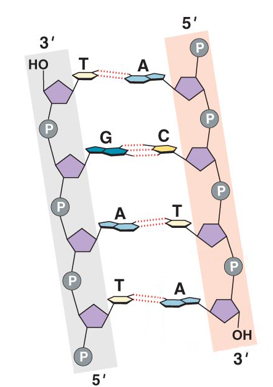
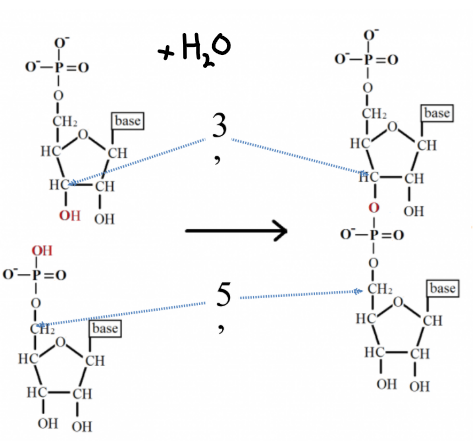
Phosphodiester Bond
Phosphate group at 5’ position on the sugar of one nucleotide is covalently bonded to the 3’ position of the sugar on the next nucleotide.
Condensation reaction (H2O is produced).
Rosalind Franklin
Produced a X-ray diffraction image of DNA.
Data is interpreted by Watson and Crick (1953).
Built a structural model that accounted for Franklin’s data.
Nucleotides linked to Polymer Strands
Strand directionality is based on orientation of the sugar molecules.
Phosphate backbone is negatively charged.
Chargaff (1940s)
Determines the nucleotide distribution across species.
Finds that chemical composition is the same.
Finds that each species has a specific quantity of DNA.
Determines that DNA found in different species shows the same ratio of bases.
i.e. the amount of A = T.
Rosalind Franklin
Produces a X-ray diffraction image of DNA.
Data is interpreted by Watson & Crick (1953).
Built a structural model that accounted for Franklin’s data.
DNA-Binding Proteins
Important for proteins to bind to the major groove.
Most DNA binding proteins recognize the major groove.
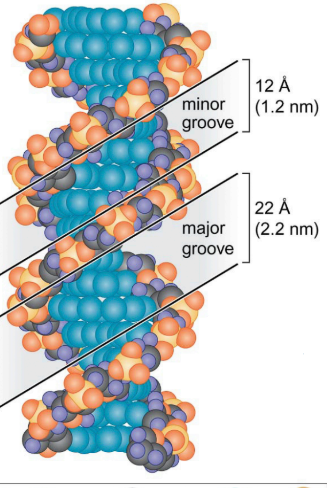
Structure of DNA
Helical molecule (2nm wide).
Made of two antiparallel nucleotide polymer strands.
Covalent bonds between phosphates and sugars connect nucleotides.
Sugar and phosphate groups face “out.”
Nitrogenous bases of two strands face the “inside” of the helix.
DNA is Antiparallel
Covalent bonds between phosphates and sugars connect nucleotides.
Sugar and phosphate groups face “out.”
Nitrogenous bases of two strands face the “inside” of the helix.
~10 base pairs per helical turn.
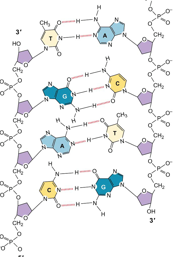
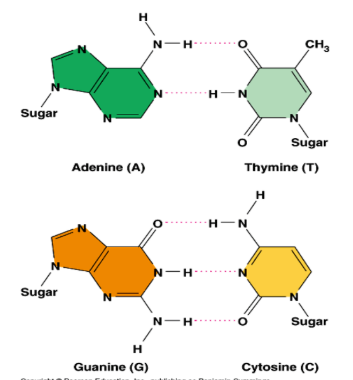
Complementary Base Pairing
Spacing led to conclusion that a purine pairs with a pyrimidine.
Chargraff’s data suggested that.
A pairs w T.
C pairs w G.
H-bonds connect the base pairs.
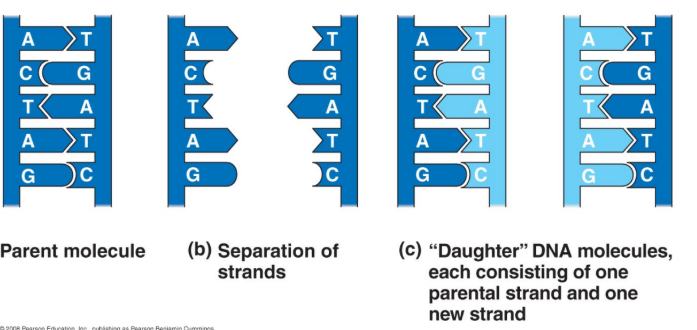
DNA Replication
Structure of DNA predicts a mechanism for how it is copied (relies on complementary base pairing).
Two parental strands separate and act as templates for the synthesis of new daughter strands.
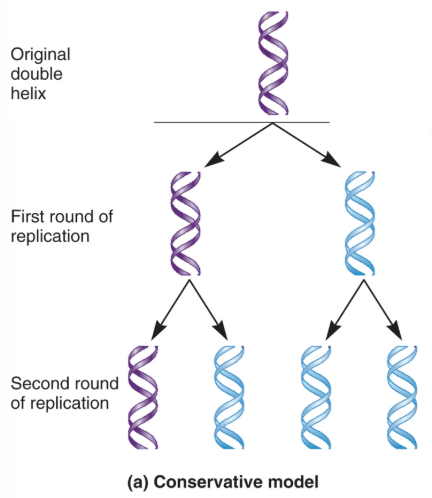
Conservative Replication Model
In each, parental strands separate and act as templates that are copied.
Parental strands reanneal and new daughter strands anneal.
If true, one expects all newly formed DNA duplexes to harbour exactly 100% new DNA.
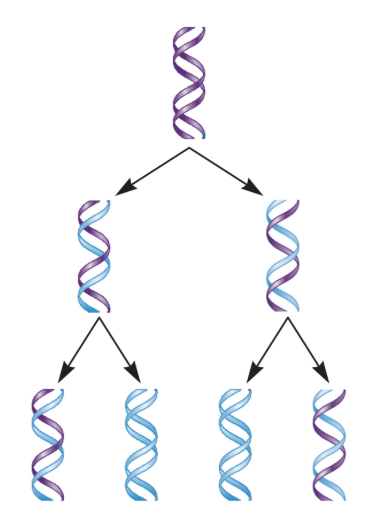
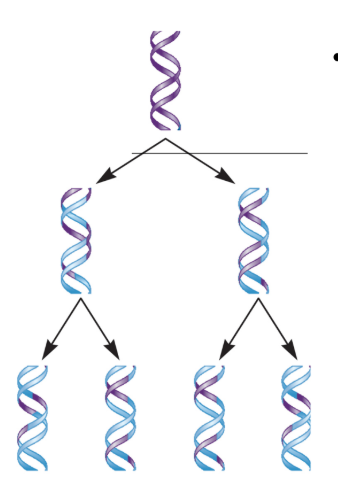
Semi-Conservative Replication Model
New DNA molecules are composed of a parental (old) and daughter (new) strand.
If true, each DNA molecule has exactly 50% from the original (parental) DNA molecule.
Dispersive Replication Model
Proposes strand breaks at every node (~5 bases) where new DNA joins parental DNA.
Results in alternating segments of ~5 bases parental DNA and ~5 bases of new daughter DNA, where new DNA base pairs with parental DNA.
If true, each DNA strand harbours slightly varying amounts of old and new DNA, but each contribution would be close to 50%.
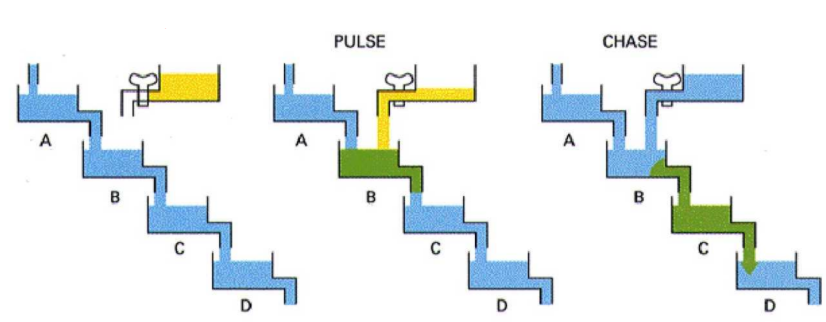
The Pulse-Chase Experiment
In living organisms, metabolism results in constant turnover of biomolecules.
By adding a isotope-labelled nutrient (i.e. sugar or aa) for a limited time (the “pulse”) one can follow the fate of the isotope over time (the “chase”).
Before and after the pulse (= unlabelled) nutrients are provided.