Immunology MICI 3115- Final/ Unit 2
1/259
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
260 Terms
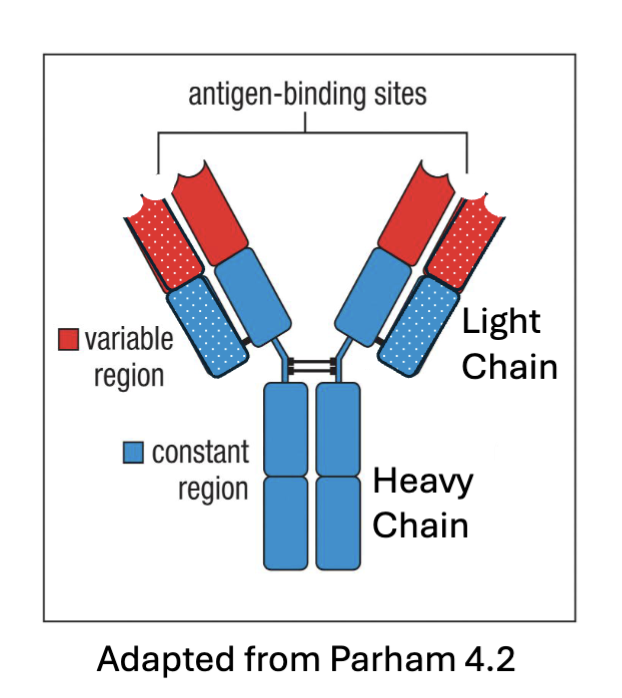
What are the features of antibody structure?
polypeptides – combination of 2 heavy chains and 2 light chains linked together
Variable regions – recognize antigens
Heavy chain constant region – determines function of the antibody
Different classes of immunoglobulin (antibodies) are expressed depending on activation and development of the B cell
How do we get recognition of different antigens? (What are the two theories? Which is correct?)
A1:Germ line theory
A2: Rearrangement of germline gene segments (somatic recombination) during B cell development -Correct
What are the features of Germ line theroy?
genetic info for each immunoglobulin encoded directly in genome
→wrong bc: Genome doesn’t contain enough DNA for this
What are the features of Rearrangement of germline gene segments (somatic
recombination) during B cell development (historically speaking/ it’s discovery)?
1965: Theory proposed by Dreyer and Bennett
1976: Tonegawa and Hozumi demonstrate that separate gene segments encode the V and C regions of immunoglobulins
Individual B cells had different sequences at the BCR locus and mature B cells were often missing parts of the constant locus
1987: Tonegawa won the Nobel Prize
(rearrangement: increase specificity w/out enormous genome)
What are the features of rearrangement of germline gene segments?
Immunoglobulins are generated from 3 gene loci: heavy chain (Ch 14), with 𝛋 light chain (Ch 2) or 𝜆 light chain (Ch 22)
Undergo rearrangement of germline gene segments (somatic recombination) during B cell development
Recombination of Variable (V), Diversity (D), and Joining (J) segments → variable regions with different specificity
How does rearrangement of germline gene segments work? (process)
Recombination is mediated by the Recombination Activating Gene (Rag1/Rag2) complex (Rag1/2 forms dimer)
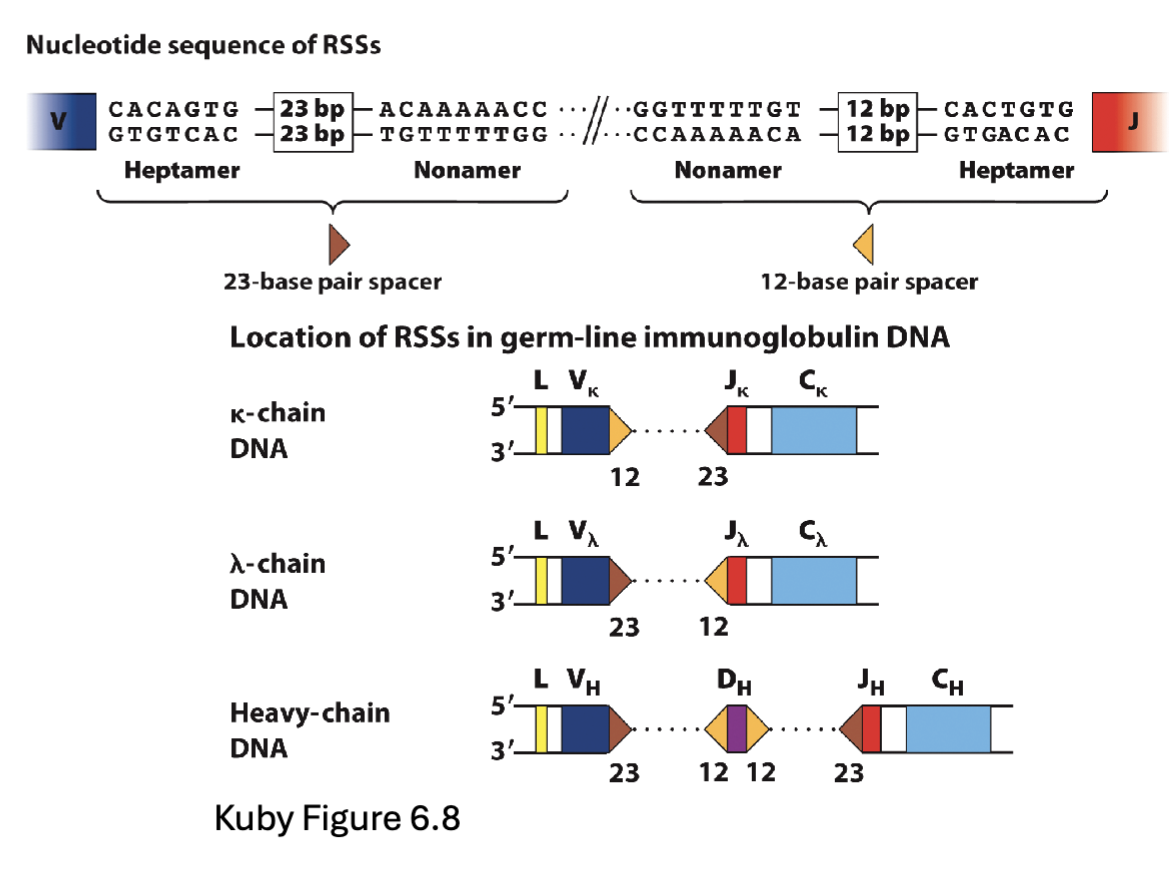
Recognize recombination signal sequences (RSS) flanking the gene segments – bind conserved Nonamer (N) and Heptamer (H) sequences (Rag1 recognizes certain sequences downstream of gene sequences that have RSSs (have few diff. domain, N and H activate: conformational change)
Endonuclease activity cleaves at RSS and DNA repair enzymes join the DNA ends (Rag1 has endonuclease activity)
Excised DNA material is lost from the cell ... no going back
Rag mutations → severe combined immunodeficiency (SCID) or Omenn syndrome (hypomorphic RAG)
(large chunk of DNA is cut out (thrown out- at single cell level) happens w/in individual B cells)

What makes sure you rearrange the right segments?
Recombination signal sequences (RSS) flanking the gene segments have different spacer sequences between the N and H sequences – 12 or 23 base pairs (12/23 rule)
Recombination only occurs between segments that have different spacers
Cannot recombine with genes lacking an RSS
(this is how the regulation of the junctions is done)
Spacer: between N and H there is a spacer sequence (12 or 23bp) added into gene segment and can rearrange between signal sequences that are different need a 12 and 23
23’s can’t join together
if gene segment lacks spacer can’t recombine
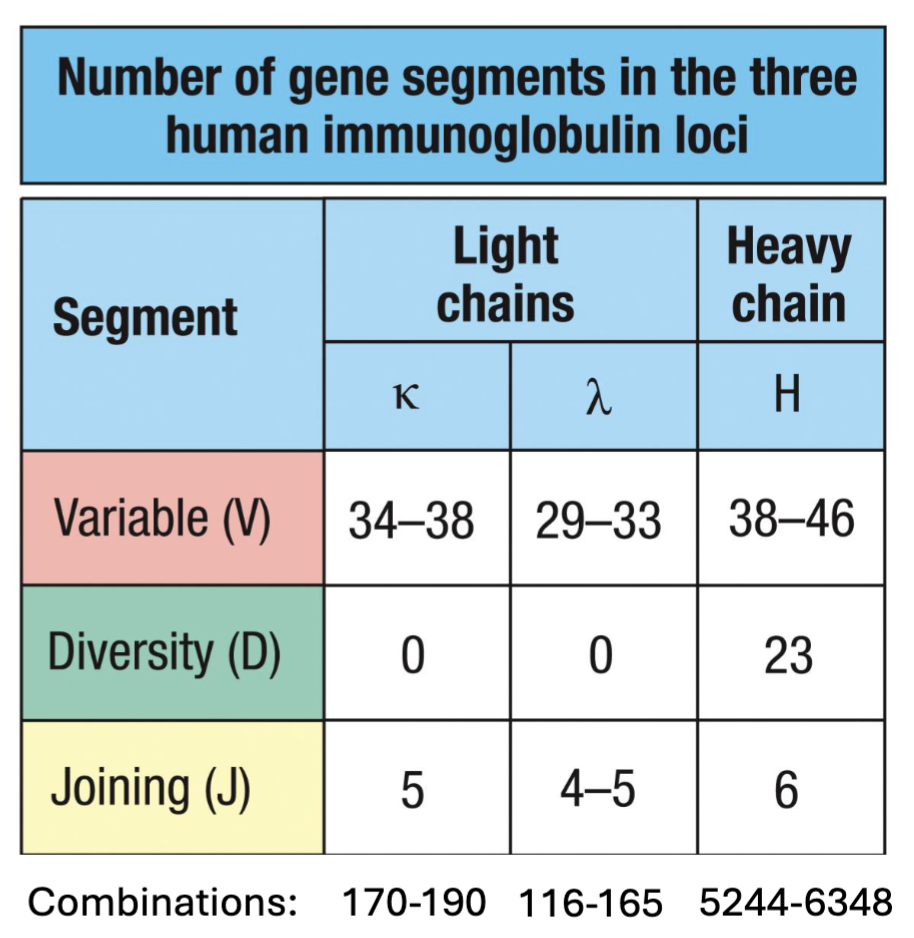
What is combinatorial diversity?
shuffling use of V, D, and J gene segments generates different variable regions
(190 k-chains + 165 𝝀-chains) X 6348 H-chains = ~2.25 Million combinations from 156 gene segments
(don’t need to know the numbers just know which has most and know more V > D, J)
Immunoglobulin repertoire is generated by ____&____?
combinatorial diversity & junctional diversity
What is junctional diversity?
Immunoglobulin diversity is further increased by the DNA repair process during recombination:
Excision (by Rag) forms DNA hairpin loops → unfolding adds palindromic sequences (P-nucleotides)
Mediated by Artemis (endonuclease)
Terminal deoxynucleotidyl transferase (TdT) adds random non-templated nucleotides (N-nucleotides)
Nonhomologous end joining (NHEJ) ligates DNA ends
Increases repertoire diversity by factor of 3 X 107 → Total diversity ~ 5 X 1013 (increase diversity adding more sequence)
(DNA repair enzymes cleave the hairpin and create a nick, cleaved 2bp in (hairpin opens up= 2 extra a.a that weren’t there before))
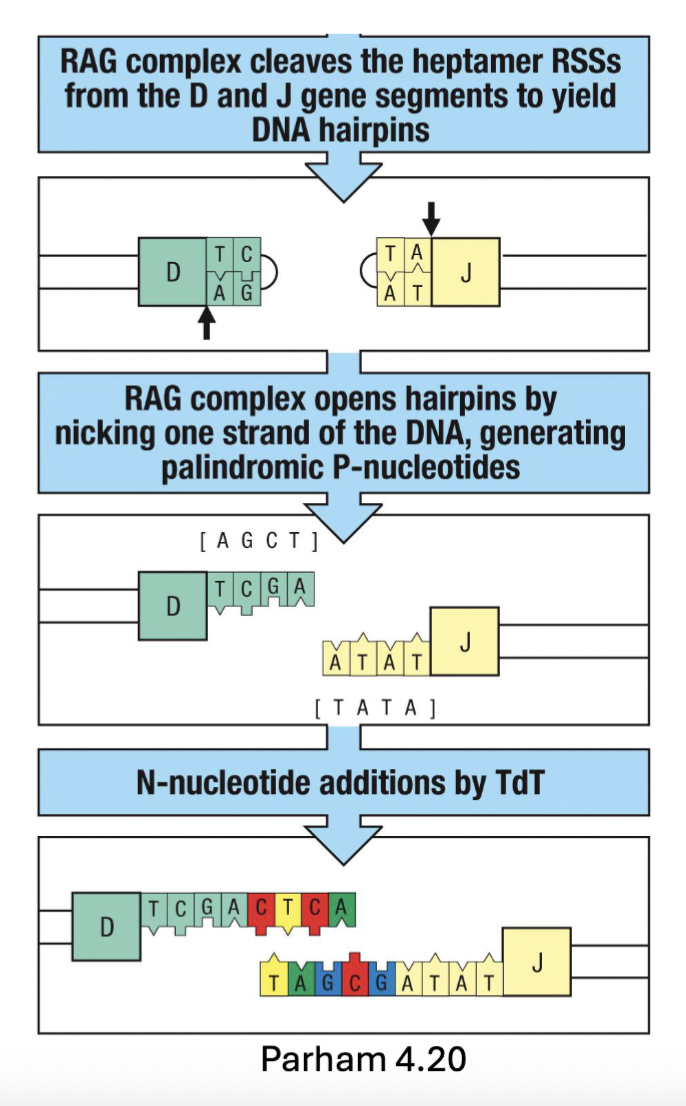
What do non-template nucleotides do?
randomly adds nucleotides into cut space, then anneals and refills, sequence that is repaired and has extra nucleotides (increase diversity)
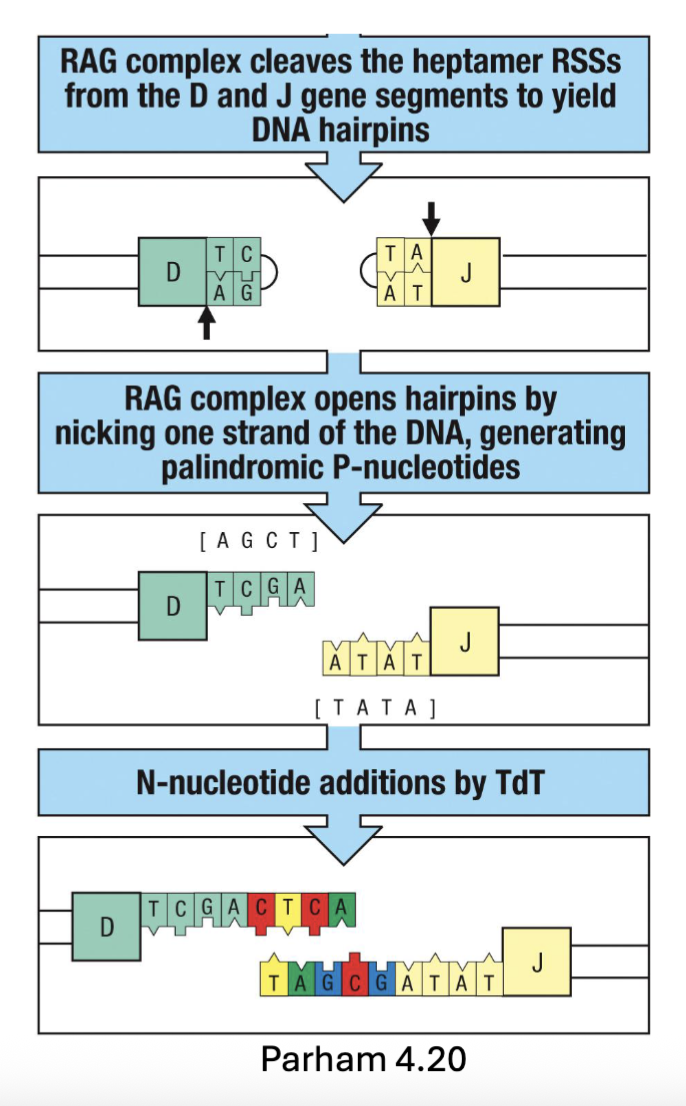
What is the finished product of Immunoglobulin gene recombination? What happens next?
Heavy chain recombines D-J segments before recombination with V segment
Each B cell only rearranges one heavy and one light chain → generates one
specificity (allelic exclusion)Recombination brings promotors and enhancers close → transcription
Unstimulated B cells are ready to express B cell receptors (BCR)
(when the rearrangement happens have allelic exclusion, brings everything closer and can induce transcription)

What are the features of B cell expression?
Cμ (mu) and Cδ (delta) are available downstream of rearranged VDJ segments
mRNA splicing generates transmembrane forms of IgM > IgD (common on immature B cells)
hypdrophobic C-terminal → membrane interaction in ER
IgM/IgD need to associate with Ig⍺ (CD79a) and Igβ (CD79b) for transport to the cell surface
Ig⍺ and Igβ are important for BCR signaling (bring it to receptor)
(downstream of VDJ is the constant region, happening at splicing level not genomic can’t signal on its own need Igalpha and beta to mediate)

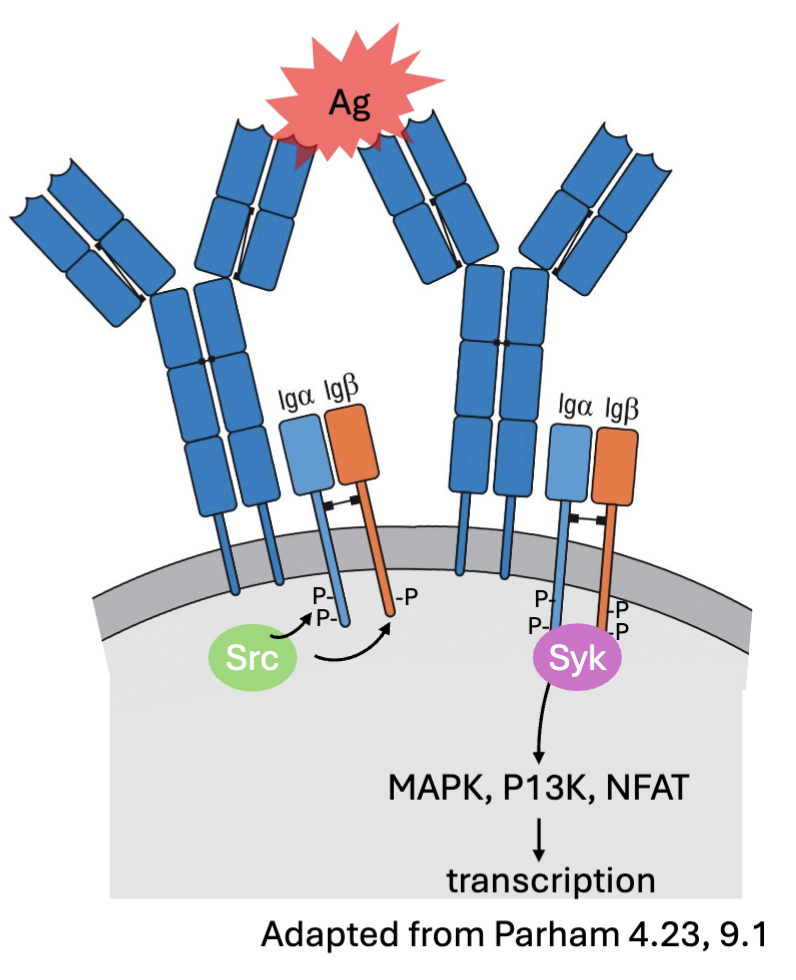
What are the features of B cell receptor signalling?
Antigen binding → IgM/IgD clustering →
Phosphorylation of ITAMs in Ig⍺ (CD79a) and Igβ by Src kinases (Blk, Fyn, Lyn) →
Recruitment of kinase Syk → downstream signaling (MAPK, P13K, NFAT) →transcription activation → survival, proliferation, differentiation
Outcome influenced by strength and duration of binding (affinity), co-stimulatory interactions (CD19:CD81,CD21/ CR2), and T cell help
(can crosslink the antigen, when cluster→activation, recruitment of certain kinases, binds strongly= longer signalling, costimulatory signals: other molecules)
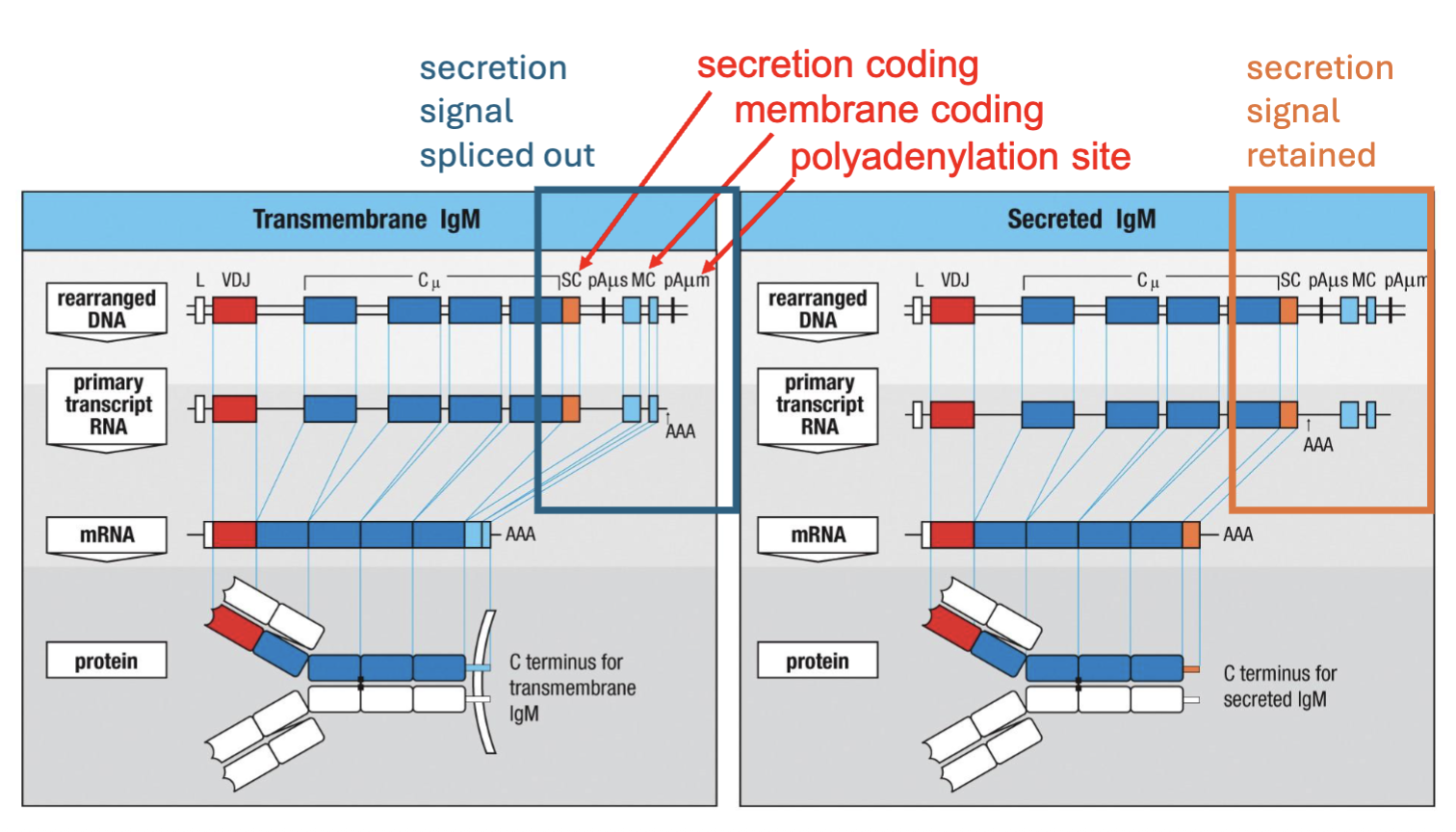
What is the difference between a BCR and an antibody?
alternative RNA splicing
Antibodies are made at higher concentrations than BCR
The difference between the membrane bound and secreted forms comes down to a hydrophobic or hydrophilic C terminus, respectively
(B cell receptor on surface when cell is activated goes from making receptor to making antibody couple of diff. sequences, secretion, membrane and polyadenylation)
What makes a better B cells (survival of the fittest)?
Rearranged V-region sequences are further mutated in activated B cells (Somatic Hypermutation)
Mediated by activation-induced cytidine deaminase (AID) generated in proliferating B cells
Induces mutations during transcription when DNA strands are separated → repair errors
Single nucleotide mutations induced in variable regions (CDRs) at ~ 1 mutation per dell division (> 1 million times normal mutation rate)
Ig variable regions enriched in sequence motifs susceptible to AID (mutational hot spots) (MHS:allows for selection of antibodies that bind better)
B cells with mutant immunoglobulins undergo competition and are selected for higher antigen binding (affinity maturation) → go on to form plasma cells and memory B cells
(when B cell activating make AID→ causes mutations in antibody gene sequence, can create a nick and needs to be repaired not always repaired properly
high mutation rate, tend to have mutation every time dell divides because of the repair error
the antibodies are competing with one another, if increased affinity of the B cell receptor: more signalling
strongest binding to antigen will survive and bind: affinity maturation (selected for really high affinity these will make plasma B cells or memory B cells)
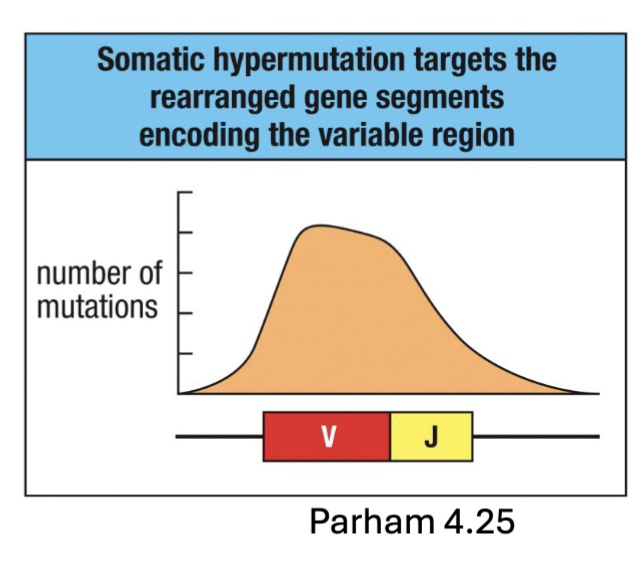
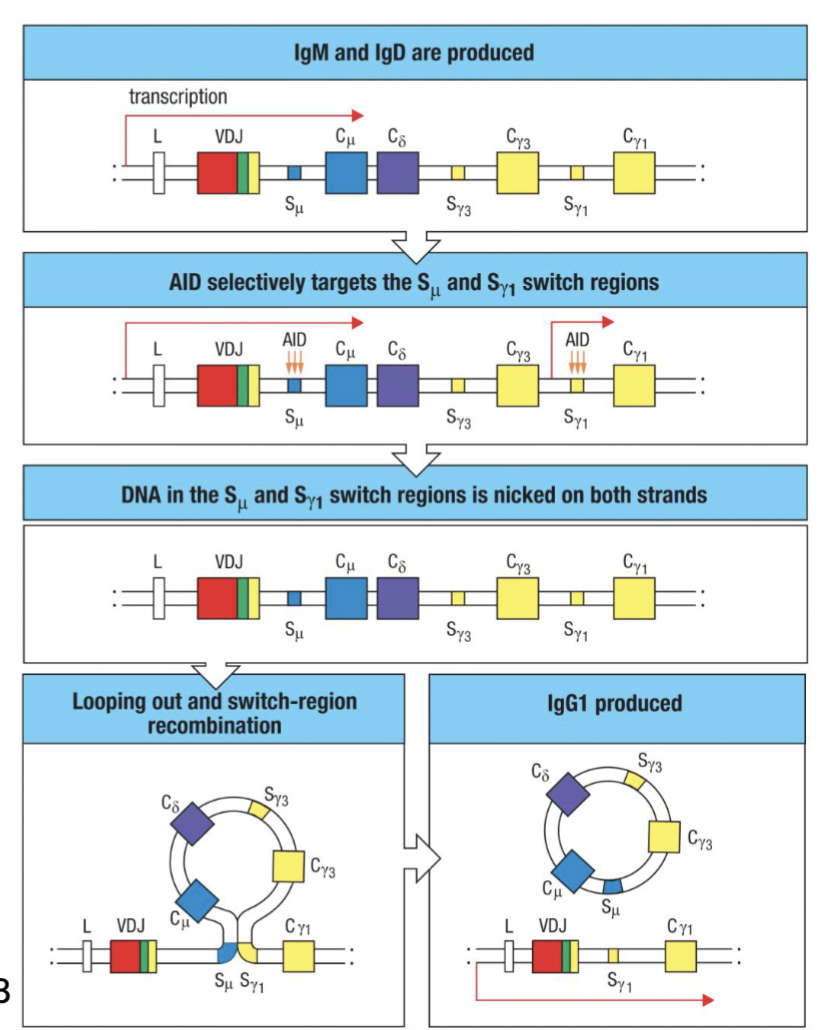
What is class switch recombination?
Activated B cells undergo recombination of heavy-chain C gene segments (Class Switch Recombination)
Requires transcriptional activity in C region to be switched to – regulated by cytokines and environment (e.g. IL-4 → IgE, IgG1; TGFβ/ IL-5→IgA; IFNɣ→IgG2a, IgG3)
Mediated by activation-induced cytidine deaminase (AID) in proliferating B cells
Single nucleotide mutations induced in switch regions (S) flanking C regions →
Nicks on both strands facilitate homologous recombination that excises upstream C regions ... no going back
AID deficiency associated with hyper-IgM syndrome (increased infections, and cancer risk)
(requires active transcription at the sites, depending on cytokine will get signals from the environment)
(in presence of certain cytokines will have transcription (DNA opens up) AID create nicks in the sequences (switch regions) and then homologous recombination occurs→ cleaves out the sequence can no longer make IgD and IgM (irreversible) creates different antibody)
What is the mechanism and the nature of change to the B cell’s genome in V-region assembly from gene fragments?
Mechanism: Somatic recombination of genomic DNA
Nature of change to the B cell’s genome: Irreversible
What is the mechanism and the nature of change to the B cell’s genome in generation of junctional diversity?
Mechanism: Imprecision in joining rearranged DNA segments adds non-germline nucleotides (P and N) and deletes germline nucleotides
Nature of change to the B cell’s genome: Irreversible
What is the mechanism and the nature of change to the B cell’s genome in assembly of transcriptional control elements?
Mechanism: Promoter and enhancer are brought closer together by V-region assembly
Nature of change to the B cell’s genome: Irreversible
What is the mechanism and the nature of change to the B cell’s genome in transcription activated with coexpression of surface IgM and IgD?
Mechanism: Two patterns of splicing and processing RNA are used
Nature of change to B cell’s genome: Reversible and regulated
What is the mechanism and the nature of change to the B cell’s genome in synthesis changes from membrane Ig to secreted antibody?
Mechanism: Two patterns of splicing and processing RNA are used
Nature of change to B cell’s genome: Regulated
What is the mechanism and the nature of change to the B cell’s genome in somatic hypermutation?
Mechanism: Point mutation of genomic DNA
Nature of change to B cell’s genome: Irreversible
What is the mechanism and the nature of change to the B cell’s genome in isotype switch?
Mechanism: Somatic recombination of genomic DNA
Nature of change to B cell’s genome: Irreversible
What gene segments come together in the heavy chain what genes come tofether in the light chain?
Heavy: D→J and then V→DJ
Light: V→J
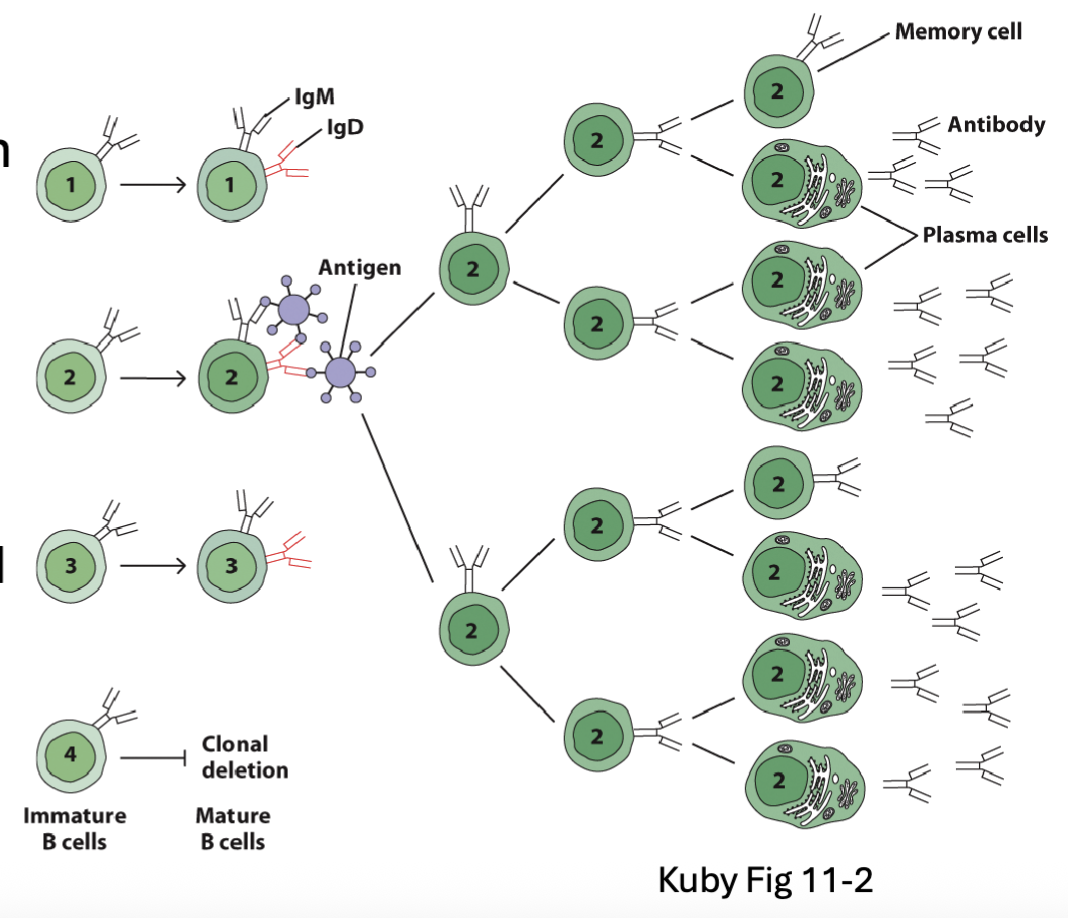
What is the Clonal Selection Hypothesis?
research that led to the discovery of B cell development
Frank Macfarlane Burnet (1957):
Predicted one Ig specificity for each developing cell
Activated cells would replicate and produce offspring with same Ig specificity as parent (partially correct-there are hypermutation later on so not exactly the same but come from the same progenitor)
Memory B cells would allow enhanced secondary response
Self-reactive cells would be deleted
predicted the BCR rearrangement
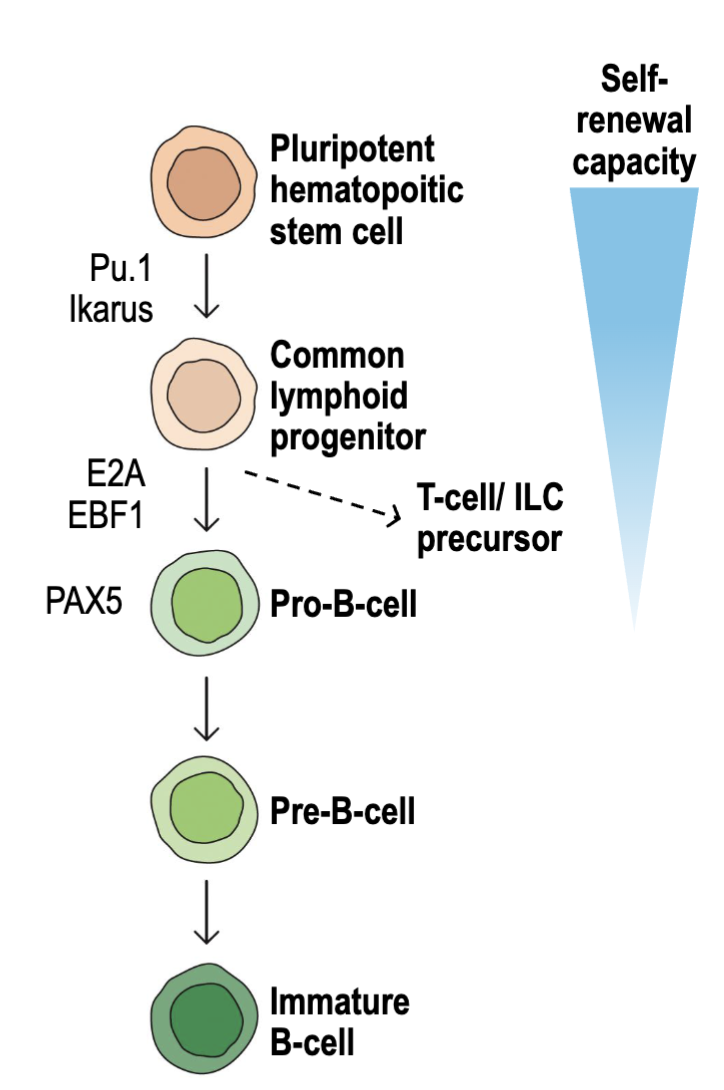
What are the general features and influences of B cell development?
Human bone marrow generates ~60 billion B cells per day (35-55 billion of them will die)
B cells are derived from the common lymphoid progenitor (CLP)
Transcription factors E2A and EBF1 drive B cell commitment and BCR rearrangement
PAX5 critical for B cell development
Ig rearrangement and selection take place during pro- and pre- B cell stages
Immature B cells migrate through secondary lymphoid tissues and undergo
additional selection and differentiation
E2A and EBF1 commit to B cell lineage, will undergo BCR rearrangement
PAX5 major factor of rearrangement
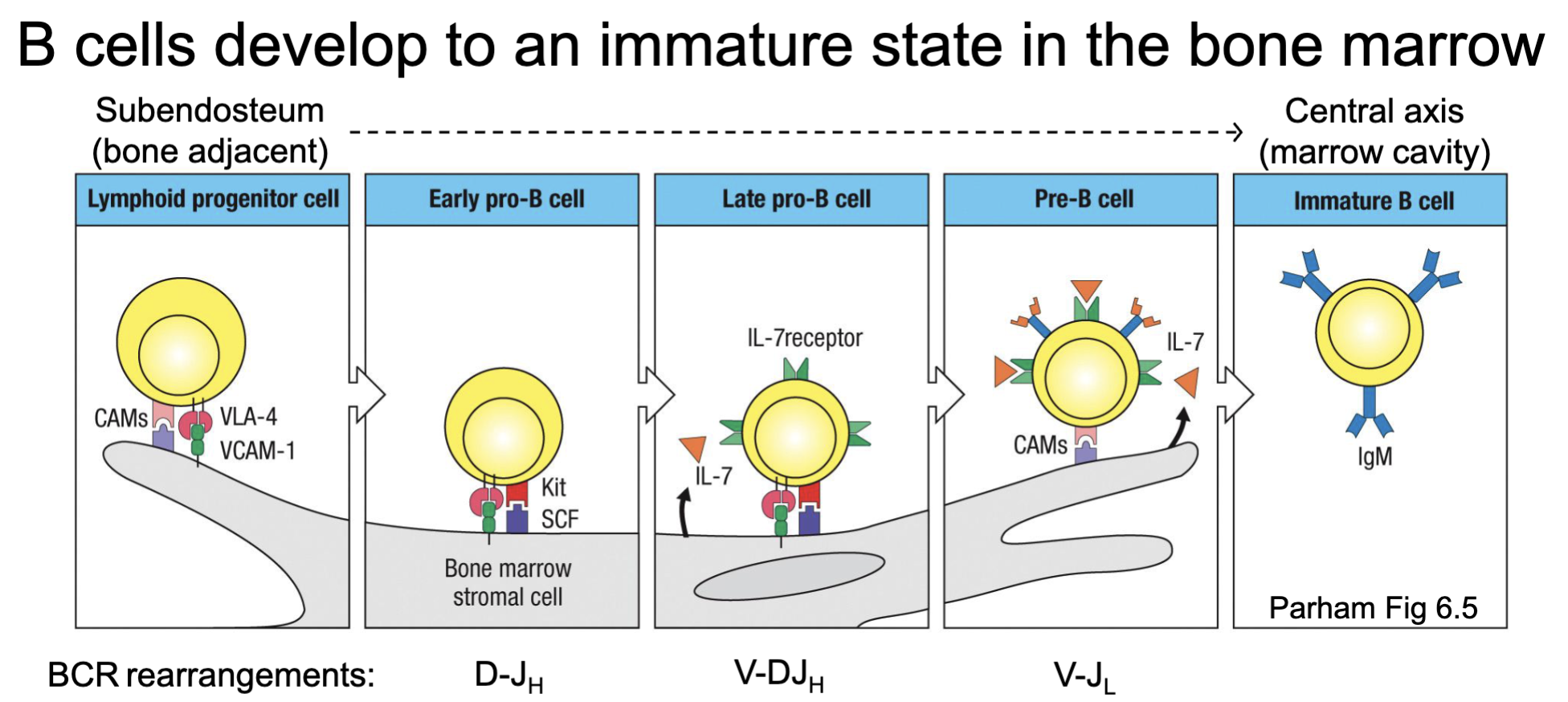
What are the features of B cell development to an immature state?
Happen in the bone marrow
As B cells develop, they move along the stroma from bone edge to central marrow
Receive different signals in different regions
Pro- and Pre-B cell stages correspond with B cell receptor rearrangements

What are the features of rearrangement in early pro-B cell and late pro-B cell development?
early pro-B cells starts to have D-J rearrangement
late pro-B cell will have some selection
What are the features of pro-B cell: Heavy chain rearrangement?
E2A and EBF1 transcription factors:
Open chromatin at heavy chain locus
induce Rag1/2 and PAX5
PAX5 transcription factors:
B cell master regulator
induces V-DJ recombination (not needed for D-J), expression of
Iga/IgB and VpreB/𝜆5, and downstream development
Two steps lead to heavy chain rearrangement
Apoptosis as a default in absence of positive survival signal (but this
happens at pre-B cell stage
Vprebeta and Vprelamda5 important for development stage
2 chances of rearrangement (bc of 2 chromosomes)
first checkpoint
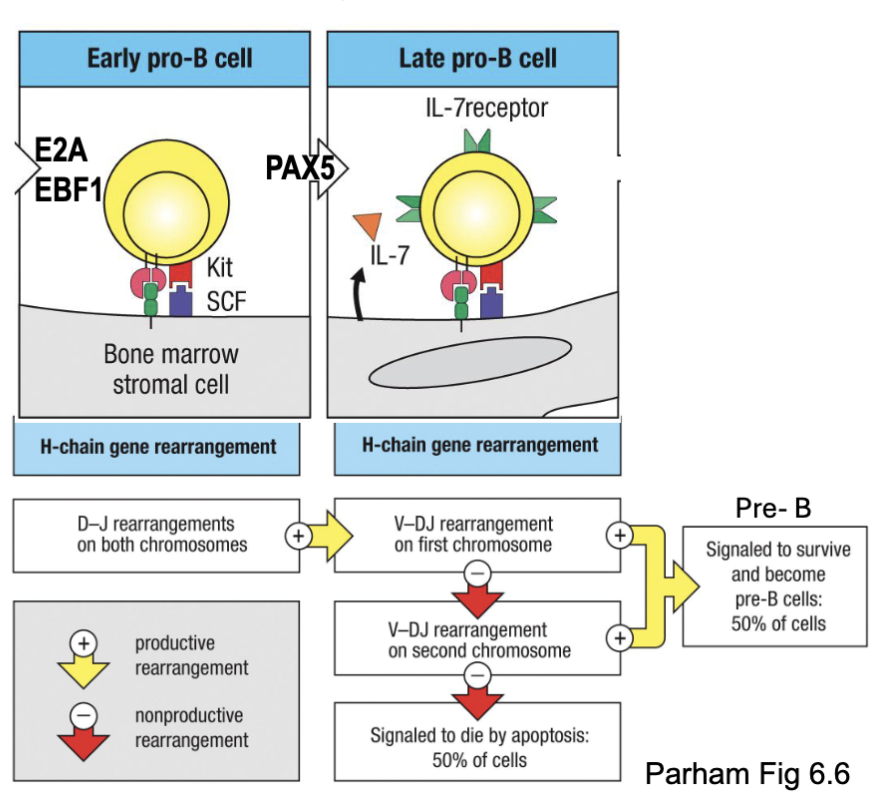
What are the features of the Pre-B cells: pre-BCR checkpoint?
Transition from late pro-B cells to early (large) pre-B cells marked by upregulation of the surrogate light chain (SLC) – made up of VpreB and λ5 subunits
SLC associates with rearranged heavy chain → expression on cell surface as pre-BCR complex
SLCs cluster → pre-BCR signaling and survival (positive selection/ 1st checkpoint)
Cells lacking productive heavy chain rearrangements do not express pre-BCR → apoptosis
Pre-BCR signaling → clonal proliferation, transient downregulaion of RAG1/2 and chromatin condensation at unused heavy locus (allelic
exclusion)
if signal, can’t go to surface cell will die

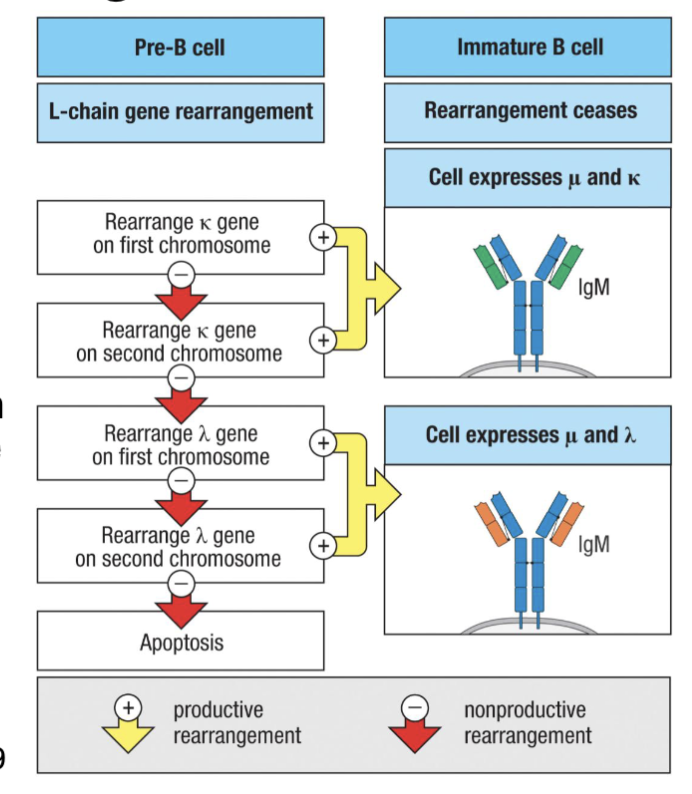
What are the features of Pre-B cells: Light Chain Rearrangement?
Following 1st checkpoint, pre-B cells proliferate and become late (small) pre-B cells
Pre-BCR signaling feedback turns off SLC expression, RAG genes reactivated (via IRF4 and 8) → light chain rearrangements
Mice rearrange KL-locus before λL-locus, random in humans (50/50 in humans)
Successful light chain rearrangement pairs with heavy chain in ER→ expression on cell surface as BCR complex
Low level (tonic) BCR signaling → survival (positive selection/ 2nd checkpoint), turns off further rearrangements
Success ... we now have immature B cells
signalling feedback turn of gene for surrogate light chain
What are the features of Immature B cells: Tolerance Induction?
Developing B cells were positively selected for BCR activity → need to do something about autoreactive clones (~75% will have some self-reactivity)
Three central tolerance (at site of primary development) mechanisms
Export of autoreactive immature B cells can result in peripheral tolerance (elimination at other sites) or potential autoimmunity (i.e. SLE)
want to eliminate self recognition
75% removed bc autoreactive
central tolerance getting rid of cell where it was developed (bone marrow)
B cells look for surface antigen but don’t eliminate anything that targets the interior
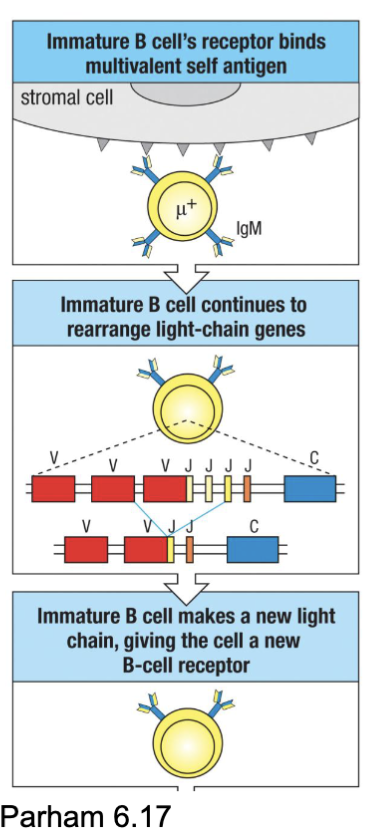
What are the three central tolerance mechanisms of immature B cells?
Clonal deletion → apoptosis of highly reactive clones (IgM cross-linking induces death of immature but not mature B cells)
Reactivate RAG genes and undergo light chain receptor editing → changed antigen specificity
Anergy: induced unresponsiveness to antigen (usually soluble/monovalent Ags: no cross- linking) → short life
2nd form tries to save some of the B cells
anergy: turns off the cell (prevent signalling and have a shorter half life)

Where does B cell maturation happen?
in the periphery (peripheral lymphoid organs- spleen, lymph nodes)
-enter T cell zones of lymphoid tissue and become transitional B cells
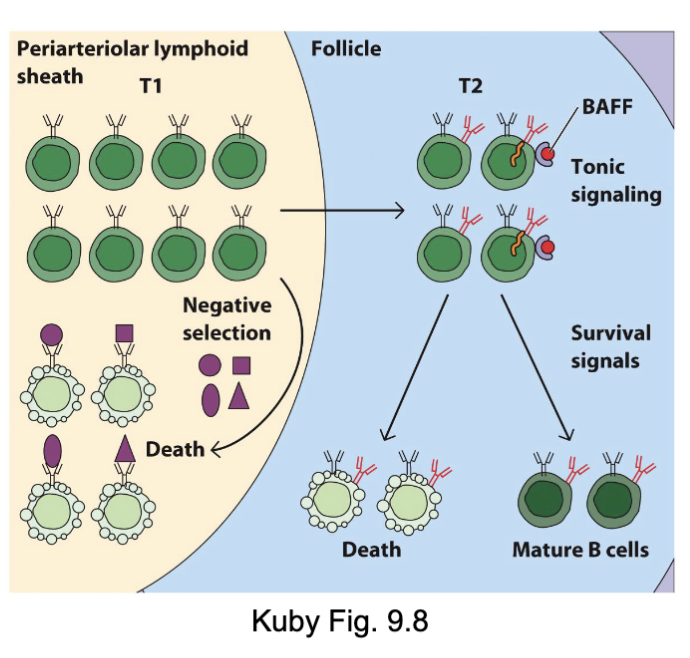
What are the features of B cell maturation in the periphery?
Immature B cells enter circulation in response to Spingosine1-phosphate (S1P) in blood
Localize to spleen and lymph nodes where they become known as transitional (T) B cells:
T1 B cells (mIgMhi/mIgDlo) enter T cell zone
Undergo negative selection if Ag encountered
upregulate IgD, CD21/CR2, CD23, BAFFR→ T2 B cells
T2 B cells (mIgMhi/mIgDint)→ enter follicle and interact with FDC → receive survival signals (tonic BCR, BAFF) → mature follicular B cell
Follicle niche size is limited and most B cells will die
Congratulations ... you made it to a mature
B cell (mIgMlo/mIgDhi)
T1 B cells: see other antigens (new recognition and deletion of them in the T cell zone)
T2 B go into the follicular zone, switch from IgM to IgD (transcriptional switch)
signal from BAFF (B cell activating factor)
more IgD gets upregulated
(issue: resources in follicule limited, not enough BAFF to go around)

What are the features of B cell recirculation?
Mature B cells survive ~100 days without antigen encounter
Mature naïve B cells recirculate through lymphoid tissues → enter LN primary follicles via HEV or afferent lymphatics
CXCL13 draws them into B cell zone
Interact with network of follicular dendritic cells (FDC) looking for antigens
No Ag → follow S1P gradient and exit via efferent lymphatics
leave lymph node will eventually return to the bloood
if they encounter antigen: B cell stimulation

What are the two things that can cause stimulation of B cells?
Antigen encounter → signaling via:
BCR complex (BCR, Ig⍺, Igβ)
B cell co-receptor complex (CR2, CD19, CD81): recognizes complement deposited on target → increases signaling 1000-10,000-fold
antigen with complement:
complement receptor recognizes it
activated by two things
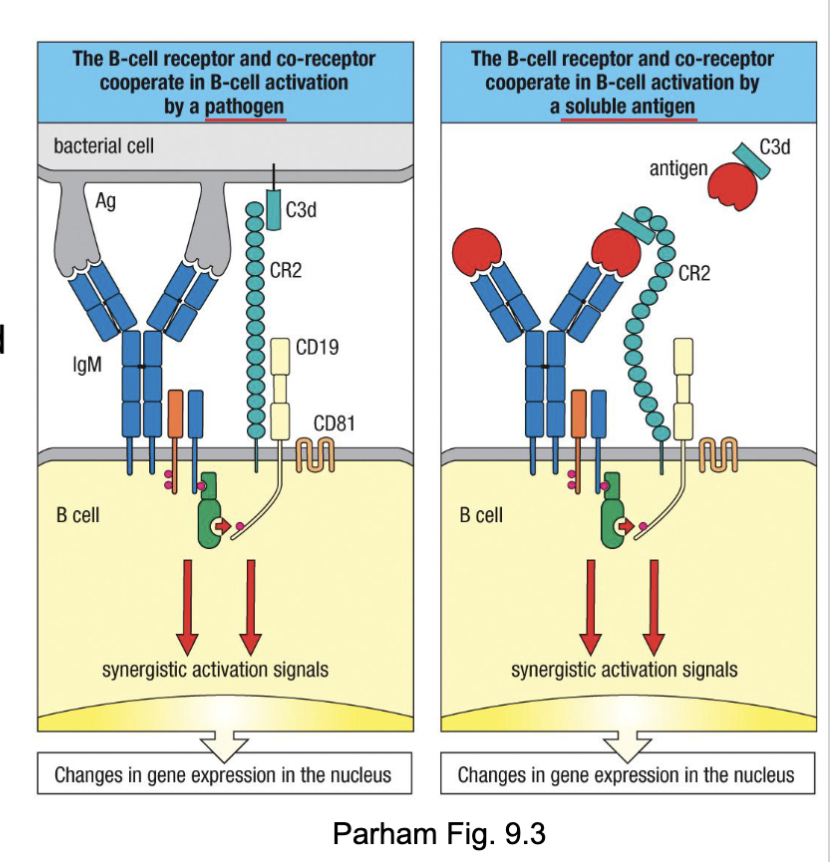
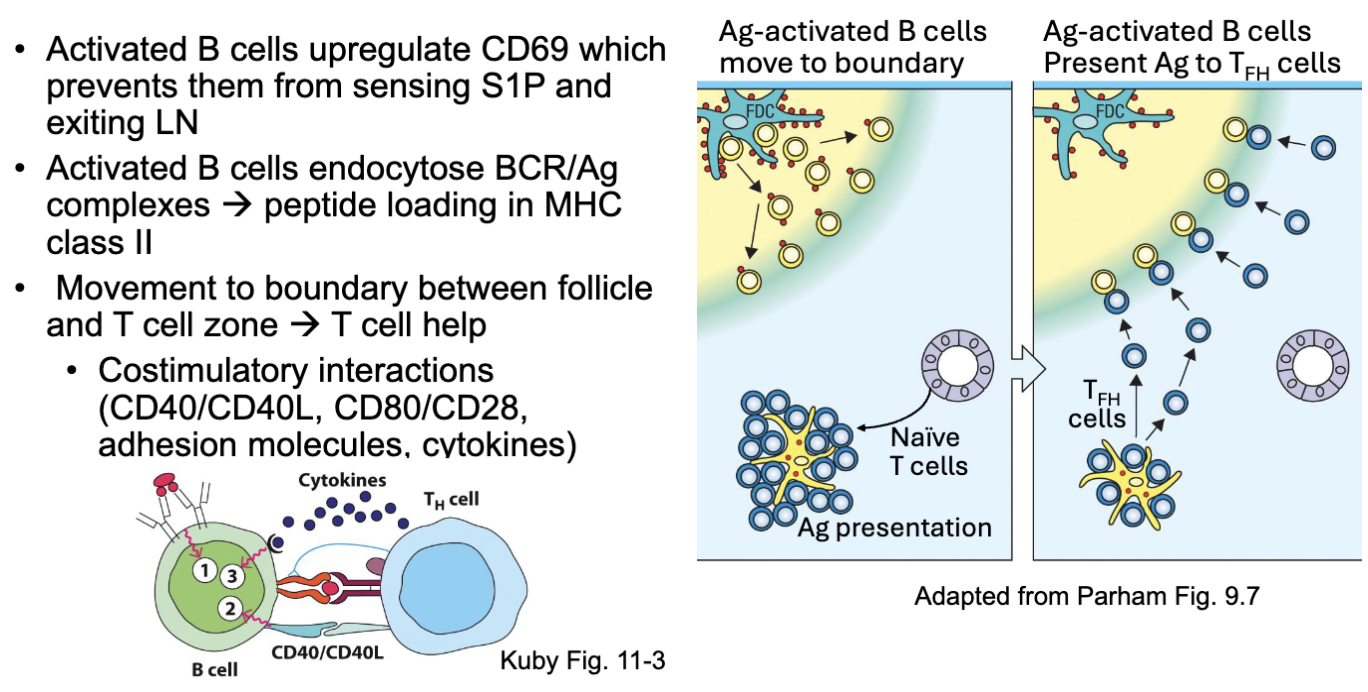
What are the features of B cell stimulation?
Activated B cells upregulate CD69 which prevents them from sensing S1P and exiting LN
Activated B cells endocytose BCR/Ag complexes → peptide loading in MHC
class IIMovement to boundary between follicle and T cell zone → T cell help
Costimulatory interactions (CD40/CD40L, CD80/CD28, adhesion molecules, cytokines)
start to recognize signals from T cell zone and migrate there
T cells interact with DC cells that will cause them to differentiate (some into T helper ) go to border junction and interact with the B cells
present antigen to T cell: interactions where T cell helps B cell (if T cell recognizes the antigen) →red and purple binding in left image (BCR signal 1)
costimulatory molecules (2) and make cytokines (signal 3)
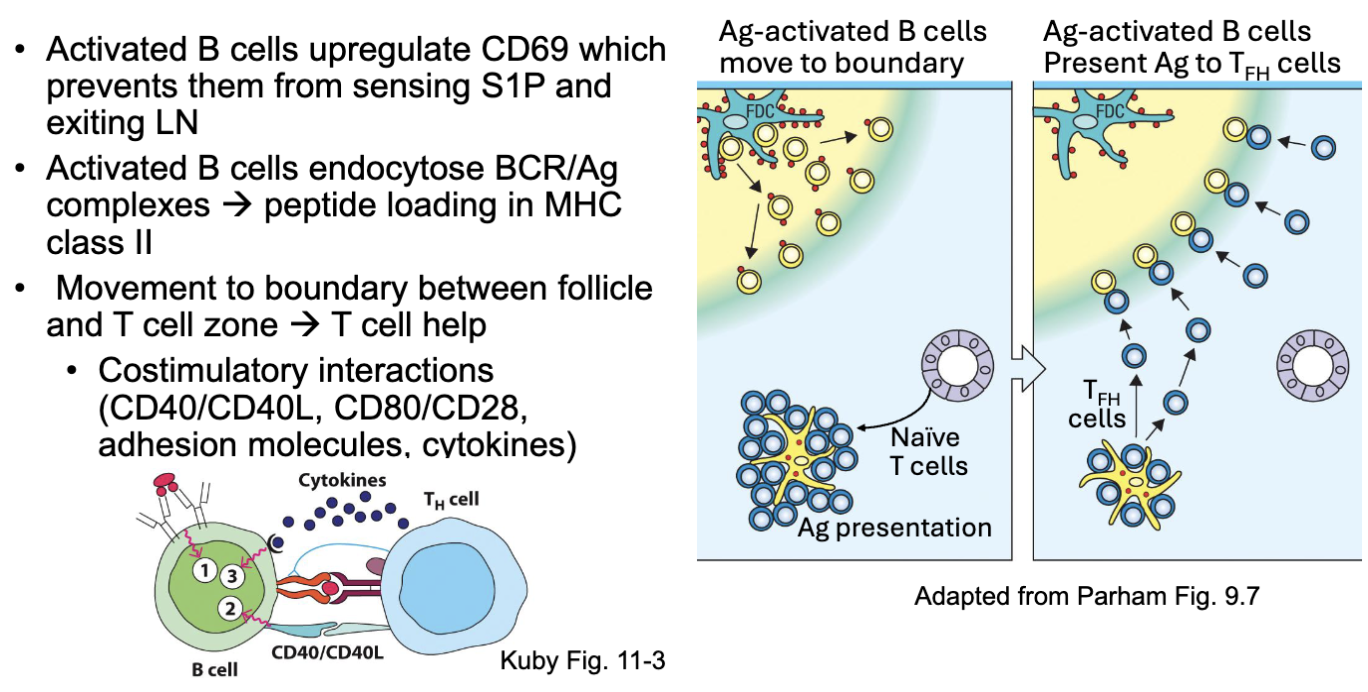
What prevents B cells from exiting the lymph nodes once activated?
when stimulated don’t want it to leave lymph node: CD69 (prevents them from leaving associates with sphingosine 1 receptor)
S1R (what lets it out via a gradient)
What are the features of the primary focus (medullary cords) and the germinal centres in relation to B cells?
B cell-TFH pairs move to medullary cords (primary focus) → proliferate (plasmablasts)
Primary focus forms in the medullary cords
Upregulation of BLIMP-1 and IRF4 causes differentiation into plasma cells →secrete low affinity IgM (early, short-lived)
Some B cell-TFH pairs upregulate Bcl6, move back to a primary follicle and form a germinal center (secondary follicle)
Expansion of Ag-activated B cells in primary follicle creates germinal center
haven’t undergo affinity maturation yet
some migrate back to the follicle (a lot of proliferation) creates germinal center
What are the features of B cell stimulation in the germinal centre?
Dark zone: Centroblasts undergo massive proliferation and upregulate AID
→ somatic hypermutationLight zone: Centrocytes proliferate less and interact with TFH →
affinity maturation – compete for antigen on FDCs → selection
class switching (regulated by cytokines and other signals)
Memory vs. plasma cell differentiation
depends on interactions with TFH cells
plasma cells develop later but at higher frequency
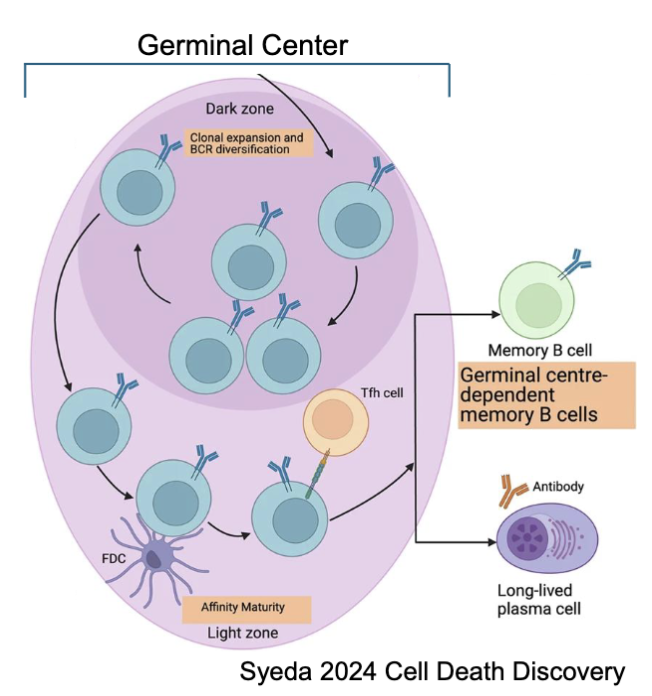
B Cell Memory vs. Plasma Cell Fate. Which model is accurate in the differentiation in the type of B cell made?
Integrative Fate model: includes both quantity of signal and quality of the signal
higher affinity cells that underwent class switch now either become memory or plasma cell
follicular helper cells mediate what type of B cell it becomes
memory cells made early and plasma accumulates later
Asymmetric model (wrong): one cell upregulate one type of transcription factor whilst the other upregulates diff. transcription factors
Instructive (partly right) :need more signalling to be plasma cell
Decreasing potential (partly right): over time loose the ability to make memory cell

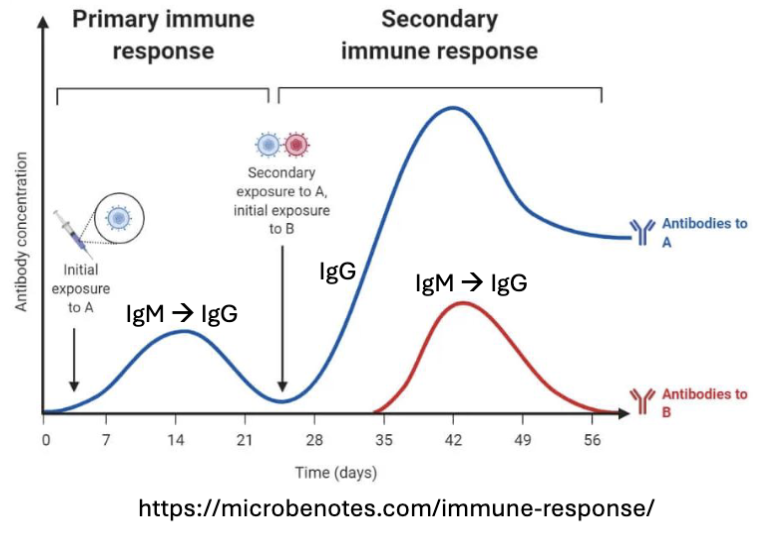
What are the features of Memory B Cell Recall Response?
Transcriptionally poised to respond → faster response
Enhanced signaling response (Ca2+ mobilization) → larger response
Higher levels of costimulatory molecules (CD80, CD40)
Less reliant on T cell help (less bottleneck)
Already class switched and high affinity Igs (IMAGE)
What are the features of B-1B Cells?
B-1 B cells: arise early in embryonic development (fetal liver) → localize to
peritoneal and pleural cavitiesLimited diversity: Use VH genes closest to D segments; TdT not expressed in fetus
self-renewal (requires IL-10) and don’t need T cell help
Low affinity natural antibodies (mostly IgM)
recognize multiple carbohydrate/ lipid antigens (polyspecific) → clearance of certain pathogens, dying cells (self), and debris
Not the normal B cell referenced that is B-2 B cells (usually B cell made in later development)
limited diversity: use genes close together
polyspecific: recognize more than one antigen
antibodies can bind and mediate complement and phagocytosis of the bacteria
antibodies important for homeostasis recognize cells undergoing apoptosis, antibodies mark the cells undergoing apoptosis so that they can be phagocytosed

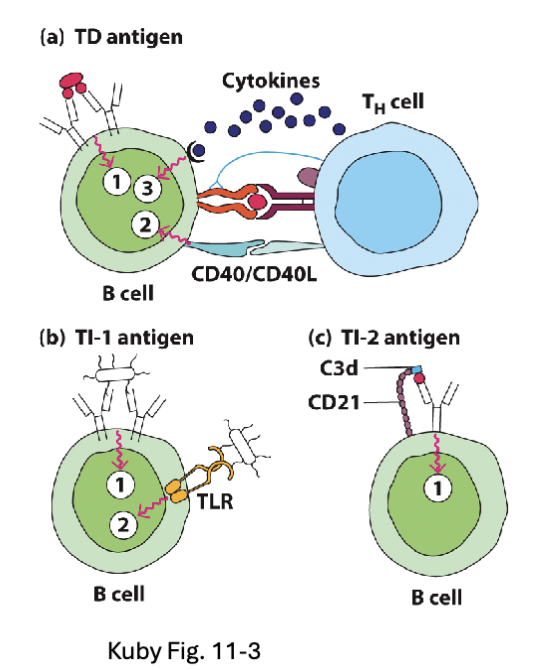
What is the difference between T-dependent vs. T-independent Antigens?
T-dependent Antigens (TD):
Soluble protein antigens that cross-link BCR
Require T cell help to provide signal 2 and 3
T-independent Antigens (TI):
TI-1: Bacterial cell wall/membrane components (e.g. LPS)
TI-2: Polymeric proteins (e.g. flagellin) and capsular polysaccharides
TD: need T cell help
exam don’t need to know diff between TI-2 and TI-1 just different between T dependent and T independent
What is the history of the discovery of the T cell receptors?
1961-2 Jacques Miller – T cells are generated in thymus and
are important for adaptive immunity (thymectomy experiments)Hypothesis that T cells used clonal recognition structure similar
to B cells1981-3: Alan Harris and Jim Golding, Ellis Reinherz, Jim Allison,
– work characterizing structure of TCR1984-5: Mark Davis, Tak Mak – isolated and mapped -chain
TCR genes in mice and humans
discovered later than B cells
What are teh features shared by TCR and BCR?
Polypeptides containing linked chains:
TCR resembles a membrane bound Fab made up of TCR⍺ and TCRβ (or TCRɣ and TCRδ)
Variable regions – recognize antigens
Constant regions – structure and function
Both undergo RAG-dependent somatic gene recombination to generate variable region
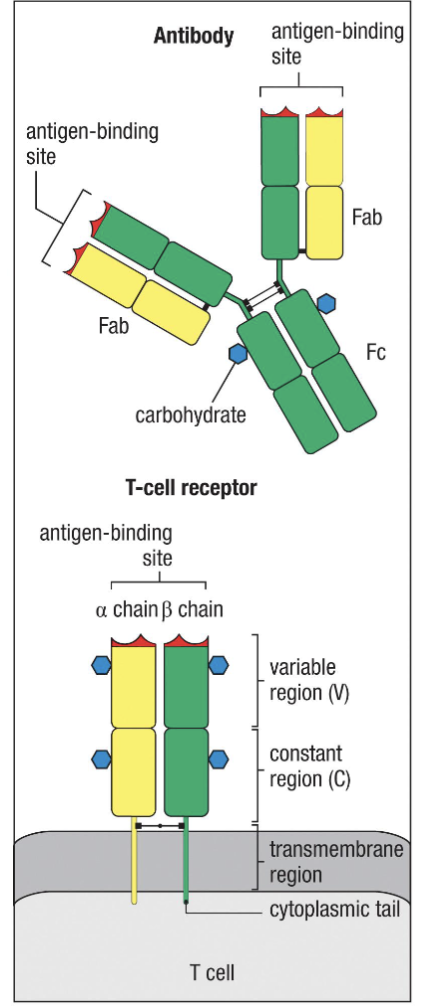
What are the differences of T receptors when compared to BCR?
Only contains one antigen binding site
Does not generate soluble form
Does not undergo somatic hypermutation
Does not undergo class switching

What are the two classes of TCR?
⍺βTCR
ɣδTCR
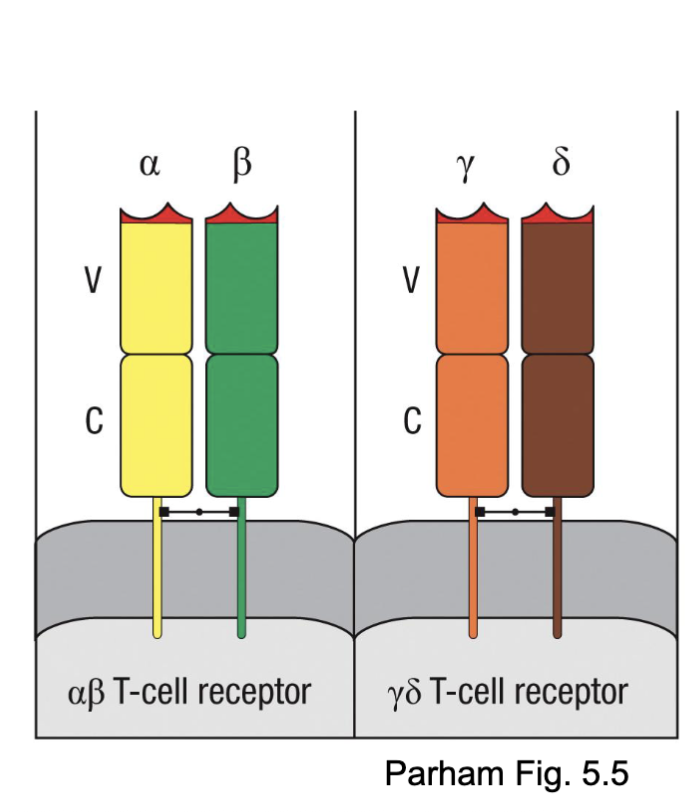
What are the features of ⍺βTCR?
Most diverse antigen recognition repertoire
Typically recognize MHC with short peptides
Most common in the human and mouse T cell repertoire
alpha/beta most common (very diverse repertoire, more segments for rearrangement)
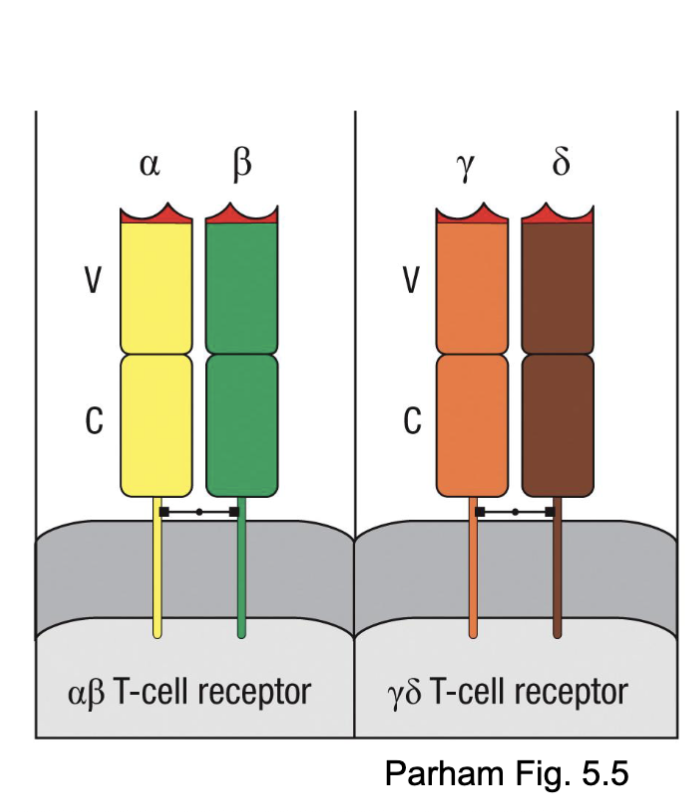
What are the features of ɣδTCR?
Less diverse (fewer gene segments)
Recognize a wider variety of antigens (lipids, phosphoproteins), with less requirement for classical MHC
Most common in the bovine T cell repertoire
gamma/delta recognize wide diversity mostly lipids and carbs (less need MHC)

What are the features of TCR⍺β rearrangement?
Account for ~90% of T cells (85-95)
TCRs are generated from gene loci encoding TCR⍺ chain (Ch 14) paired with TCRβ chain (Ch 7)
Recombination of Variable (V), Diversity (D), and Joining (J) segments T cell receptors →variable regions with different specificity.
Regulated by RAG-1/2 complex acting on flanking RSS in segments
TCRβ chain recombines D-J segments before recombination with V segments.
P- and N- nucleotides added (TdT)
Ch= chromosome
same RAG complex with RSS
Rag cuts out sequence Artemis cuts open hairpin,
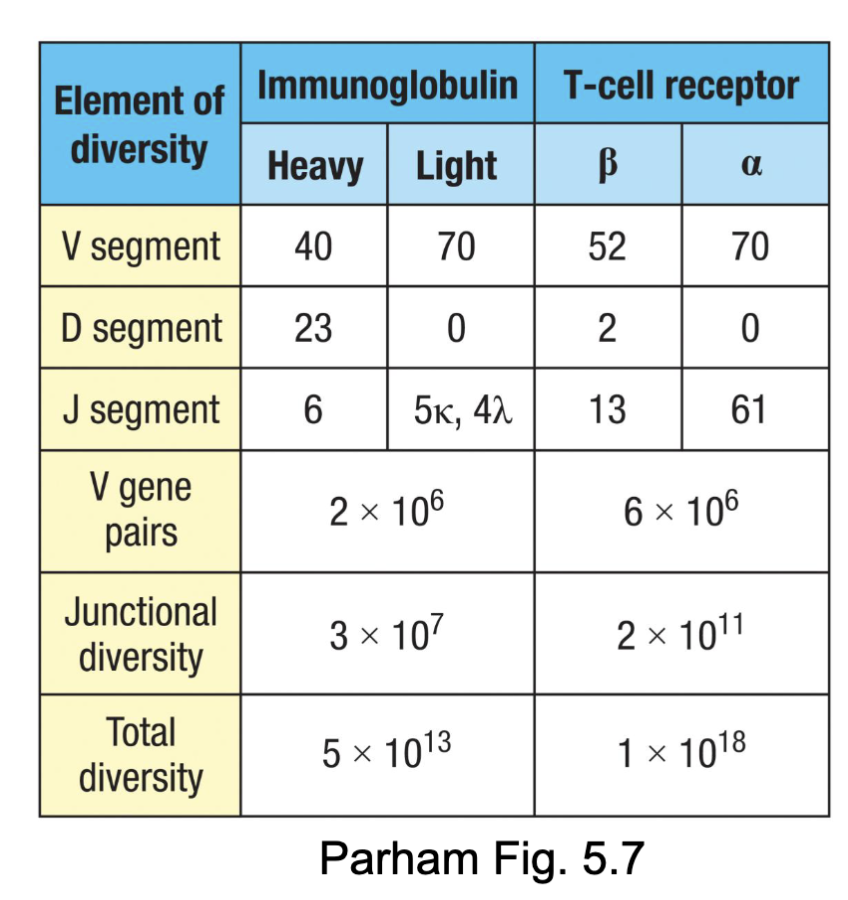
What are the features of TCR⍺β Diversity?
Compared to immunoglobulins:
Greater number of gene segments to recombine (combinatorial diversity)
Greater junctional diversity potential (N- and P-nucleotides)
TCR: both chains have N- and P-nucleotides added
BCR: light chains have few N- and P- nucleotides (TdT
downregulated at time of rearrangement)
Lots of diversity without taking up a lot of space in the genome
What are the features of TCRɣδ rearrangement?
Account for ~10% of T cells
TCRs are generated from gene loci encoding TCRɣ chain (Ch 7) paired with TCRδ chain (Ch 14)
TCRδ chain sequence are contained in the TCR⍺ locus → rearrangement of TCR⍺ locus deletes the TCRδ genes.
TCRδ chain can recombine incorporating 2 D segments → increased diversity
P- and N- nucleotides adde
delta -equivalent to the heavy chain in BCR

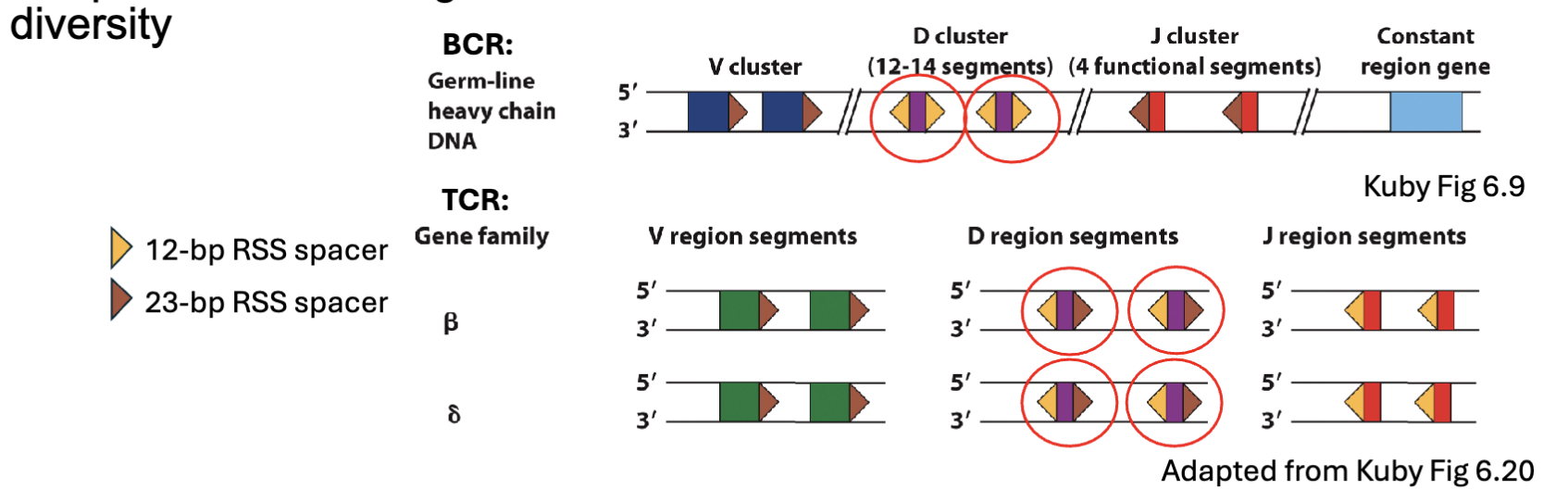
What are the features of Recombination Signal Sequences (RSS)?
Recall: Heavy Ig chain had same RSS spacer sequence flanking D gene
segments → 12/23 rule ensures recombination with V and J segmentsTCRβ and δ have different RSS spacer sequences flanking D gene segments→potential to recombine without D segment or more than one
Not actually seen at TCRβ locus
Frequent D-D rearrangements found at TCRδ locus → increased
diversity
same = cannot recombine
beta and delta chain have the diff. bp so have potential to recombine
What are the features of T Cell receptor expression?
Allelic exclusion prevents rearrangement of more than one successful TCRβ or ɣ chain in mature T cells, TCR⍺ expression regulated after selection.
TCR chains expressed as transmembrane disulfide-linked polypeptides (no soluble form)
TCR chains associate with CD3 complex proteins: CD3ɣδε (linked genes on Ch 11), CD3ζ (Ch 1)- dimers of ɣε, δε and ζζ
Required for TCR transport out of ER
Contain transmembrane signalling domains: immunoreceptor tyrosine-based activation motifs (ITAMs)
only one TCR per cell
need to reach the surface to do that needs to associate with diff. protein: CD3 chains (gamma-epsilon or delta-epsilon chain and zeta chains

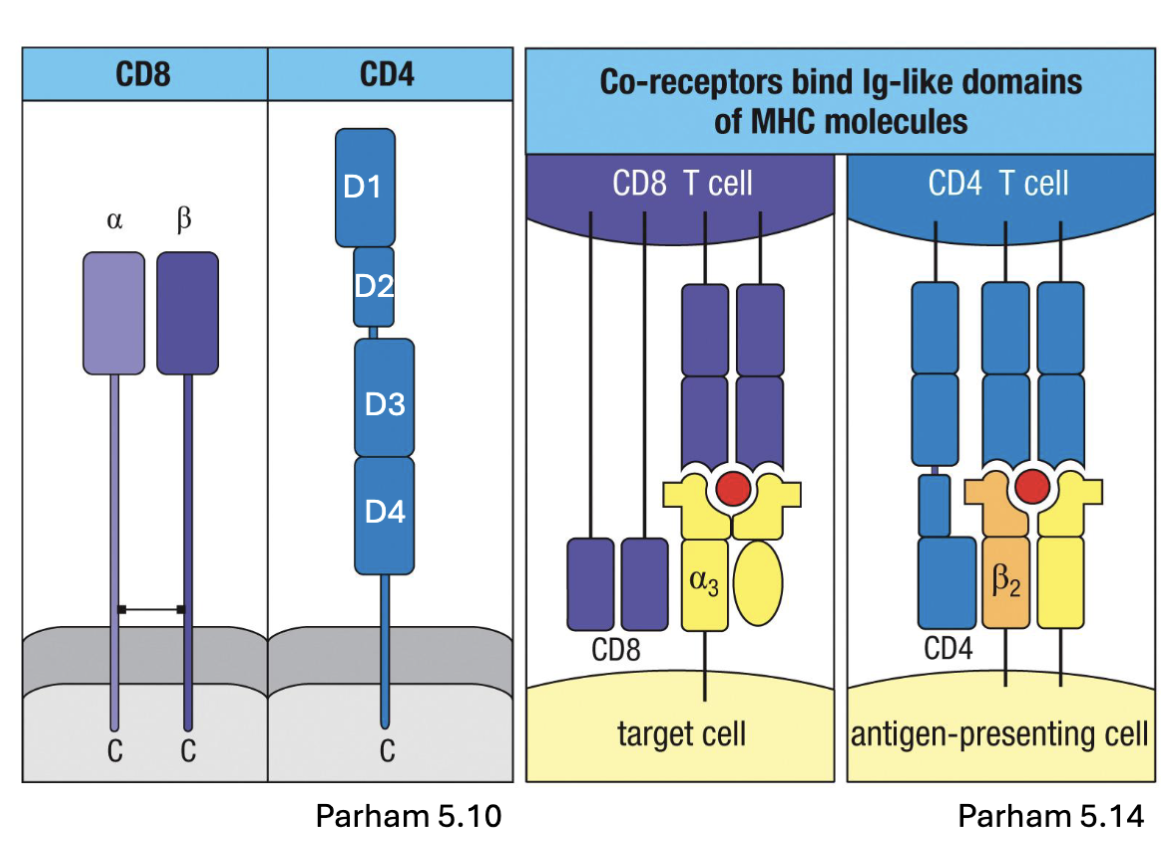
What are the features of CD4 and CD8 of the TCR?
TCR complex also includes CD4 or CD8 glycoproteins:
CD4: one chain, 4 domains, hinge in the middle
CD8: linked ⍺ and β chains
co-receptors that aid in binding MHC II or MHC I, respectively
What are the differences between MHC class I and MHC class II?
MHC I: widely expressed (except erythrocytes)
MHC II: primarily APCs (DC, B cell, macrophage)
CD8 internal so virus infected
CD4 external
What is the first signal of TCRs?
MHC binds to TCR
Signal 1: TCR:MHC clustering with CD4/CD8 stabilization→ recruitment and activation of Lck
Lck phosphorylates ITAMs in CD3 complex cytoplasmic tails
ZAP70 is recruited and activated (phosphorylated) → downstream signaling via PLCɣ → transcription factors (AP-1, NF𝛋B, NFAT-1) →survival, proliferation
CD4 enters and binds has LcK (tyrosine kinase) associated with it and it will phosphorylate ITAMs (can now act as docking site)
LcK can phosphorylate ZAP70
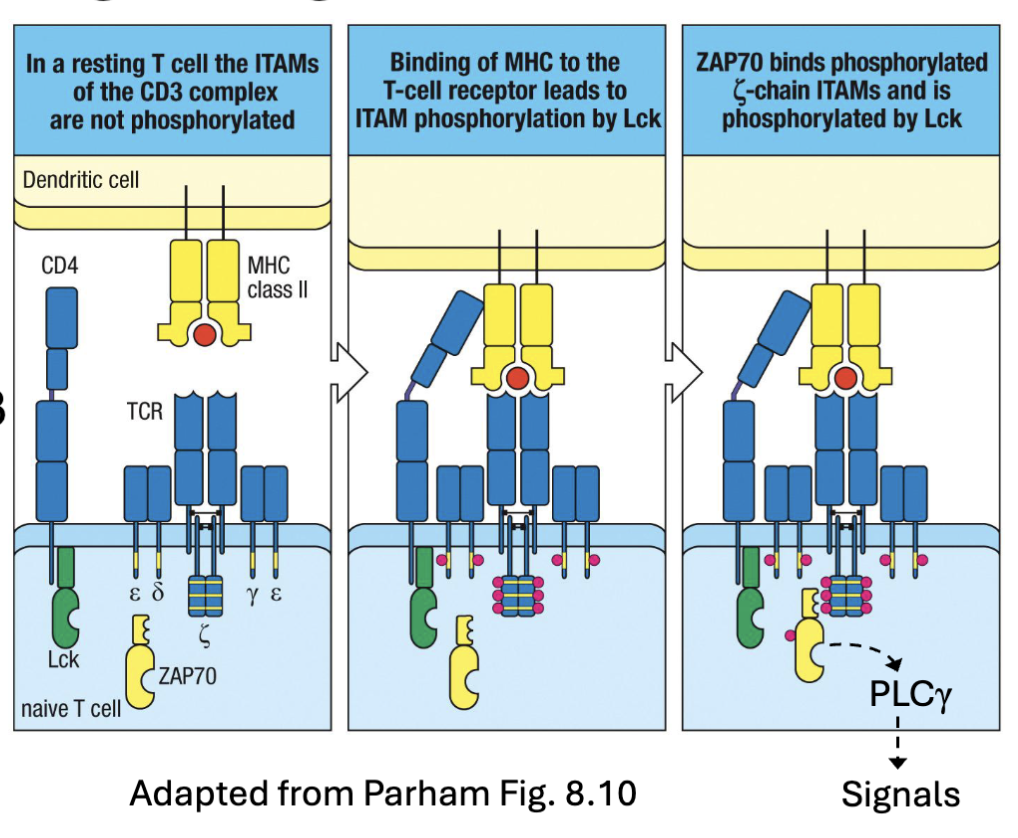
What are the second and third signals of the TCR?
TCR:MHC signaling is insufficient for full activation → anergy
Costimulation (signal 2): B7 molecules (CD80, CD86) upregulated on activated APCs→ bind CD28 on T cells→ enhance signal 20-30 fold
Src kinases (Lck, Fyn) phosphorylate cytoplasmic tail→ PI3K recruitment
AP-1/NFkB/NFAT-1 signaling → upregulation of IL-2 and CD25 (IL-
2Rα chain)Cytokine (signal 3): IL-2 is a critical survival and proliferation factor
signal 2 and 3 to reinforce activation
signal 2: costimulation CD80/86 of activated APCs
IL-2Ralpha (high affinity receptor- better than IL-2Rbeta)
signal 3: IL-2 signal
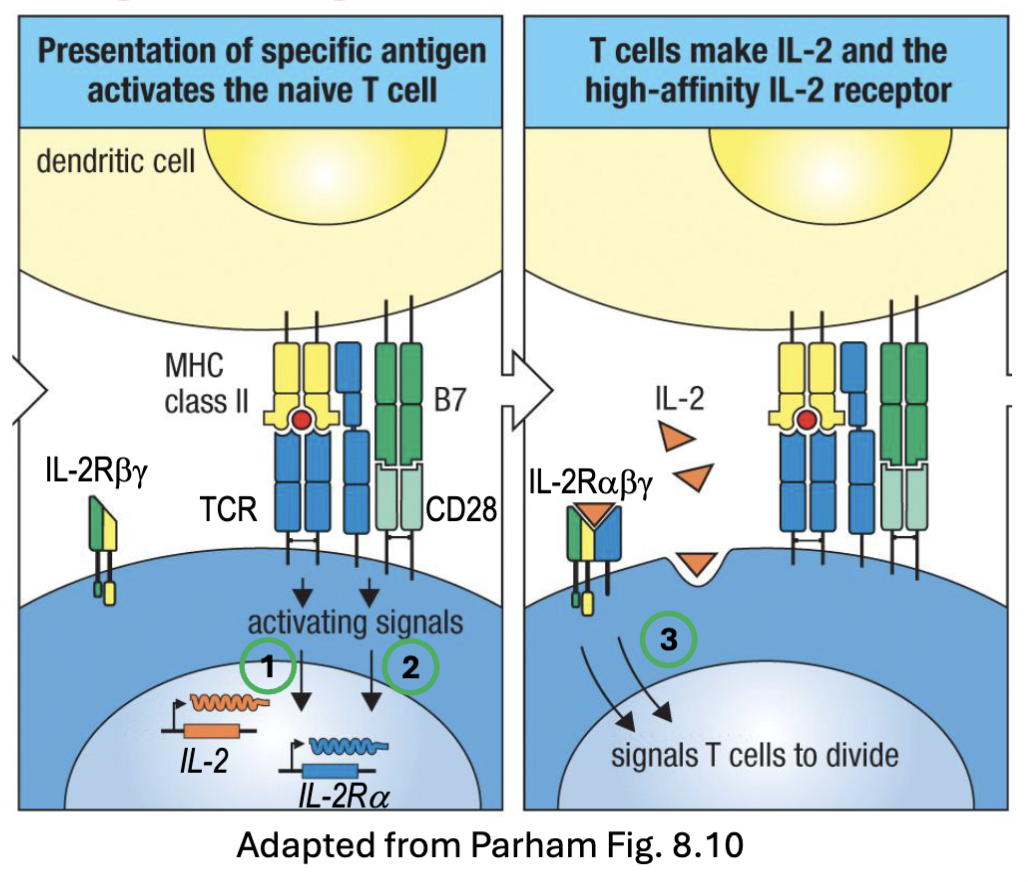
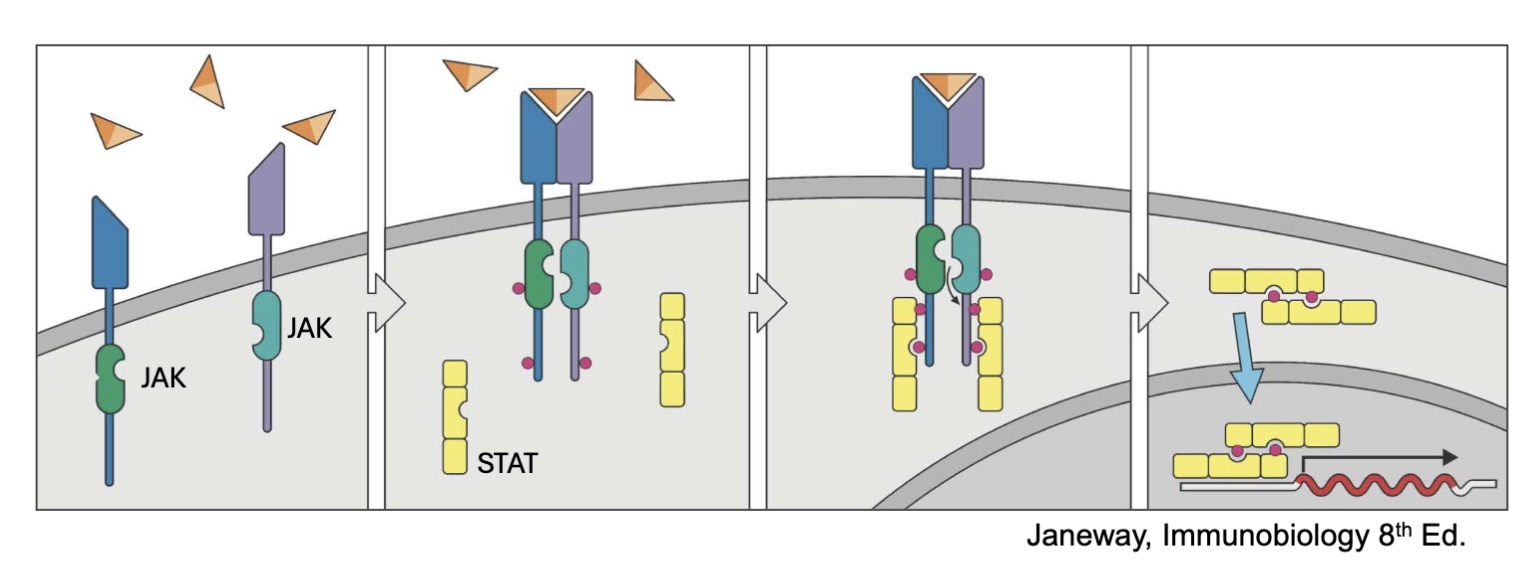
What is JAK/STAT pathway? (recall)
Signaling mechanism for many cytokines (IL-2, IL-4, IL-5, Il-7, IL-12, Il-23, IFNɣ, etc.
IL-2R associates with JAK1 and JAK3 → STAT 5 (mostly), STAT1, STAT3
when the JAKs are together autophosphorylate and the receptor C’ terminus⇒ allows docking sites (STATs)
STATs become phosphorylated by JAK can then leave and dimerize: go to nucleus cause transcription
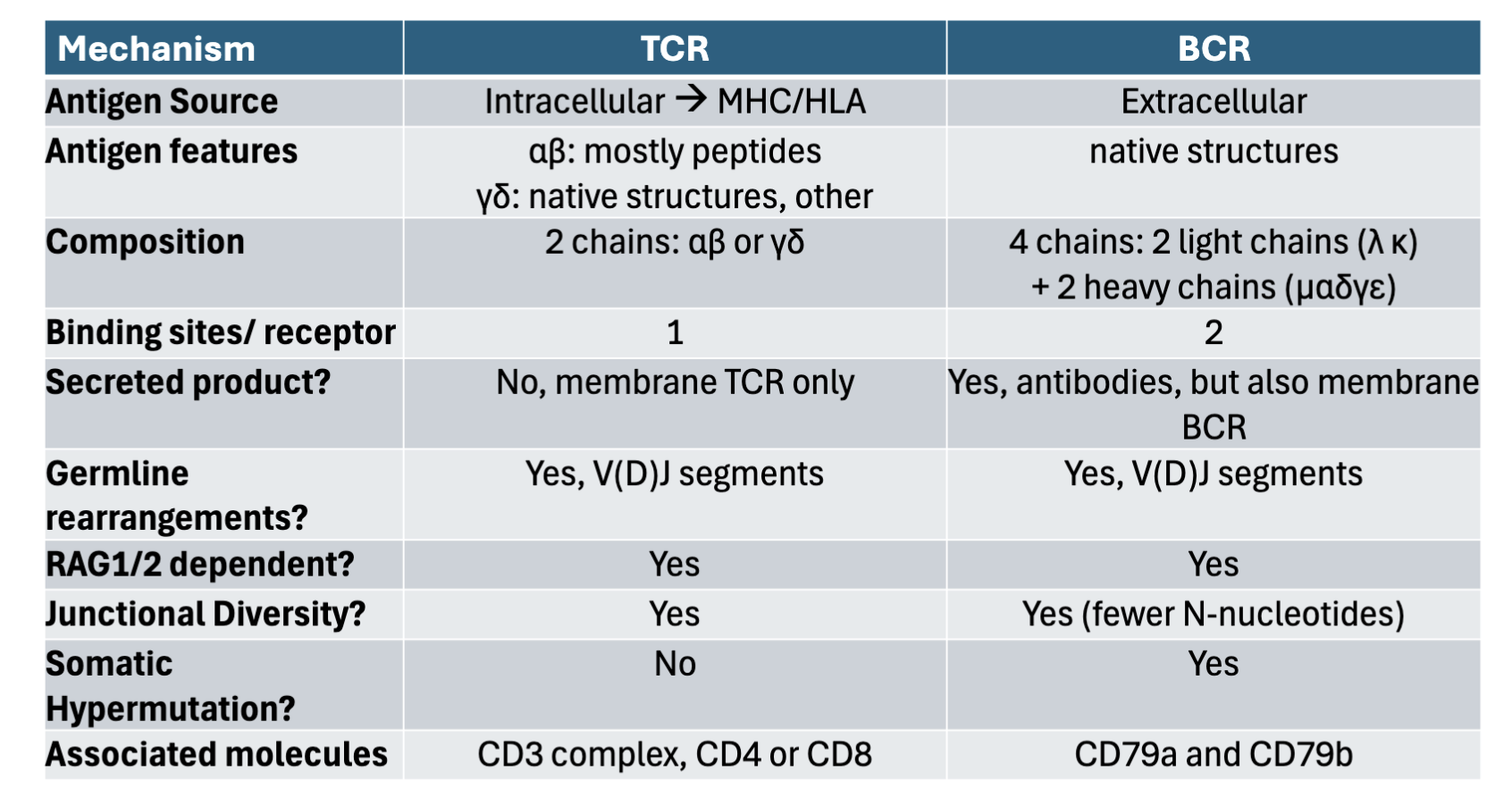
What are the differences between BCR and TCR in regards to: antigen source, antigen features, composition, binding sites/ receptors, secreted products, germline rearrangement, RAG1/2 dependent, junctional diversity, somatic hypermutation, associated molecules?
TCR BCR
antigen source: Intracellular→MHC/HLA Extracellular
antigen features: ⍺β: mostly peptides Native structures
ɣδ: native structures other
composition: 2 chains ⍺β + ɣδ 4 chains: 2 light chains (𝛋, 𝛌)
+2 heavy chains (µ,⍺, ε, ɣ,δ)
binding sites/ receptors: 1 2
secreted products: No membrane TCR only Yes, antibodies but also membrane BCR
germline rearrangement: Yes V(D) J segments Yes V(D) J segments
RAG1/2 dependent: Yes Yes
junctional diversity: Yes Yes (fewer N-nucleotides)
somatic hypermutation: No Yes
associated molecules: CD3 complex, CD4 or CD8 CD79a and CD79b
BCR: native 3D structure being recognized
What are the stages of T cell development?
Originate from CLP (common lymphoid progenitor)- from bone marrow
Differentiate to T cell progenitor: upregulate Flt3, L-selectin, CCR7, CCR9
In thymus: become early thymic precursors (ETPs) a.k.a. double-negative (DN) thymocytes bc CD4- and CD8-
T cell development from this stage onwards is in thymus (primary lymphoid tissue)
T cell pool remains stable until old age (long lifespan w/ self-renewal) → but loses function over one’s lifetime (immunosenescence)
Stomal cells provide growth/ survival signals (IL-7, Ftl3L, Notch-1) & coordinate selection of developing TCRs for self-restriction (recognizes MHC) + self-tolerance
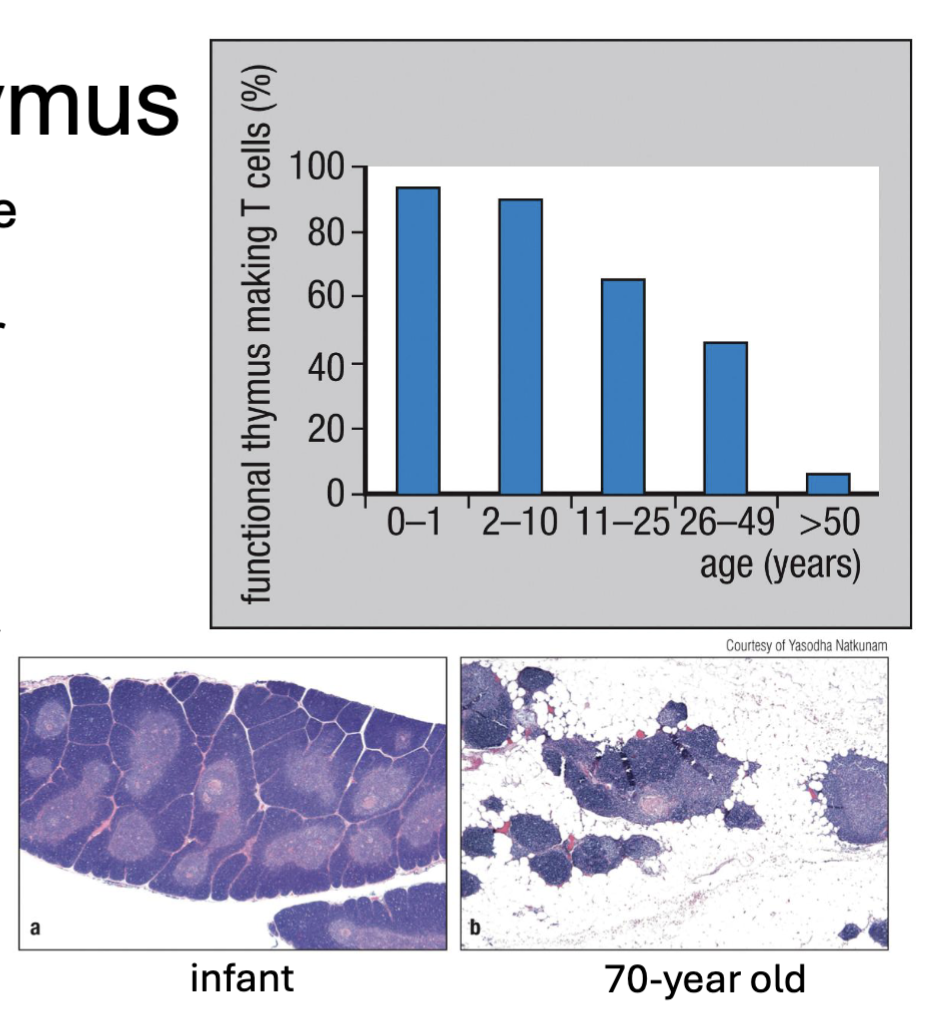
What is immunosenescence?
(thymus shrinks with age = can’t make new T cells so:)
T cell pool remains stable until old age (long lifespan w/ self-renewal) → but loses function over one’s lifetime
cells tired work less well
What are the features of T cell development in the thymus?
Thymus is a primary lymphoid tissue (site of development) lying over heart
fully developed at birth and shrinks over time
T cell pool remains stable until old age (long lifespan with self-renewal) → but loses function (immunosenescence)
DiGeorge syndrome → lack thymus → T cell deficiency
stromal cells provide growth/survival signals (IL-7, Flt3L, Notch-1)
interactions select for self-restriction
(recognize MHC) and self-tolerance
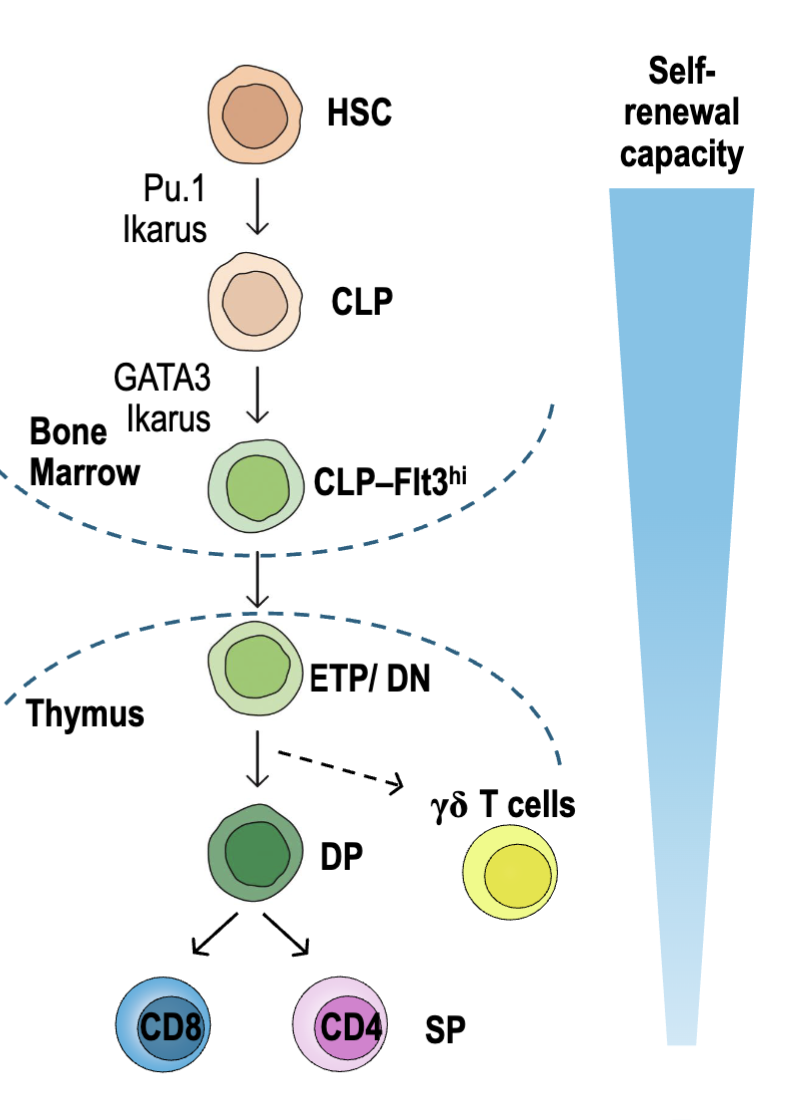
What are the features of T cell development (makes it a T cell)?
T cells are derived from a subset common lymphoid progenitors (CLP)
High levels of Flt3 (cytokine receptor)
Upregulate L-selectin, CCR7 and CCR9 (adhesion molecules)
Seed thymus and develop into early thymic precursors (ETPs/ DN): CD4-CD8-
Expansion, T cell commitment, TCRβ𝛄δ rearrangements
Double positive (DP): CD4+CD8+
TCR⍺ rearrangements expressed
Positive and negative selection
Single positive (SP): naïve CD4 or CD8 T cells → exit thymus and recirculate through lymphoid tissues
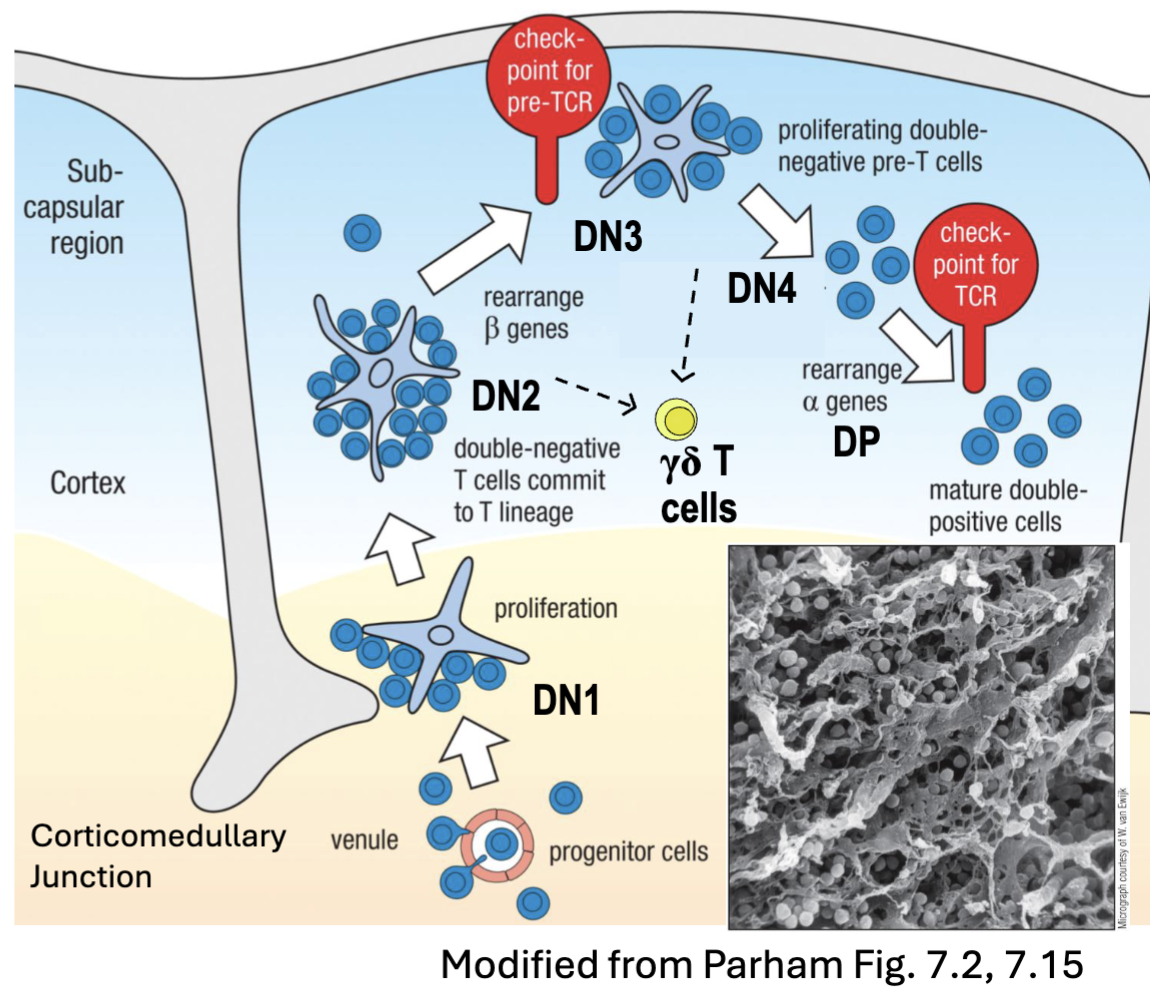
What are the features of T Cell Development: Double Negative (CD4-CD8-) Thymocytes?
DN1 (c-kit++ CD44+ CD25-): move from corticomedullary junction towards outer cortex (C44+ surface= interactions with TEC)
interactions with stromal thymic epithelial cells (TEC)
IL-7 → survival, proliferation
Notch signaling → T cell commitment
DN2 (c-kit++CD44+CD25+): simultaneous rearrangements of TCRβ𝛄δ
DN3 (c-kit+CD44-CD25+): preTCR expressed → β-selection
DN4 (Ckit-CD44-CD25-): proliferation; allelic exclusion of β-chain; 𝛾-chain silencing
DP (CD4+CD8+): ⍺-chain rearrangement and TCR𝜶β expression → positive selection
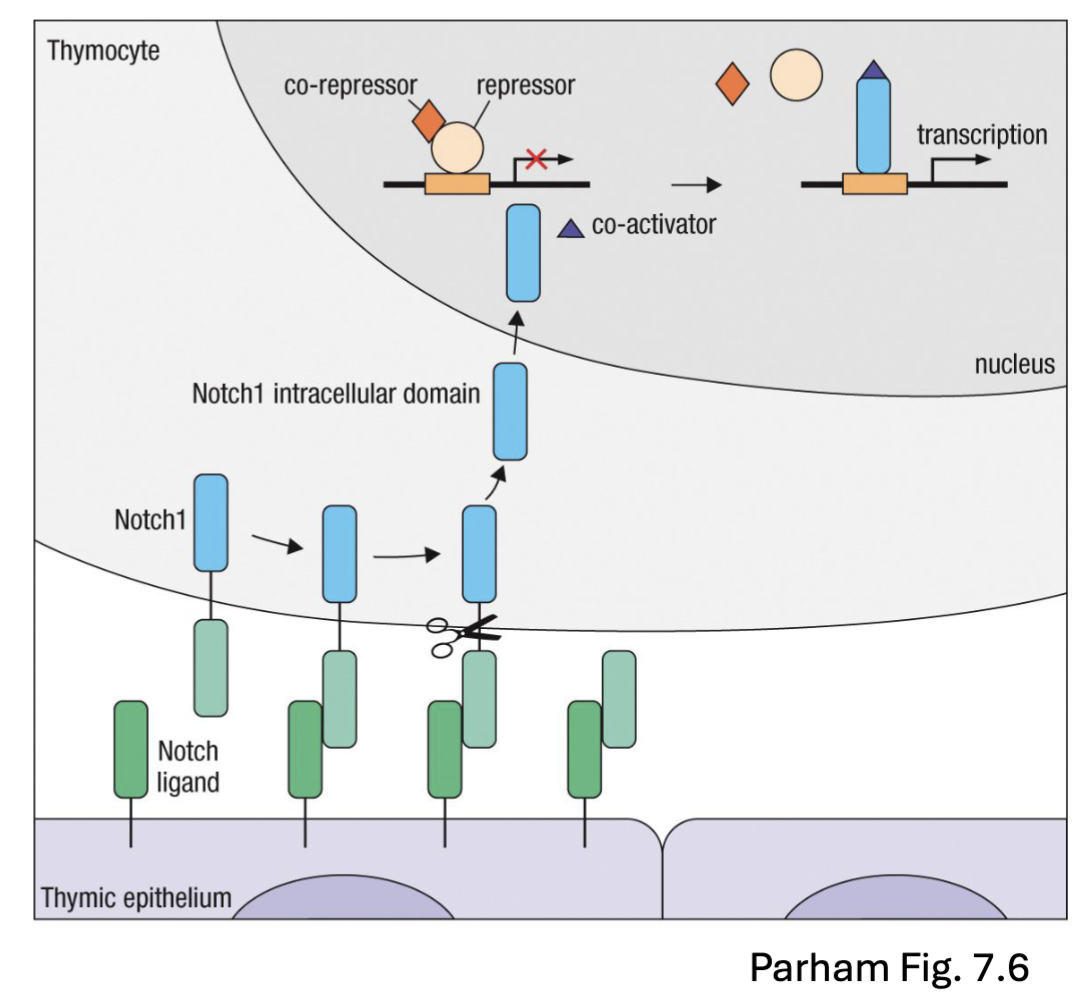
What are the features of the Notch transcription factor in relation to DN thymocyte T cell development?
Notch: transcription factor
transcription factor leaves the membrane and then enter the nucleus and affect transcription
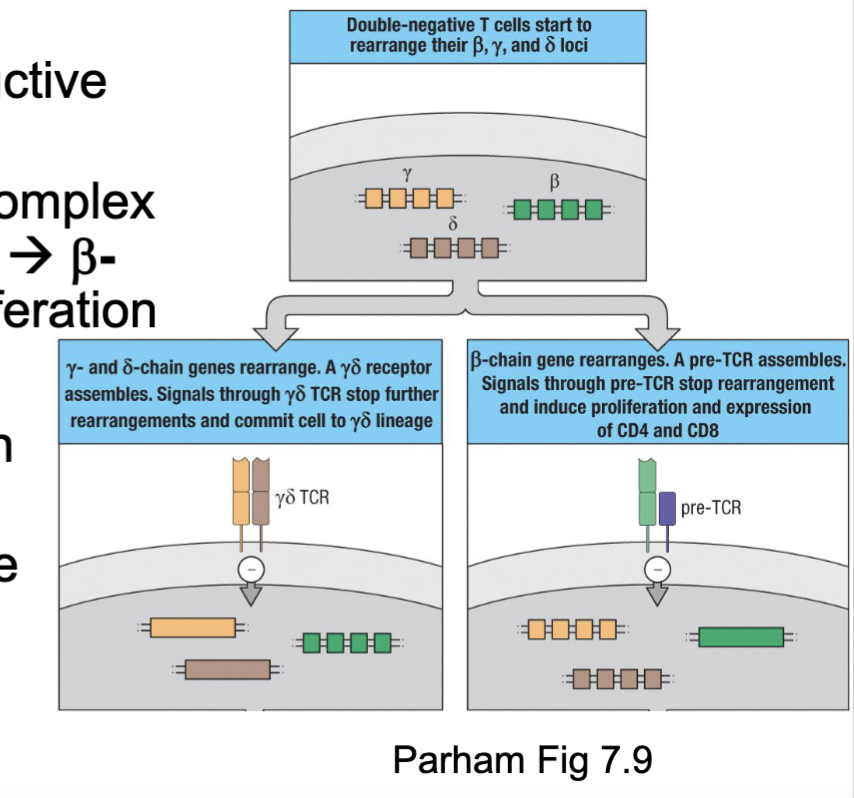
⍺β or 𝛾δ is it a race?
TCRβ, 𝛾, or δ rearrangements take place in thymocytes simultaneously
𝛾δ T cells need to successfully rearrange both 𝛾 and δ chains→ TCR signalling → commitment
Cells fated to be ⍺β T cells only need a productive TCRβ rearrangement
β chains associates with preT⍺ and CD3 complex → pre-T cell receptor (preTCR) signalling → β-chain selection → commitment & proliferation
⍺β T cells favoured due to:
Lots of V⍺ and Vβ segments and β-chain with two D-J-C regions → multiple attempts
β-selection checkpoint → proliferation of large
pool for trying out ⍺-chain rearrangements⍺-chain rearrangements deletes the δ-chain gene segment
(gamma delta need to rearrange both at the same time for the lineage commitment
alpha beta only need to upregulate beta chain)
When are 𝛾δ T cells most regulated?
𝛾δ T cells seem to be regulated during development
Come out of the thymus in early waves before birth
Likely regulated by transcription factors and gene segment accessibility within chromatin
𝛾δ T cells at this point appear to be mature and exit the thymus without further education
Most recognize atypical antigens (e.g., lipids and phosphoantigens)
independent of classical MHC

What are the features of T Cell Development: Double Positive (CD4+CD8+) Thymocytes?
DP thymocytes (CD4+CD8+): represent >80% of cells in thymus
have undergone ⍺-chain rearrangement
upregulate CD4 and CD8
start to express mature ⍺βTCR
Migrate from subscapular region deeper into cortex
Interactions with cortical thymic epithelial cells (cTECs), macrophage →
positive selection (MHC restriction) → survival (~ 2% are successful)CD4 and CD8 ensure DP cells can engage with either MHC II or MHC I →
differential signaling will commit to CD4 or CD8 SP thymocytes
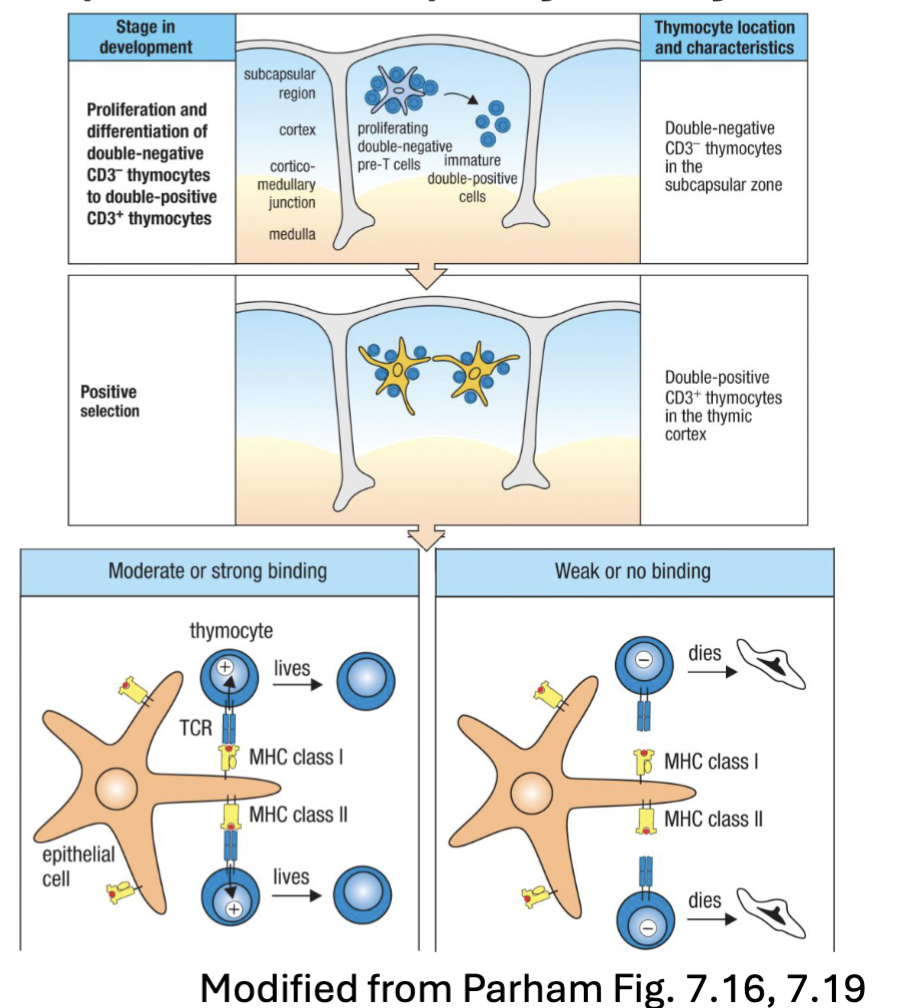
What is positive selection in DP thymocytes?
positive selection, if the cell can recognize MHC = positive selection
if not recognized the cell don’t get a signal and undergo apoptosis
(Positive selection is an immunological process that ensures lymphocytes, such as T cells, have receptors capable of recognizing self-MHC molecules)
What are the features of T Cell Development: Negative Selection?
Negative selection eliminates autoreactive thymocytes binding to MHC/peptides
Results in central tolerance via apoptosis of autoreactive thymocytes → clearance by macrophage
Can take place in either the thymic cortex (DP cells) or medulla (SP cells)
Medullary thymic epithelial cells (mTEC) express AIRE (autoimmune regulator) and FEZ transcription factors
Bind histones with closed chromatin →recruit transcription factors and RNA polymerase →expression and MHC processing of proteins found outside thymus (i.e. insulin, MBP)
(presented peptides from self loaded by MHC)
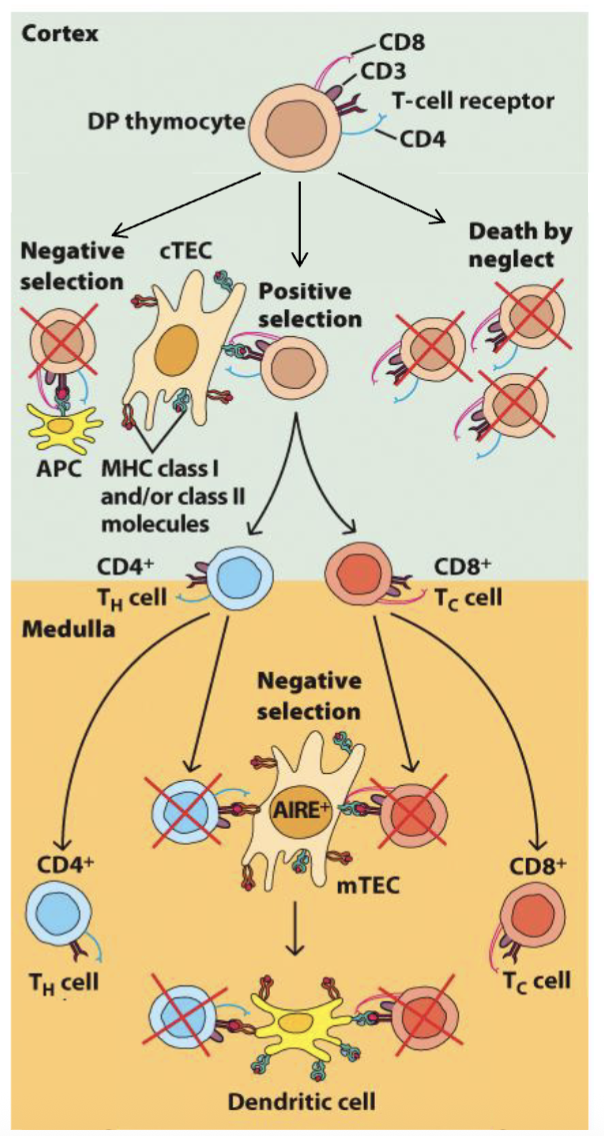
What is the selection paradox that arises during T cell development?
Positive selection: required to recognize self-MHC
Negative selection: eliminates autoreactive thymocytes
Q: Why aren’t positively selected thymocytes eliminated by negative selection?
Affinity model of selection: fate determined by binding/signal strength
No/low signal → death but neglect
Low/int signal → positive selection
High signal → negative selection
Altered peptide model: cTEC process different peptides for positive selection
Unique catalytic subunit in thymic proteosome → altered low affinity
peptides
mix of both models, looking for intermediate zone (positively selected and not deleted)
Altered Peptide model: proposes thymic epithelial cells process peptides differently
thymic protoesome has different catylic region
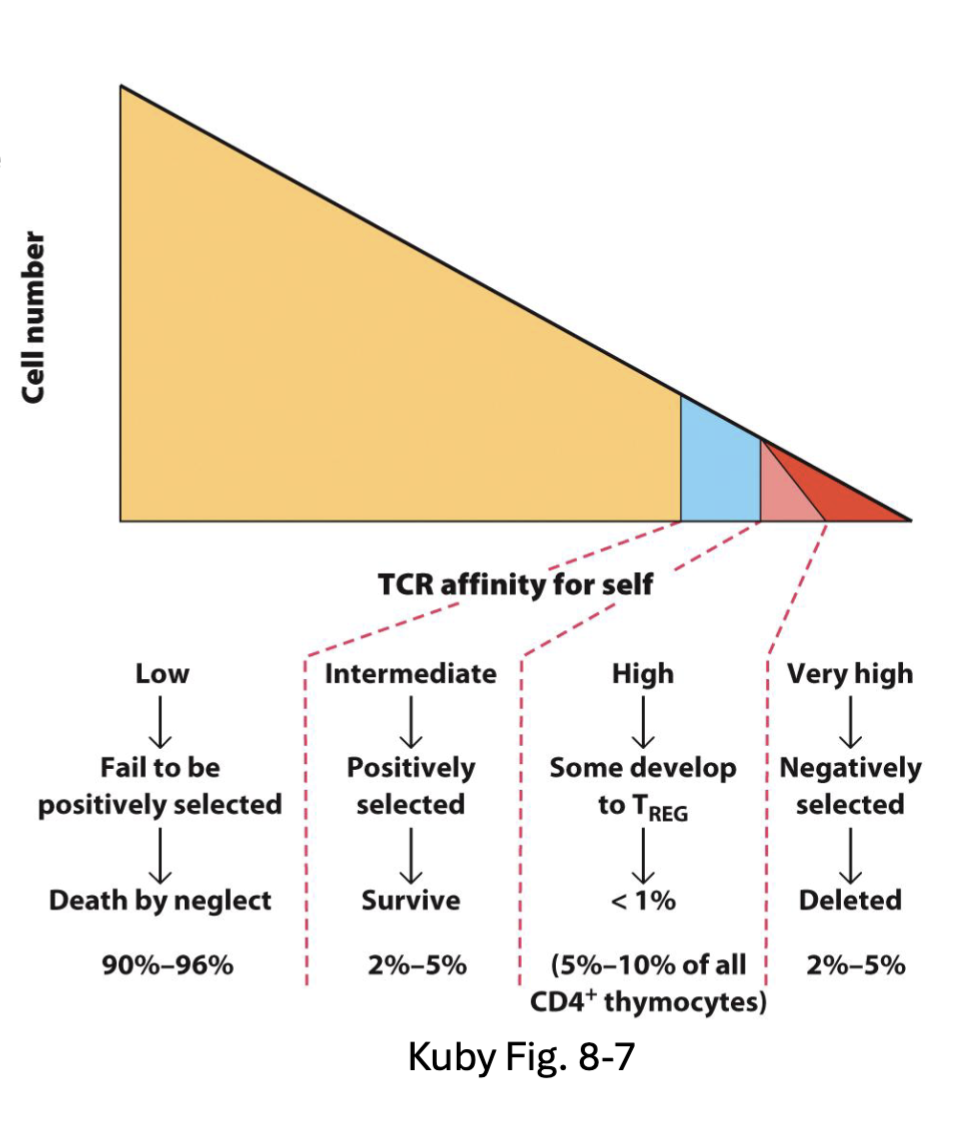
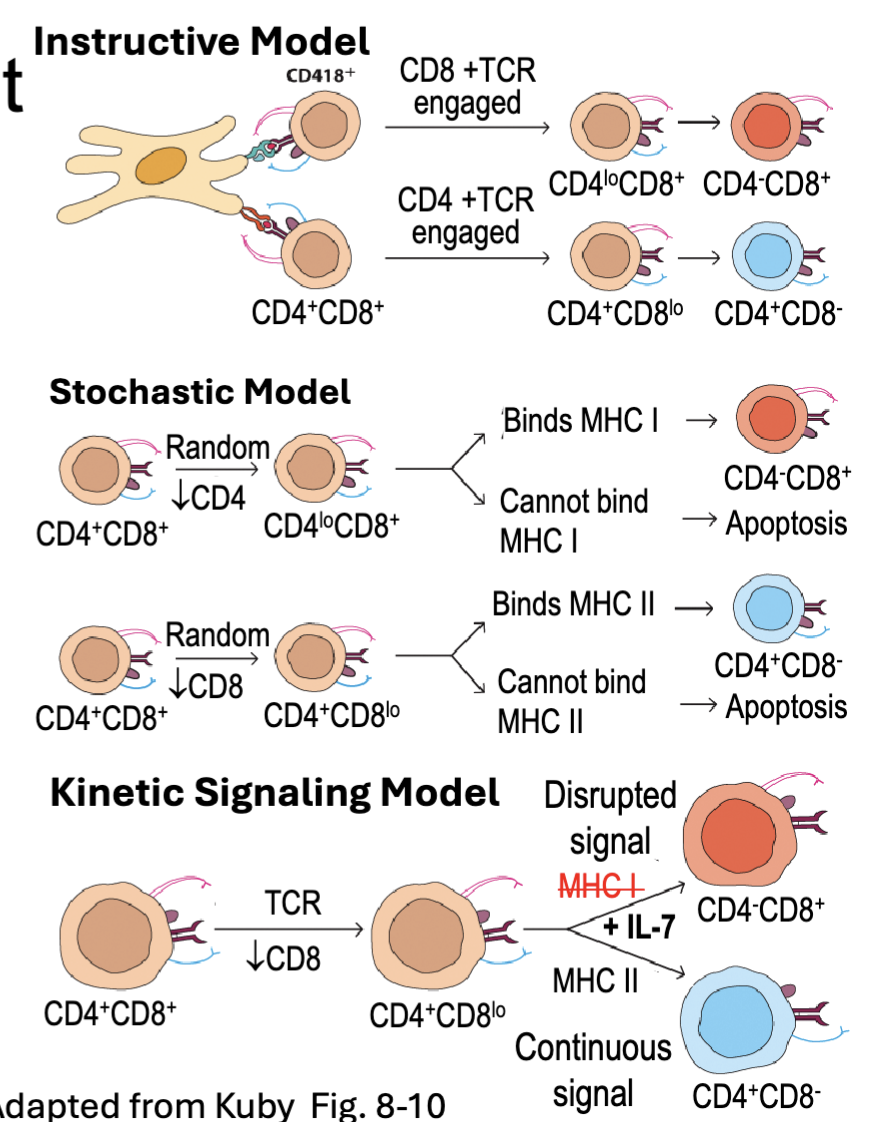
What are the features of CD4 and CD8 T Cell Commitment?
CD4 and CD8 ensure DP cells can engage with either MHC II or MHC I
Differential signaling commits DP cells to CD4 (Th-POK) or CD8 (RUNX3) program
Instructive Model – MHC restriction signals directly for cell fate
CD4/MHCII engagement → CD4 program
CD8/MHCI → engagement CD8 program
Stochastic Model – positively selected cells randomly select CD4 or CD8 program → survival only if TCR signal maintained
Kinetic Signaling Model – positively selected cells transiently downregulate CD8 → commit to CD4 if they receive continuous signal, commit to CD8 if signal interrupted (IL-7 maintains survival) (continuous signal commits to Th-POK)
after positive selection down regulate CD8
Kinetic is most consistent with experimental evidence
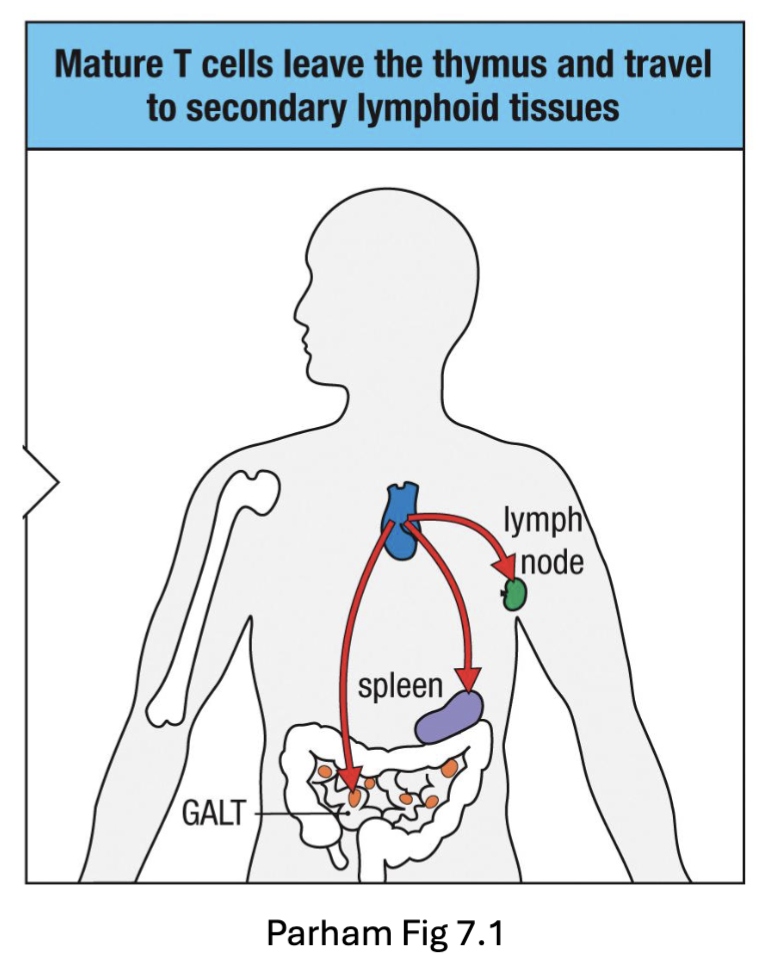
What are the features of Thymic Exit of T cells?
CD4 and CD8 SP T cells migrate back to corticomedullary junction (CCR7)
Exit thymus into blood (S1P)
Circulate and home to secondary lymphoid tissues (lymph nodes, spleen, Peyer’s patches)
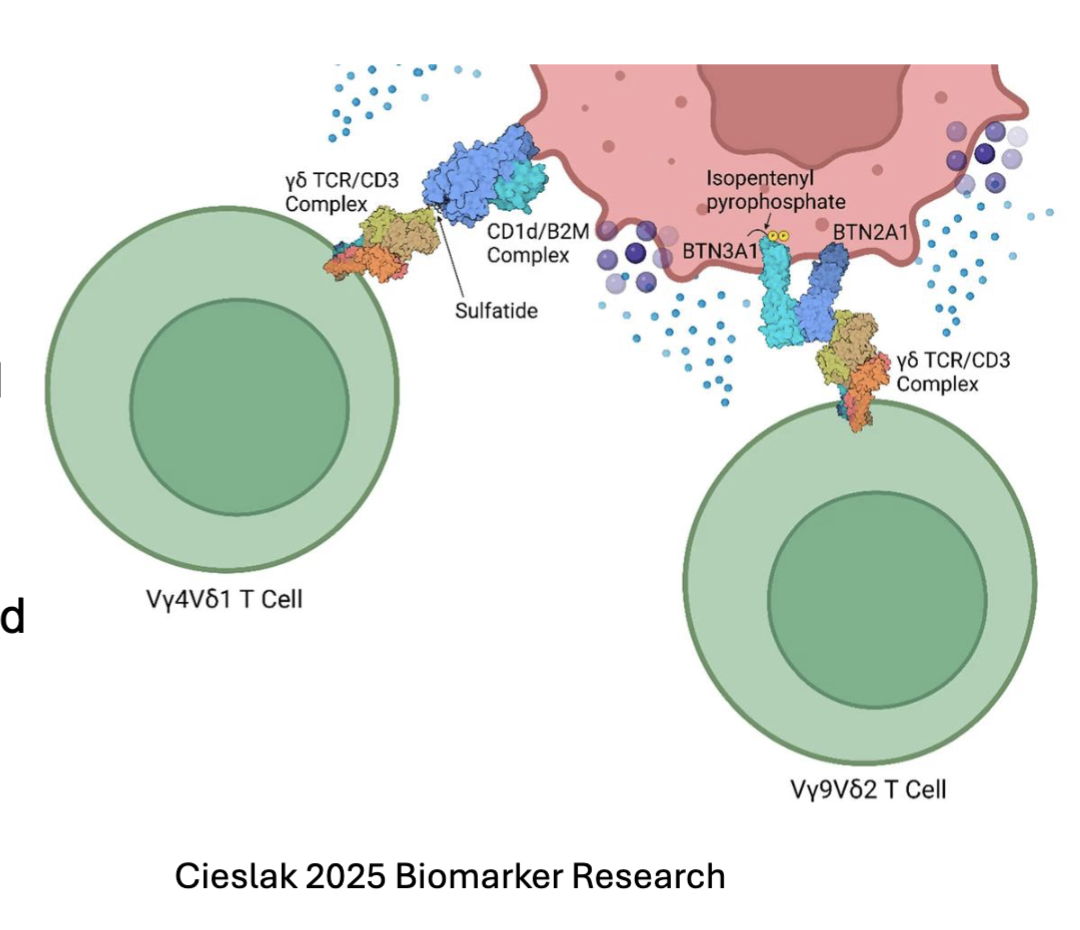
What are the features of atypical T cell Populations 𝛾δ T cells?
𝛾δ T cells exit thymus at DN2-3 stage
Limited TCR usage
Don’t require MHC for selection during thymic development
NOTCH signaling required for development
Accumulate in mucosal and epithelial tissues
recognize lipids and phosphoantigens upregulated by stress or infection:
TCR V𝛾4Vδ1 : recognizes lipids loaded on MHC-like molecules CD1c and CD1d
TCR V𝛾9Vδ2: binds BTN21/BTN3A1 complex after that forms after sensing phosphoantigens
phosphoantigen binds to BTN3A1= conformational change
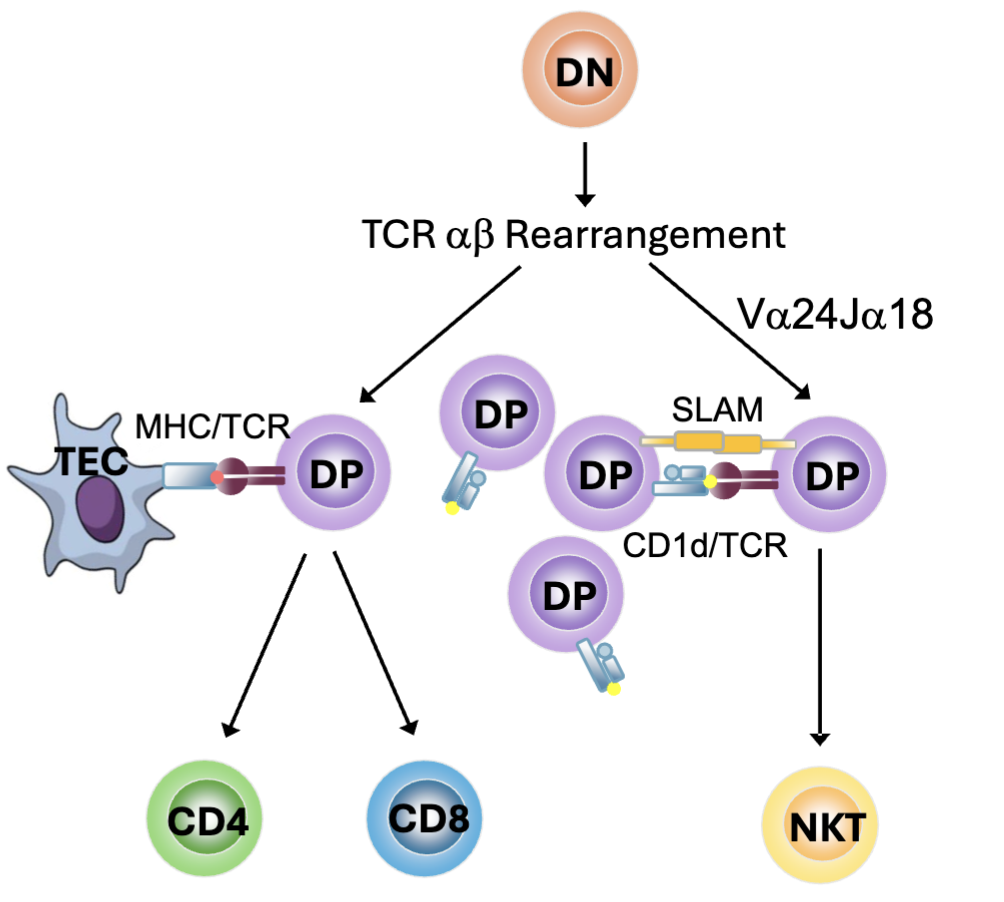
What are the features of Atypical T Cell Populations: NKT cells?
Natural killer T (NKT) cells develop from DP thymocytes
Restricted TCR usage: V⍺24J⍺18 paired with Vβ11
Selected by CD1d on other DP cells rather than MHC I or MHC II
SLAM signals contribute to selection
Transcription factor PLZF drives development
recognizes lipids and glycolipids loaded on MHC-like molecule CD1d
self-glycolipids upregulated as stress response
foreign glycolipids from pathogens
CD1d widely expressed and don’t need the thymic epithelial cells for activation
SLAM (signalling lymphocyte activating molecule) make signals that help with survival and transcription factor PLZF
What are the features of Atypical T Cell Populations: MAIT cells?
Mucosal-associated invariant T (MAIT) cells develop from DP thymocytes
Restricted TCR usage: V⍺7.2J⍺33
Selected by MHC-related protein 1 (MR1) on DP thymocytes
SLAM signals contribute to selection
Transcription factor PLZF mediates development
Recognize vitamin B (riboflavin)-related antigens produced by yeast and bacteria loaded on MR1
MAIT cells do not develop in germ free mice → Ag needs to be transported to thymus
MR1 is not expressed on surface in absence of antigen
specific TCR that is rearranged
selected by atypical MHC related protein: MR1
recognize Vitamin B (can’t be generated by us)
antigen goes from gut to thymus for positive selection

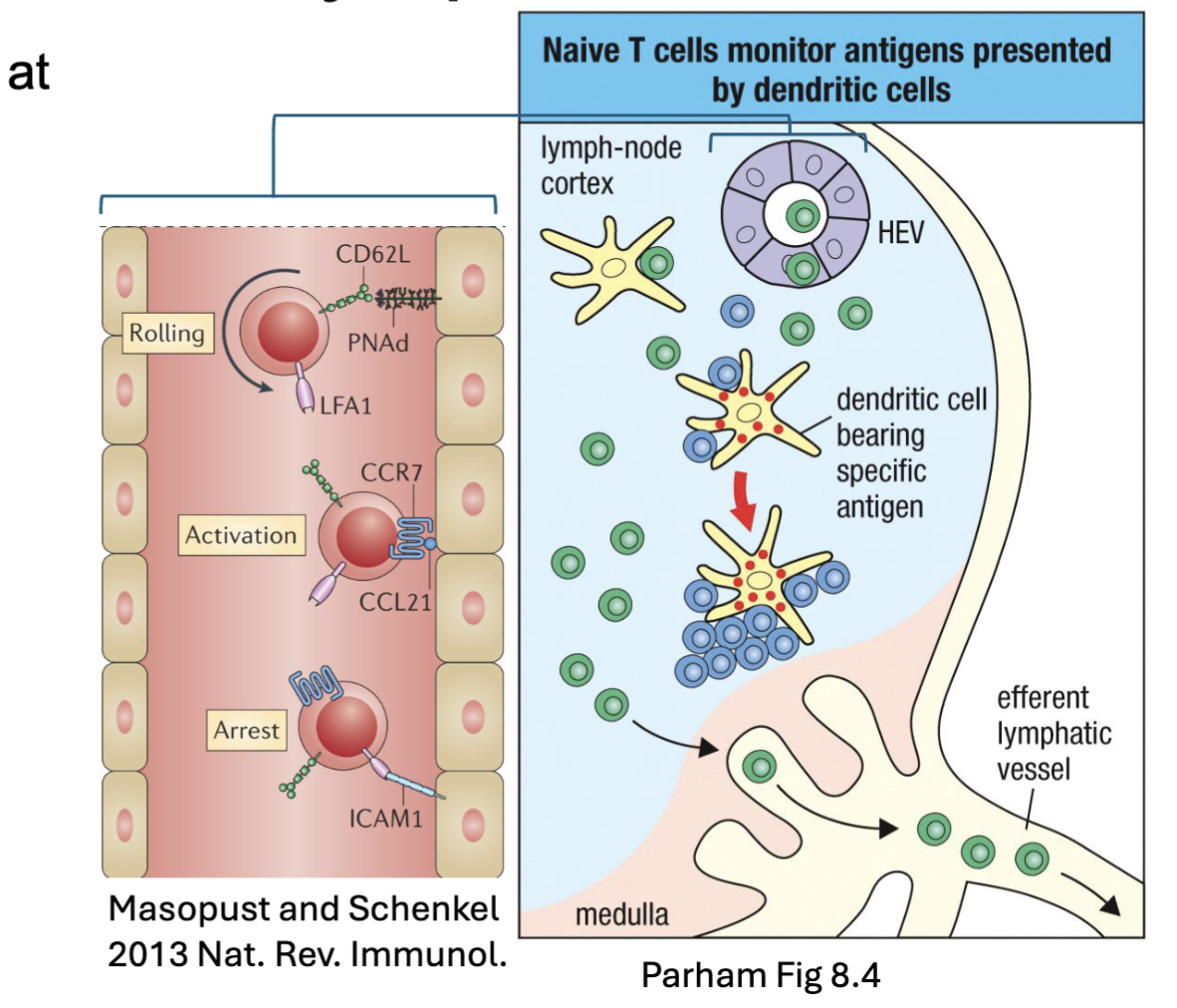
What happens in the lymph nodes with naive T cells?
Circulating naïve T cells enter LN at high endothelial venules (HEV)
L-selectin/ PNAd → rolling
CCR7/ CCL21 → activation
LFA-1/ ICAM-1 → arrest
Scan antigens presented by DCs
No Ag → Exit via efferent lymphatics (S1P)
Ag → Activated cells retained (CD69) → proliferation and differentiation
CD69 (inhibit the responsiveness to S1P)
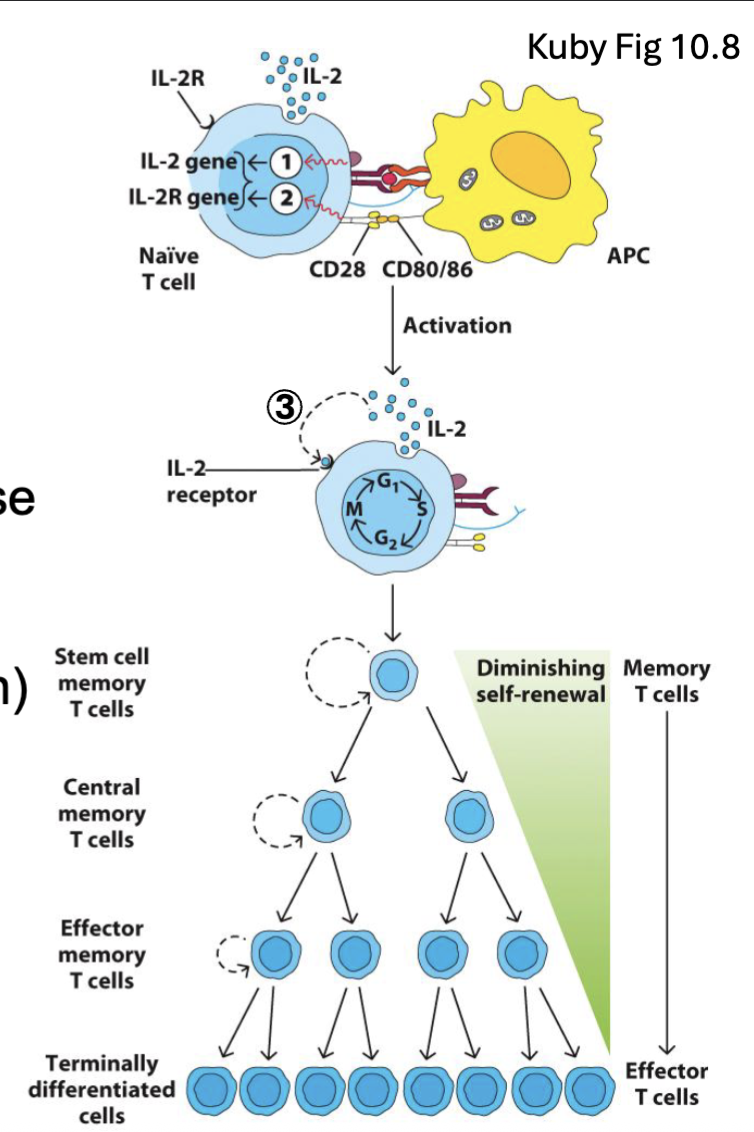
What is the T cell response (generally)?
Activated T cells proliferate and differentiate into different functional populations:
Memory T cell pools retain long term memory for recall responses
Stem cell and central memory T cells (TCM) recirculate through lymph nodes → recall response
Effector memory T cells (TEM) alter adhesion molecules → tissue homing
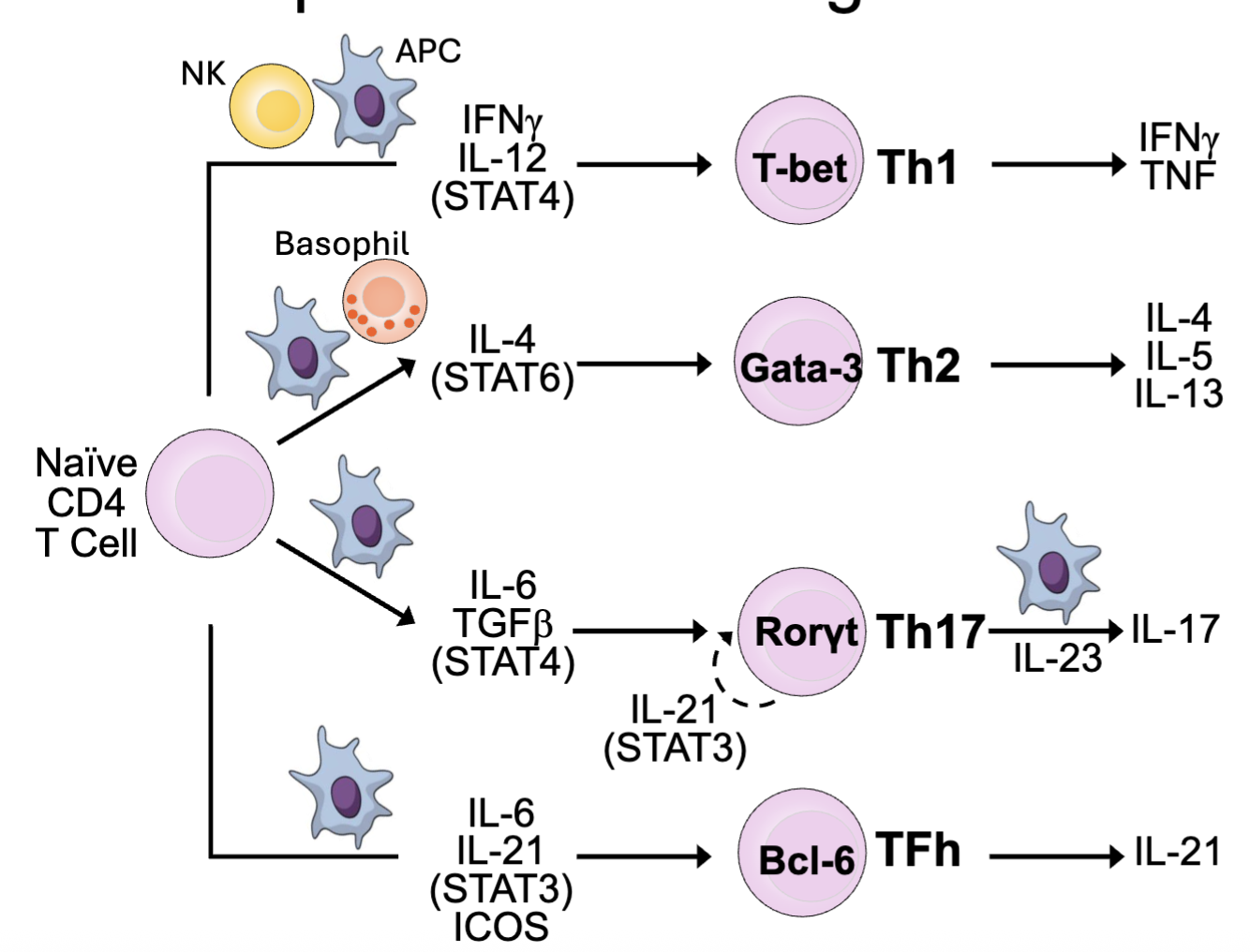
CD4 T cells differentiate into different T helper (Th) subsets: Th1, Th2, Th17, TFH
Depends on type of pathogen, cytokine environment, innate responses, type of APC
CD8 T cells differentiate into cytotoxic effectors
(CTLs)
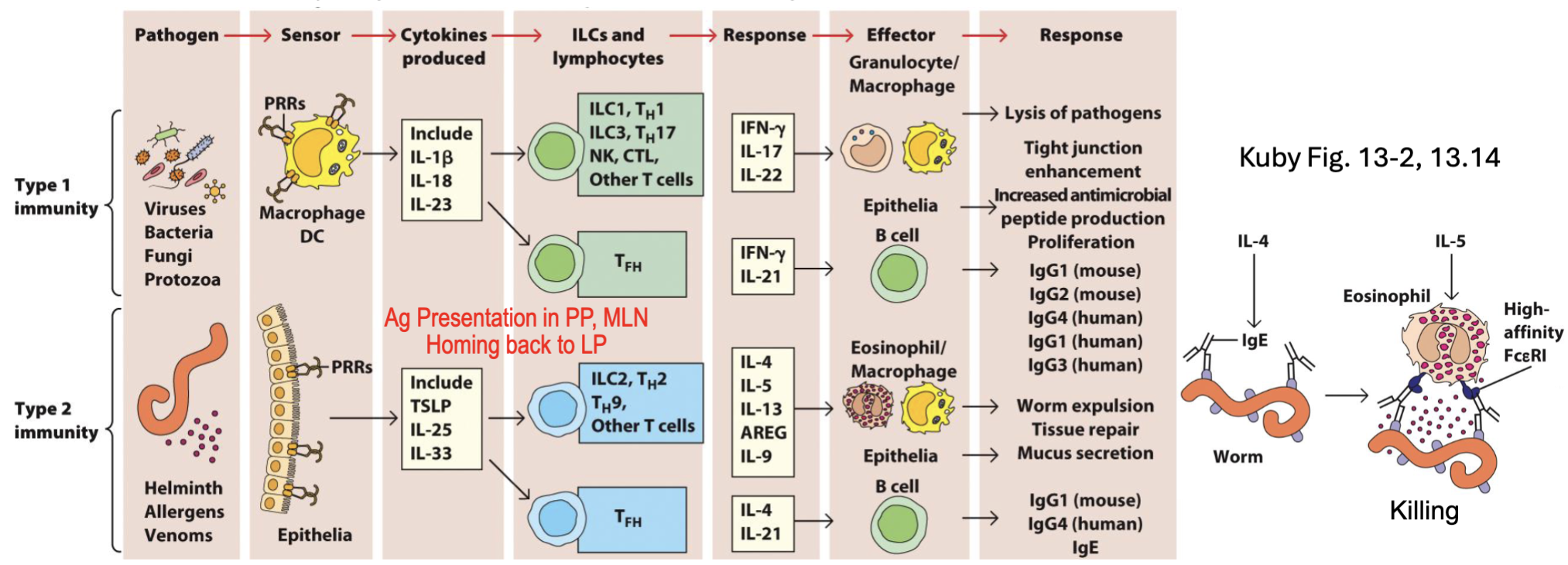
What are the T Helper Subsets: Programs for Different Responses?
TH1: Protection vs. intracellular bacteria, viruses
→Macrophage, NK, CTL
TH2: Protection vs. parasites
→Eosinophils, ILC2
TH17: Protection vs. fungi, extracellular bacteria
→Epithelium, neutrophils
TFh: B cell help
in pink: master transcription factor
need IL-23 to make IL-17
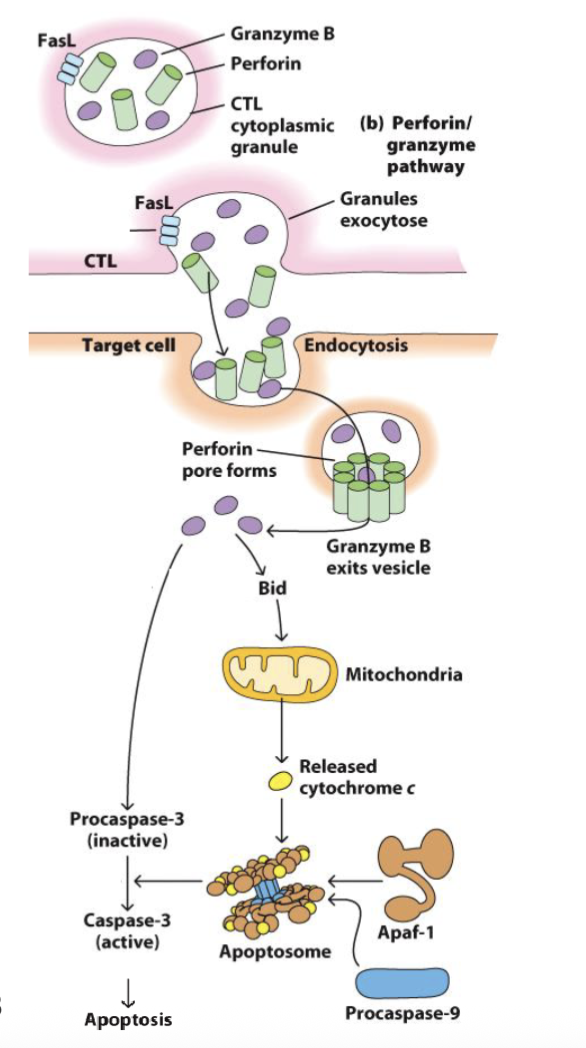
What are the features of CD8 Cytotoxic T Lymphocytes (CTLs): Serial Killers?
We can’t target pathogens hiding inside cells → kill the cell instead
Barrier to activation of naïve CD8 T cell is higher → need strong antigen or CD4 T cell help (IL-2)
Memory CD8 T cells need less costimulation
Memory CD8 T cells upregulate cytotoxic molecules (stored in granules):
Perforin / granulysin → pore formation
Granzyme → apoptosis via Bid-induced release of mitochondrial cytochrome-c and caspase activation
CTLs form synapse with targets and direct granular release
Each CTL can kill up to 16 target cells in succession
What are barrier tissues?
Barrier tissues: interfaces separating our “insides” from the outside world (while allowing productive interactions)
Skin
Mucosal tissues: intestinal tract, respiratory tract, urogenital tract, lactating mammary gland
Combination of physical (epithelium, keratin, cilia) chemical (mucus, enzymes, peptides), and cellular mechanisms (immune cells) to protect against pathogens
Hosts a commensal microbiome that exists in equilibrium with the immune system
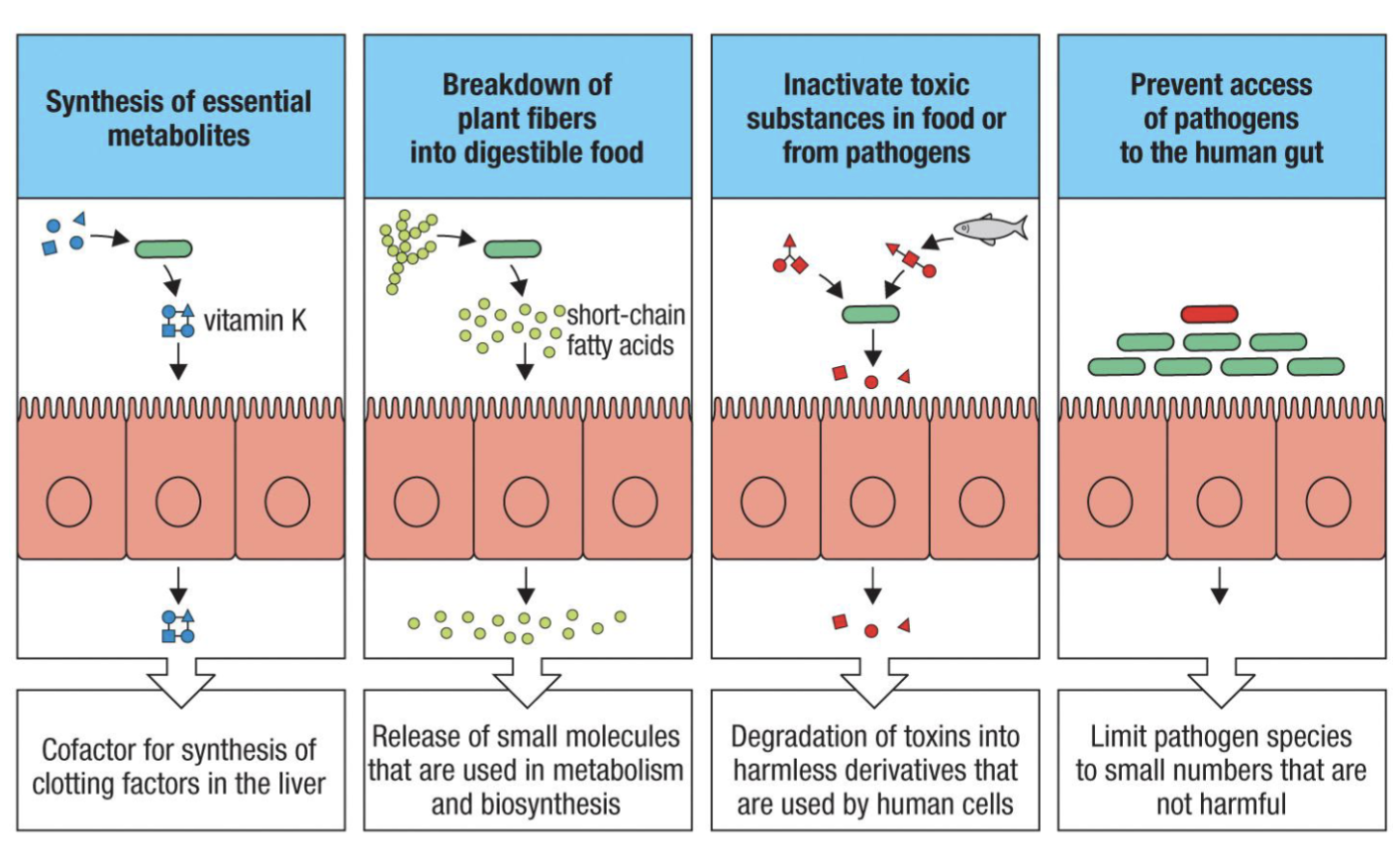
What is Commensal microflora?
The microbiome
Commensal microflora are the “friendly” bacteria, viruses, fungi, protozoa, and
worms that exist in equilibrium (symbiosis) with the host
Provide vitamins and nutrients (ex: Vitamin K, A, B-12)
Degrade toxins (inactivate toxins)
Protect from pathogens (competition) (outcompete pathogens)
*image important
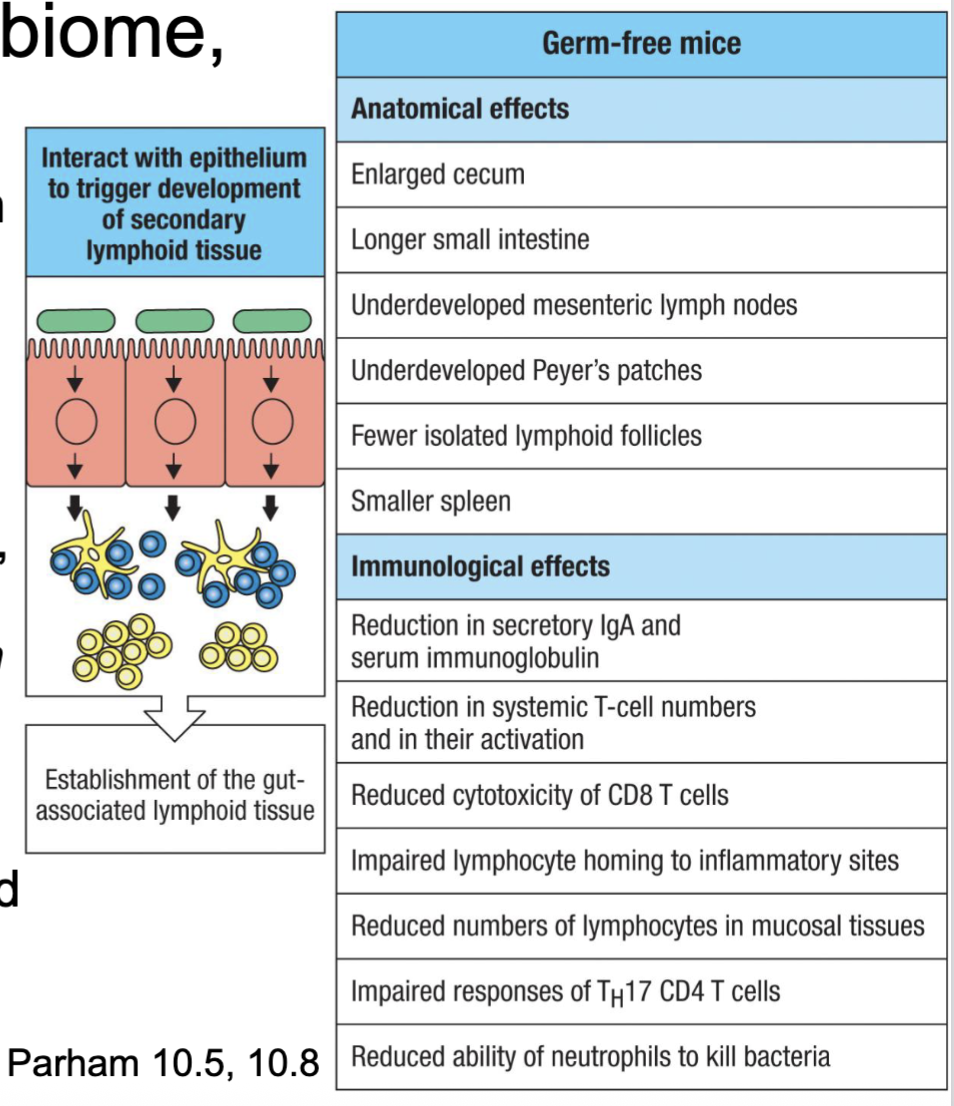
How do we codevelop with our microbiome?
We are colonized during and early after birth
Microbiome mediates secondary lymphoid development in intestine and subsequent
immune balance (figured out in germ free mice)Pathology associated with dysfunctional/ altered microbiome (i.e. antibiotics, infection,
diet, pollution, stress, genetics)Emergence of fecal transplant therapies
defect in immune development in C section children
What are the various pathologies associated with dysfunctional/ altered microbiomes? What causes these alterations?
Susceptible to infection (e.g., Clostridium difficile) and nutritional defects
Associated with autoimmune diseases (IBD, T1D, MS, SLE, RA)
Impaired immune response to cancer and immunotherapies
dysfunctional/ altered microbiome:
antibiotics,
infection,
diet,
pollution,
stress,
genetics

Why do we need to distinguish between friend and foe?
Healthy barrier in homeostatic balance (no inflammation) → tolerogenic response
Immune system samples contents from lumen (small numbers of microbes)
Trigger production of anti-inflammatory molecules (IL-10, TGFβ) → iTregs
and IgA-producing B cells
Disrupted microenvironment → barrier damage → invasion
of pathogenic microbes → inflammation
Resolution or disease?
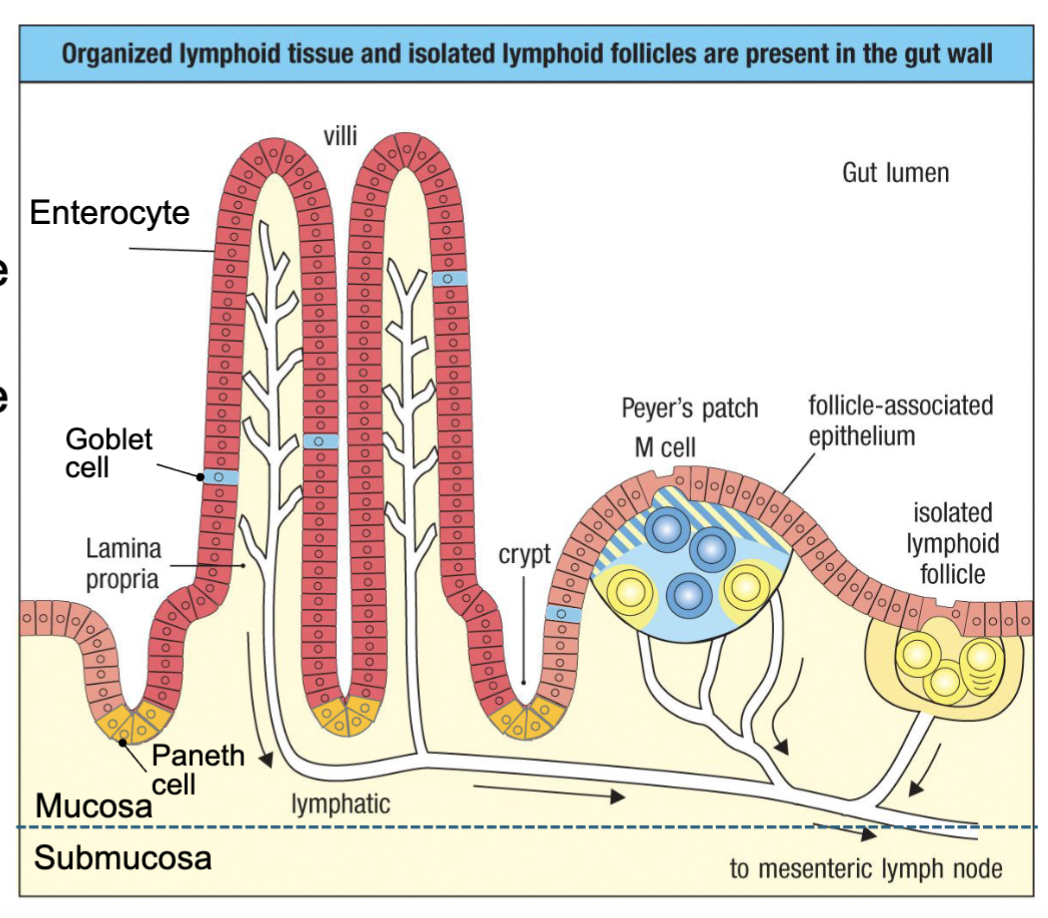
What are the features of the intestinal mucosa (epithelium and lamina propia)?
Single layer of epithelium separates host from luminal microbes:
Enterocytes – structural epithelial cells, transport nutrients
Paneth cells – secrete antimicrobial peptides (AMPs) – defensins, lysozyme
Goblet cells – secrete mucus, AMPs
Microfold cells (M cells) – transcytose antigen from lumen
Lymphoid tissues form in lamina propria directly under M cells:
Peyer’s patches – DCs, B cells, T cells
Isolated lymphoid follicles (cryptopatches) – B cell follicles
Lymphatics drain to mesenteric nodes
Peyer’s patch anologous to lymph node: B, T and DC
lymphoid follicle: mostly B cells in there
How do barriers maintain homeostasis?
Keep microbes away from epithelium
Mucus – viscous fluid formed by polymeric mucin glycoproteins → trap microbes → washed away (peristalsis)
Cysteine residues in globular heads bind each other → crosslinked networks
Covalently bind secreted IgA, IgM, and defensins
Antimicrobial peptides (AMPs) – kill microbes to keep them away
defensins (cause pores in bacterial membranes)
lysozyme (digests cell walls)
REG3 proteins (kill Gram-positives) - stimulated by IL-22 from ILC3

How is homeostasis maintained? ( healthy barriers)
Tregs
Barrier tissues
Antigen sampling
IgA plasma cells
Dimeric IgA
How does antigen sampling maintain homeostasis?
Antigens can be transported across epithelium by multiple mechanisms:
M-cells – transport bacteria and antigens and deliver to APCs (microfold cells)
FcR – IgG-immune complexes can be transported by neonatal Fc receptor (FcRn)
Goblet cells – transport small soluble antigens
APCs – can reach across and directly sample lumen
Antigens delivered into a controlled environment (no inflammation) → tolerance
FcR: bind immunoglobulin
Fc receptor transports IgG

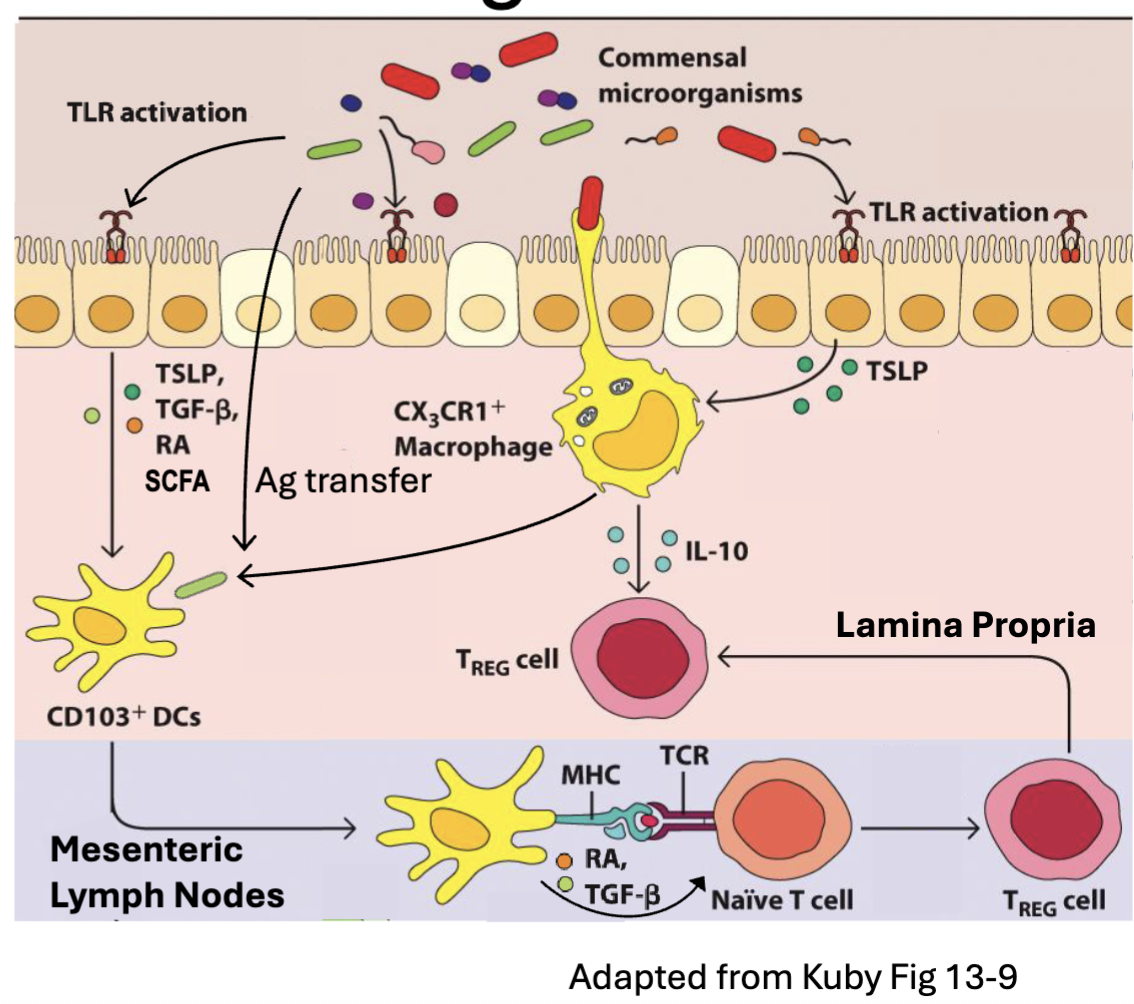
How do Tregs maintain homeostasis?
Regulatory T cells (Tregs) – CD4 T cell subset that generates TGFβ and IL-10 → inhibit immune responses
DCs primed in presence of TSLP, TGFβ, retinoic acid (RA), short chain fatty acids (SCFA) take on tolerogenic phenotype (TGFβ, IL-10, ICOSL) → Treg induction
TGFβ and RA induce the FoxP3 transcription factor (Treg master regulator)
RA imprinting induces ⍺4β7 integrin and CCR9 → T cell homing back to intestinal tissue
Tregs maintained by in tissue by IL-10 (and IL-2) → inhibit T effectors
to get tolerance need to make T regs
need DC to be primed in certain way
survival signals IL-10 and IL-2 by macrophages
How do Tregs work?
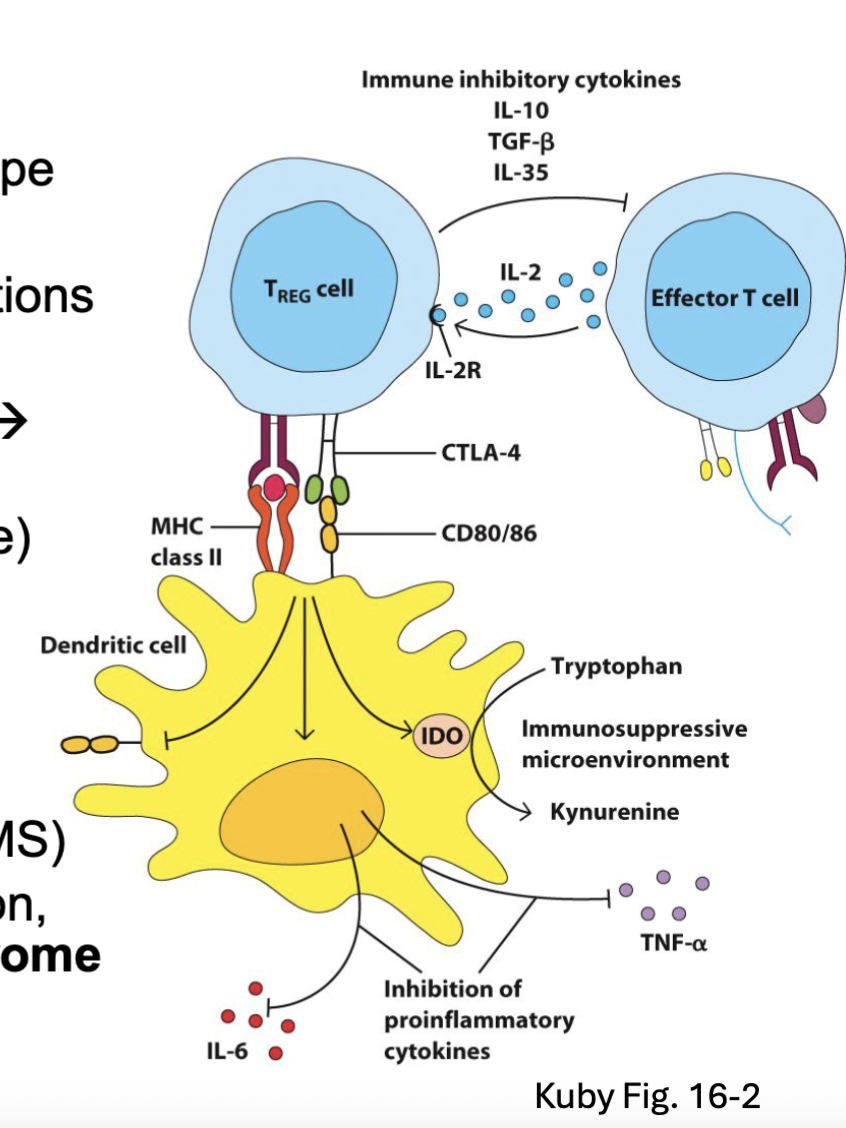
FoxP3 drives an immunoregulatory T cell phenotype
Tregs suppress immunity by several mechanisms:
TGFβ and IL-10 → inhibit APC and T cell functions
Express high levels of CD25 → soak up IL-2
Upregulate CTLA4 → binds CD80 and CD86 → inhibitory signal
Induce IDO (converts tryptophan → kynurenine)
Inhibit inflammatory cytokine production (IL-6, TNF)
Downregulates CD80/ CD86 expression
Acquired Treg dysfunction → autoimmunity (RA, MS)Mutations in FoxP3 → IPEX (Immune dysregulation, Polyendocrinopathy, Enteropathy, X-linked) syndrome →autoimmune symptoms (neonatal T1D, IBD, dermatitis)
CTLA4 is inhibitory to the DC and upregulates IDO
How do plasma IgAs maintain homeostasis?
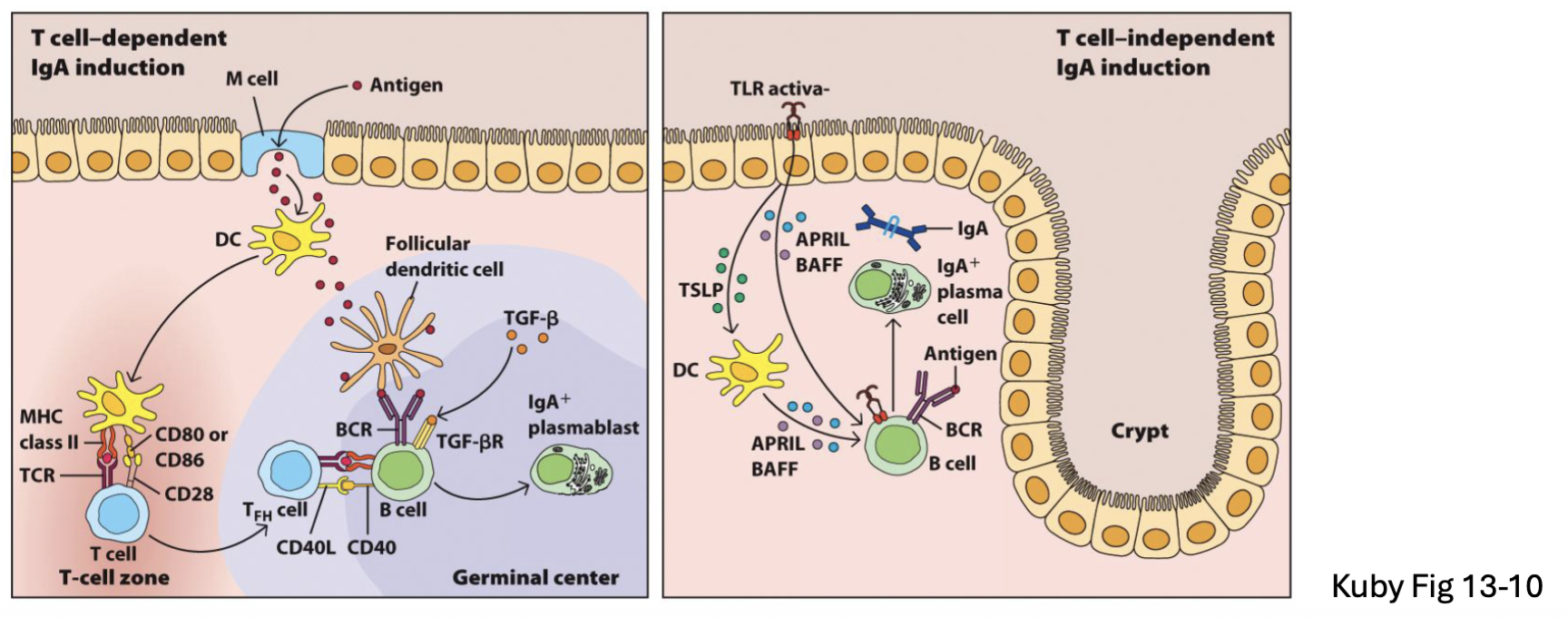
Plasma cells in intestine generate IgA (and some IgM, IgG)
B2 B cells (T-dependent): TGFβ → IgA class switch in Peyer’s patch or MLN
B1 B cells (T-independent): APRIL and BAFF → IgA in lamina propria or isolated lymphoid follicles (faster but lower affinity)
APRIL and BAFF: cytokines
T-independent lower affinity (no affinity maturation)
How does dimeric IgA maintain homeostasis?
Intestinal tissue generates 3-5g of IgA per day (respiratory and reproductive tracts also generate IgA)
IgA binds J chain and dimerizes → transported across epithelium via the polymeric immunoglobulin receptor (polyIgR) → cleavage of polyIgR leaves secretory component attached to luminal IgA (this also happens for IgM)
IgA neutralizes microbes but doesn’t cause inflammation (doesn’t activate Complement)
Secretory IgA is an important line of defense for mucosal surfaces against bacteria (e.g. Salmonella, Vibrio cholerae) and viruses (e.g. polio virus, reovirus)
IgA good at neutralizing salmonella, …

What are the features of oral tolerance?
Tolerance to food antigens appears to work the same as tolerance induction to commensal microbes
Food antigens are taken up and presented in a tolerogenic steady state environment → Tregs
and IgA
Guidance on food introduction to infants previously recommended delayed exposure to foods that
cause allergies (e.g., eggs, peanut, wheat, milk)
Changed by 2008 study showing exposure to peanut antigens during first year → 10-fold
lower incidence in peanut allergy
Epidemiological evidence suggests infection precedes onset of food allergies
Rotavirus infection preceded celiac disease (gluten allergy) onset in genetically susceptible
individuals
What are the pathogen responses of the barrier (controlled vs uncontrolled)?
Healthy epithelium samples microbes in a controlled way → tolerance
Excessive or “uncontrolled” exposure to microbes → inflammation → eradicate pathogen
and restore homeostasis
PRRs on epithelial basolateral surface or cytoplasm → inflammatory signals
Inflammatory signals > local tolerance signals → induction and effector responses
PRR’s recognize the excessive or uncontrolled
on basolateral surface means it has crossed the barrier
gauging the inflammatory signals and the local tolerance signals
What happens when there is excessive inflammation in the gut?
It is pathogenic
it can lead to IBD (inflammatory bowel disease)
there is also Celiac disease: (not IBD)
similar symptoms but represents an autoimmune response triggered by glutens in grains (antibodies when gluten appears)
medications: antibiotics can create this issue (imbalance)
