MBIO 2020 / Topic 5a: Regulation - Transcription Factors
1/31
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
32 Terms
How do microbes control the amount of an enzyme?
How do microbes control the activity of an enzyme?
Amount
They vary the amount of mRNA made, i.e. transcriptional control.
They vary the amount of protein made, i.e. translational control.
Activity
They have regulatory processes post-translation.
What is the one important reason for why microbes regulate protein function?
Cell economics: Genes will only be expressed only when products are needed in the amount needed (i.e. physiological condition of cell), which conserves energy and prevents interference between expressed products.
Are protein-nucleic acid interactions (PNA) always going to be specific? Why or why not?
Protein-nucleic acid interactions may be specific or non-specific, depending on whether the protein attaches anywhere on the nucleic acid or whether the protein binds to specific site.
If protein-nucleic acids are specific, how is specificity achieved?
Where is the usual site of PNA interactions on DNA?
Specificity is provided by interactions between specific amino acid side chains of the proteins and specific chemical groups on the nitrogenous bases and the sugar phosphate backbone.
The usual site of PNA interactions on DNA is the major grove, due to the usual relatively big size of a protein.
DNA-binding proteins are usually homodimeric, with each subunit called a domain. What is a domain, and what does it do?
Regions of the protein with a specific structure and function, interacting specifically with a region of DNA in the major groove.
What is the most common domain of a DNA-binding protein? Relate its structure to its function.
The helix-turn-helix (HTH) structure is the most common, which is composed of an alpha helix connected to a turn (made of three amino acids) connected to another alpha helix. The first helix (recognition helix) binds specifically with and in DNA, while the second helix (stabilizing helix) stabilizes the first helix with hydrophobic interactions.
Between the sugar phosphate backbone and the nitrogenous bases, what is more specific in regards to PNA interactions?
If you want to be more specific, your interactions should involve less of being generic. The sugar phosphate backbone is generic, therefore there will be less interaction specificity. What does bring more specificity are the nitrogenous bases because they are less generic - there’s four of them.
What are the two kinds of regulatory proteins in regards to how often they are expressed? Differentiate them by function.
Constitutive: Protein always present due to 24/7 gene expression.
Inducible or repressible: Protein not always present due to gene expression being regulated in response to change in physiological conditions.
What is enzyme repression?
The regulation wherein enzymes that catalyze the synthesis of a specific product (anabolism) are not made if the product is already present in the medium in sufficient amounts.
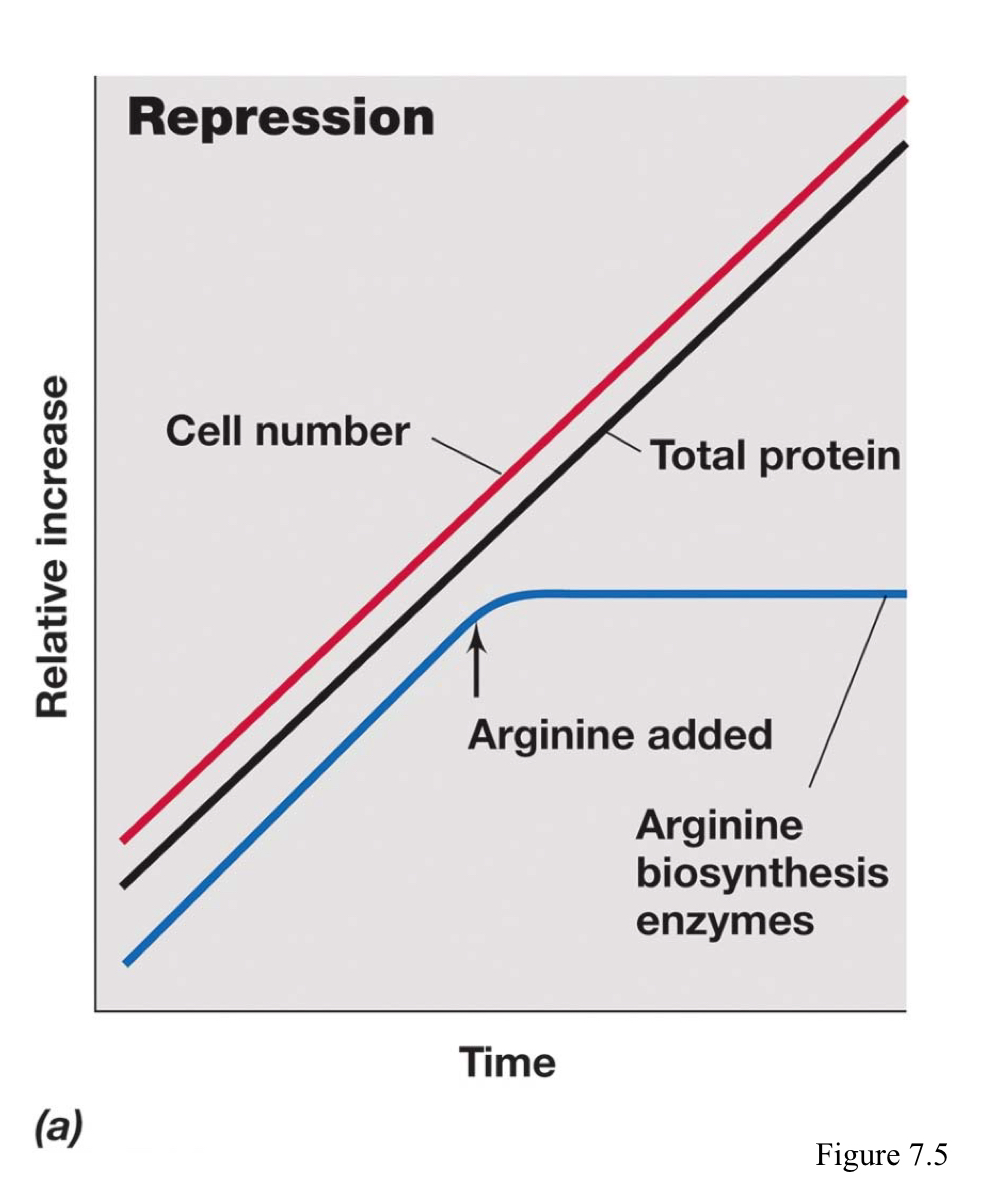
What is enzyme induction?
The regulation wherein enzymes that catalyze the break down of a specific product (catabolism) are not made if the substrate is present.
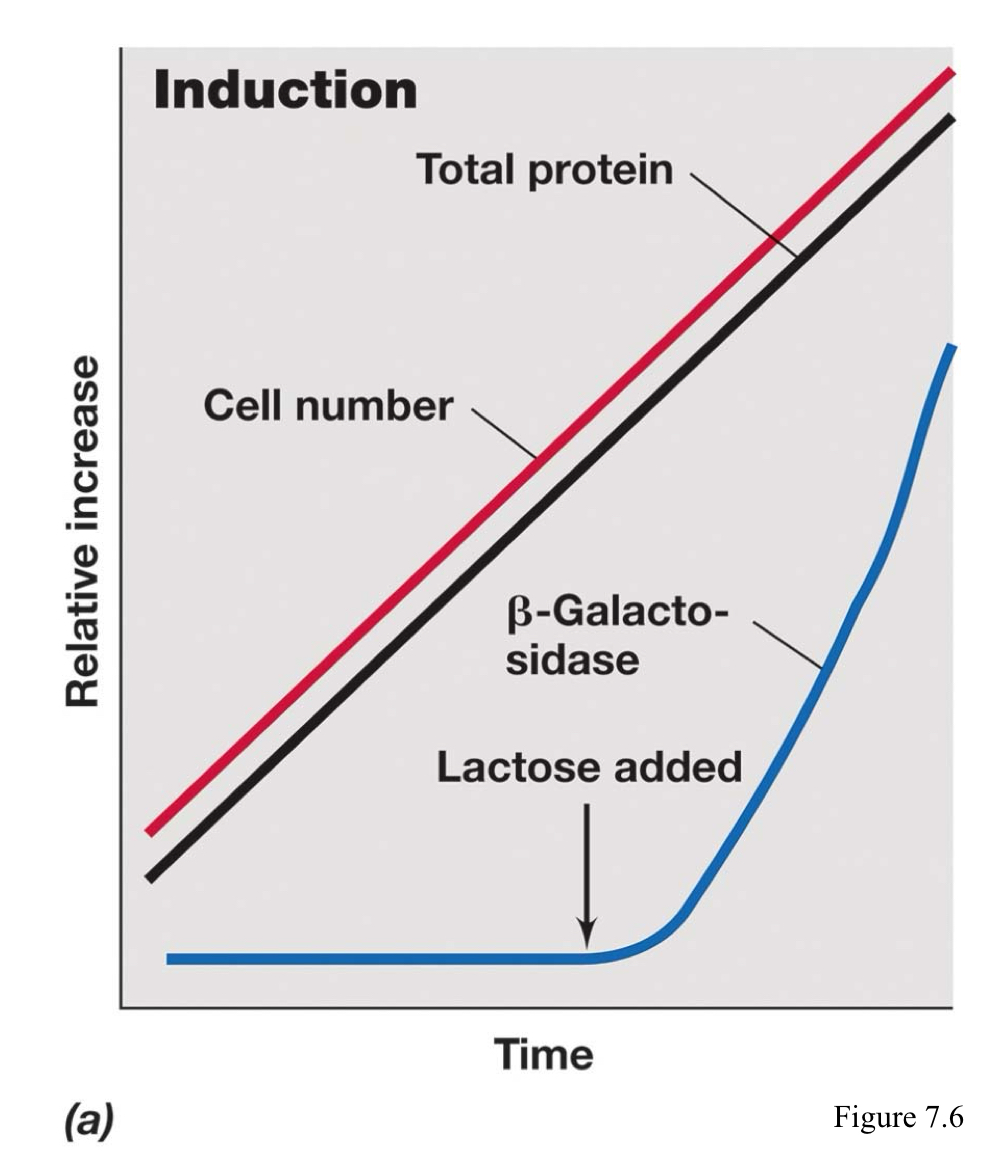
What are the two kinds of regulatory proteins in regards to their ability? Differentiate them by function.
Sigma factors: Controlling the binding of RNA pol to promoter region.
Transcription factors: Controlling transcription rate by binding to specific DNA sequences.
What are the two kinds of transcription factors? Differentiate them by: (1) the kind of regulation they do, (2) their function, and (3) where they bind to in DNA.
Activators: For positive regulation, they bind to the activator binding site located proximal of the promoter region, assisting recruitment of RNA pol holoenzymes.
Repressors: For negative regulation, they bind to the operator located downstream of the promoter region on DNA, blocking access of RNA pol holoenzymes.
What is the name of genes that encode for transcription factors?
Are transcription factors limited to regulation of expression of only one gene? Feel free to explain.
> regulatory genes
> can express more than one separate gene, a regulon, or its own gene (autoregulation)
+ regulon = group of genes controlled by one regulator, e.g. transcription factor)
When can you consider a regulatory gene to be part of an operon?
> RG is transcribed w/ other genes in operon
> RG is controlled under its own product
What are cell metabolites?
What are effectors, and how do they relate to transcription factors?
> small molecules produced/consumed in pathways
> typically cell metabolites, bind TFs → activate/inhibit ability to regulate gene expression in related pathways
What are two types of effectors? Differentiate them via function.
Inducers: Substance that induces enzyme synthesis.
Corepressors: Substance that represses enzyme synthesis.
What is allosteric regulation?
> binding of an effector to a protein’s allosteric site (not the active/DNA-binding site)
> conformational change
> alters the protein’s activity (activates or inhibits)
How does inducible expression occur together with allosteric regulation?
Allosteric change results from effector binding, allowing protein to bind to DNA for activation.
How does repressible expression occur together with allosteric regulation?
Allosteric change resulting from effector binding allows protein to bind to DNA for repression.
Is regulation of gene expression turned on and off instantly? Why or why not?
No, as proteins vary in concentration, affinity, and in specific physiological conditions.
Should the determination of an effector as an inducer or corepressor be dependent on the transcription factor it binds to?
No. It is dependent on the definitions themself:
An effector is an inducer if the final outcome in the mechanism is enzyme/protein making.
An effector is a repressor if the final outcome in the mechanism is enzyme/protein stop-making.
What’s the mechanism of repression?
Specific repressor is activated when bound to effector, and repressor thus binds to the operator, blocking the way of RNA pol and stoping transcription.
The final outcome is to stop making, so the effector must be a corepressor.
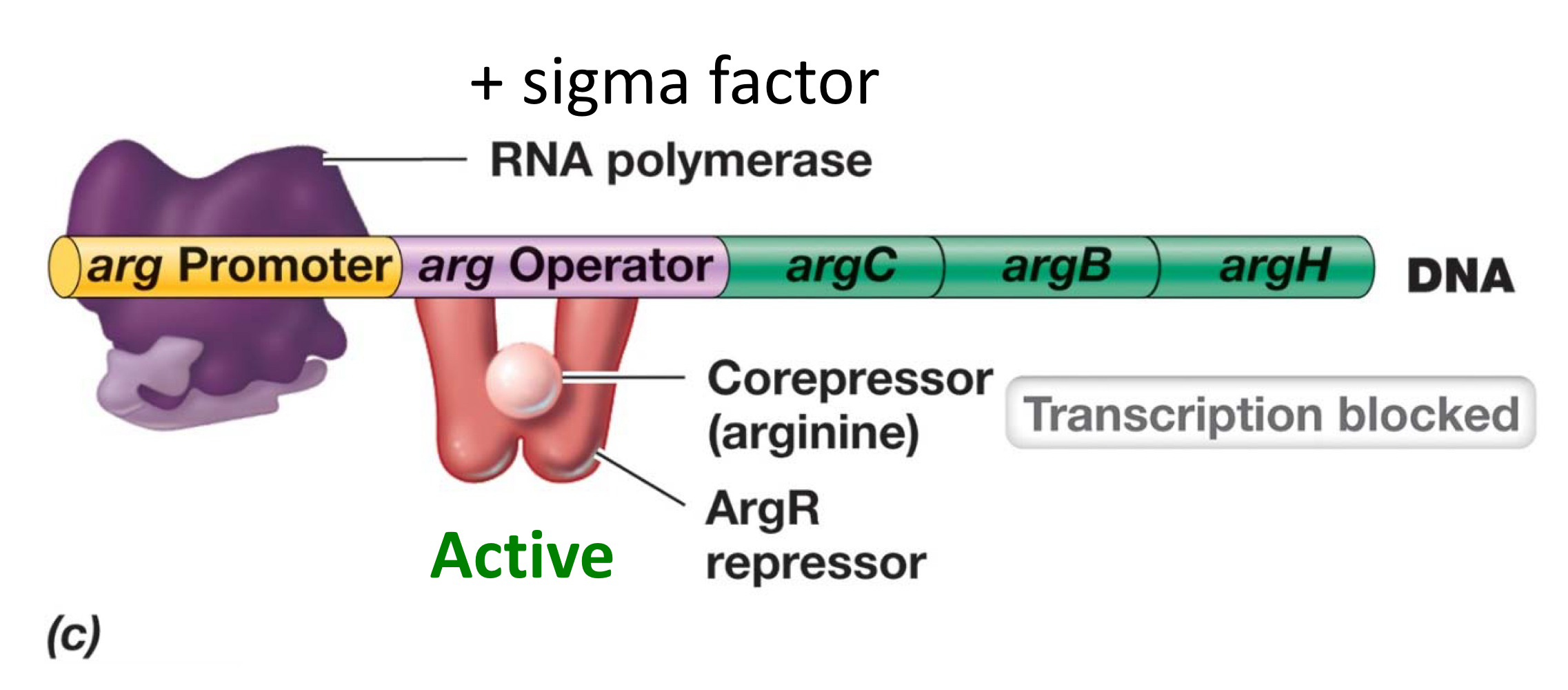
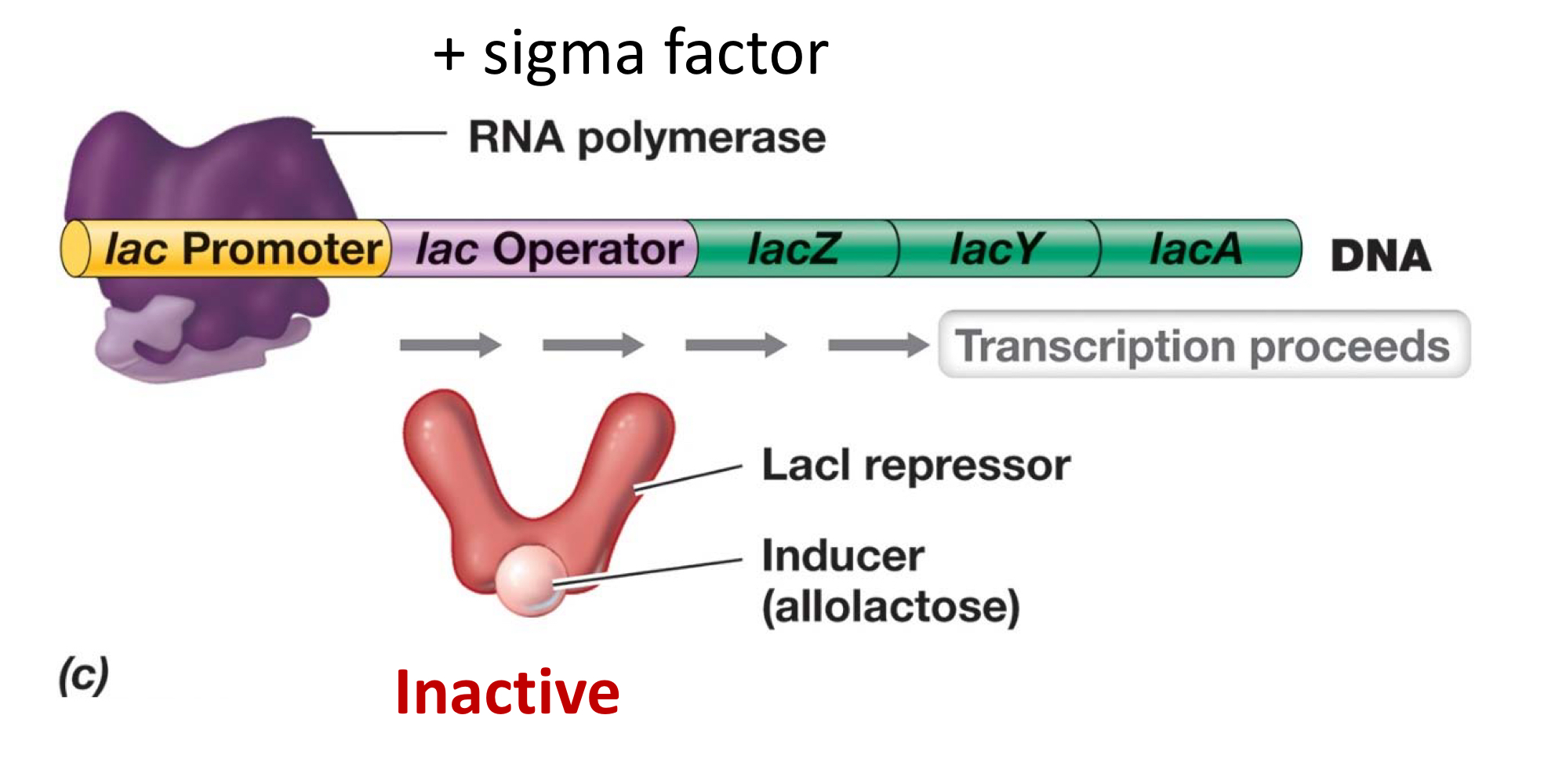
What’s the mechanism of de-repression?
Specific repressor is deactivated when bound to effector, and repressor thus stops binding to operator, continuing transcription.
The final outcome is to continue making, so the effector must be an inducer.
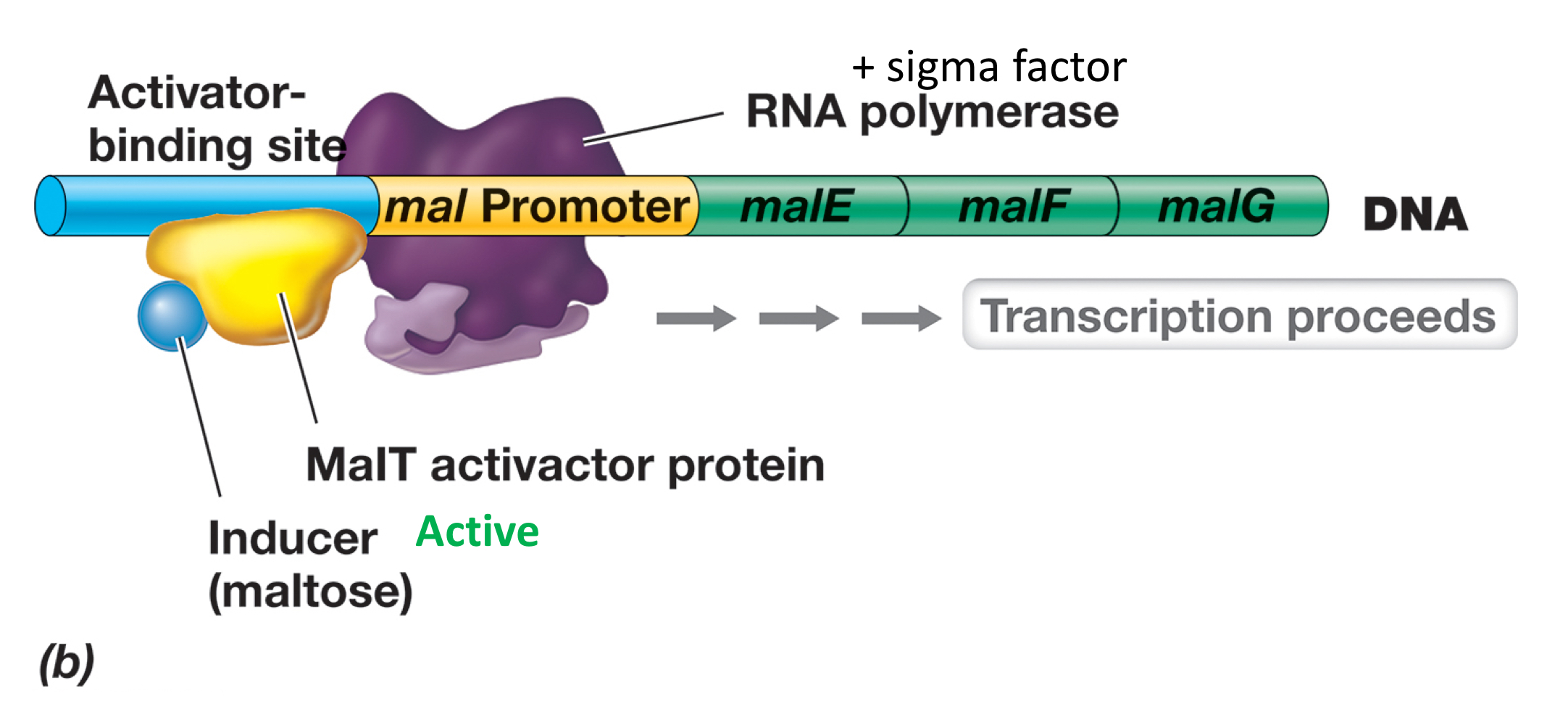
What’s the mechanism of activation in the scenario of proximal binding and binding further upstream?
Specific activator is activated when bound to effector, and activator thus binds to the activator binding site, recruiting more RNA pol.
If the binding site is further upstream by several hundred basepairs, then the specific activator-bound DNA must be looped to make contact with the RNA pol to activate transcription.
Differentiate inducible and repressible operons.
usually off → induced to make product → inducible
usually on → repressed to not make product → repressible
What does lacI code for?
What does lacZ code for?
What does lacY code for?
What does lacA code for?
> lacI = LacI = repressor for lac operon
> lacZ = ß-galactosidase → cleaves lactose into glucose + galactose
> lacY = LacY/lac permease → transports lactose into the cell.
> lacA = LacA/transacetylase → modifies lactose and lactose analogs + may prevent toxic buildup in cytoplasm
Explain the negative transcriptional control mechanism for the lac operon.
When there is no lactose in the environment, LacI is constitutively expressed and functional. LacI binds to operator, blocking the path of RNA pol, and thus preventing transcription.
When there is lactose in the environment, there will be two reactions: major and minor.
MINORS FIRST: Pre-existing molecules of ß-galactosidase converts some of the lactose into allolactose. The allolactose will act as the inducer to LacI, ultimately unblocking the path of RNA pol, allowing transcription.
MAJORS LAST: ß-galactosidase made from the now transcribed operon cleaves lactose into glucose and galactose.
Explain the positive transcriptional control mechanism for the lac operon.
Adenylate cyclase produces cAMP but is only activated by enzyme IIAGlc in the phosphoenolpyruvate: phosphotransferase system (PEP), which is involved in glucose transport into the cell.
If lactose and high glucose are present, IIAGlc won’t activate adenylate cyclase on because it is not phosphorylated. Instead, it inhibits LacY.
If lactose and low glucose (or if glucose is absent) are present, IIAGlc is phosphorylated and can activate adenylate cyclase to produce cAMP. IIAGlc will also not inhibit LacY. cAMP will then bind to a catabolite activator protein (CAP), which allows the complex to bind to the promoter, recruiting RNA pol.
Taking into account now both positive and negative transcriptional lac operon, when is optimal transcription of lac operon?
Glucose must be absent and lactose must be present.
Why is glucose a better carbon source and energy source for many bacteria?
What is diauxic growth, and how does it relate to glucose to being a better carbon and energy source?
More efficient to break down because of how much ATP is stored and the ease of breaking down one monosaccharide instead of a disaccharide.
Diauxic growth is two log phases separated by a short lag phase. For example, if the growth medium contains both lactose and glucose, there will be two log phases.
The first log phase will have the lac operon repressed because glucose is being used for the source, and lactose is not.
There will be a lag phase in between, which is where the switch of source occurs.
There will be a second log phase wherein lac operon is expressed, and lactose is now used.
What is basal transcription?
Low-level, constant transcription in the absence of additional regulatory factors.
Explain the negative transcriptional control mechanism for the trp operon.
> TrpR-encoding gene constitutively expressed but not functional
> TrpR-encoding gene upstream of trp operon
@ low trp
> TrpR does not bind to operator → > trp transcribed → polycistronic mRNA for enzymes to make trp
@ high trp
> trp = corepressor to TrpR → TrpR now functional
> TrpR binds to operator → trp operon not transcribed to stop ultimately stop making trp