module 1 lectures 1 & 2 - genes and mutations, early cancer detection via molecular screens
1/137
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No study sessions yet.
138 Terms
human genome composition
1.5% genomic DNA encodes protein
2% biologically important sequences (promoters/enhancers)
96.5% "junk," non-coding DNA
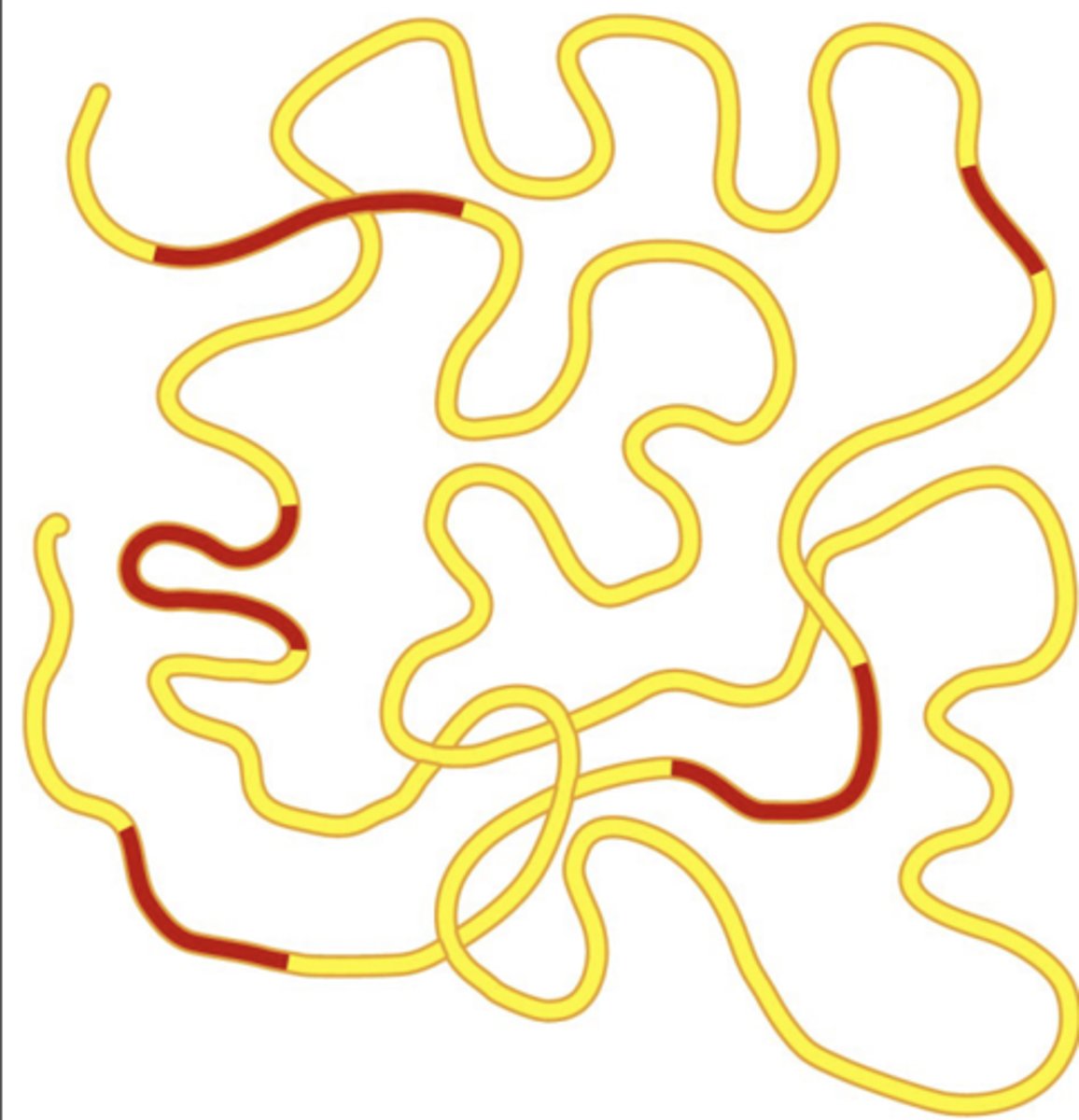
2012 ENCODE data showed that most of the genome is
composed of functional elements (not junk)
human genome project
- 2000
- 3 billion base pairs DNA sequences
- approx. 21,000 genes discovered
encyclopedia of DNA elements (ENCODE)
launched in september 2003 to identify all the functional elements in the human genome
ENCODE discoveries
demonstrated that over 80% human genome serves functional purpose
- 30 research papers published Sept 2012 from 32 labs
ENCODE by the numbers
- 147 cell types studied
- 80% functional portion of human genome
- 20,687 protein-coding genes
- 18,400 RNA genes
- 1640 data sets
- 30 papers published this week
- 442 researchers
- $288 million funding for pilot, technology, model organism, and current project
polymorphisms
- functionally silent genetic differences between individuals
- phenotypically silent, but identifiable via DNA sequencing
- "fast" evolving DNA; heterozygous locus
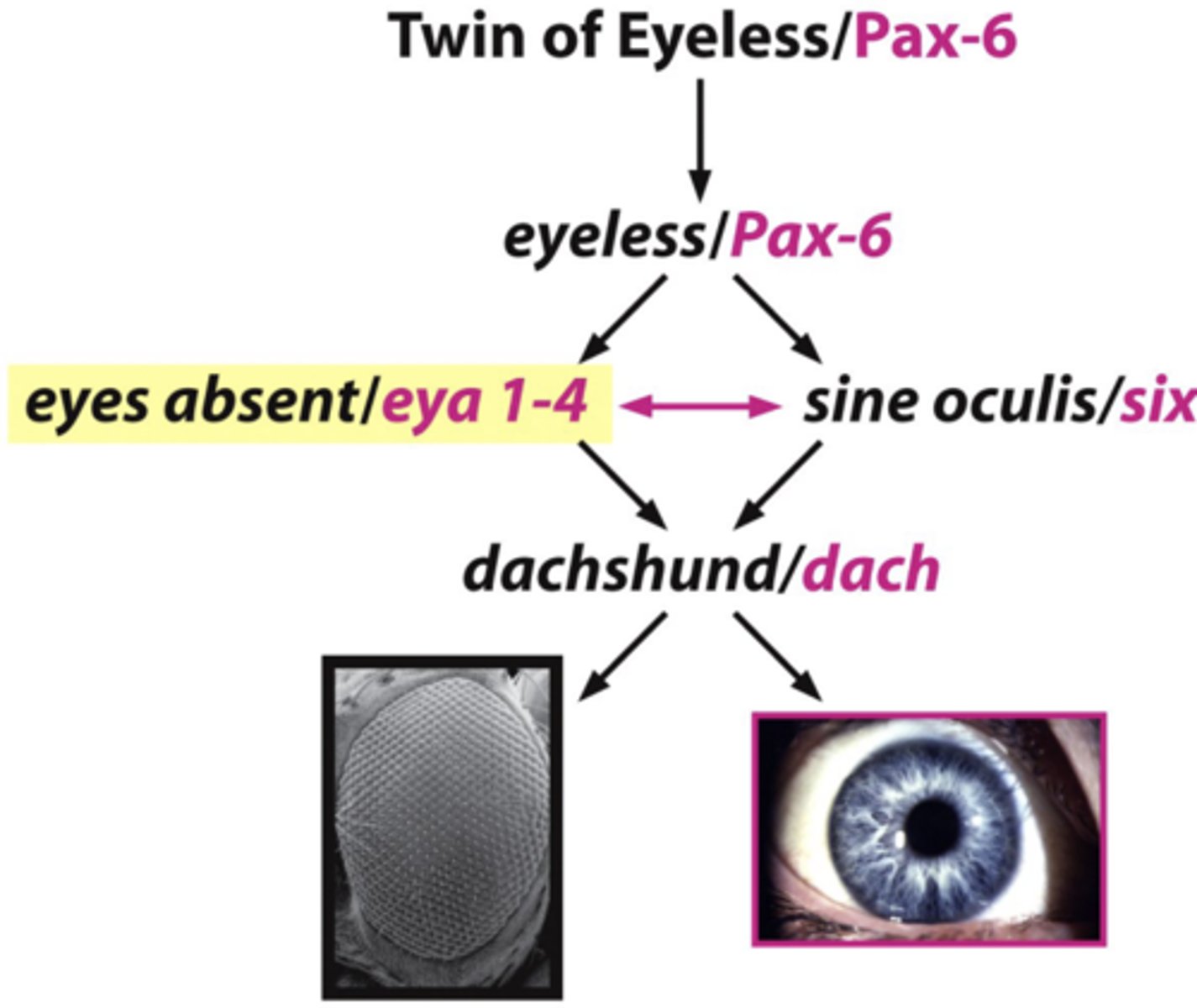
genes are conserved over millions of years and between species: example
- last common descendant of fly and mammals = 600 million years ago
- yet, genes for eye development: eyeless (fly) and Pax 6 (mammal) are highly conserved and interchangeable!
experiment: express mouse Pax6 gene in fly embryo --> result and conclusion?
result: mouse Pax 6 gene expression able to direct formation of an eye on fly leg
conclusion: interchangeability of genes
conservation of gene function lies in
amino acid sequence
- which can remain similar even when nucleotides differ

how does conservation of gene function/amino acid sequence simplifie genetic studies?
if you want to study a human gene, find it in yeast first
gene localization
finding the chromosomal region that contains the gene
- FISH mapping with fluorescent probes (short nucleotide sequences that bind to chromosomes and light up)
- identification of 6 distinct genes on chromosome 5
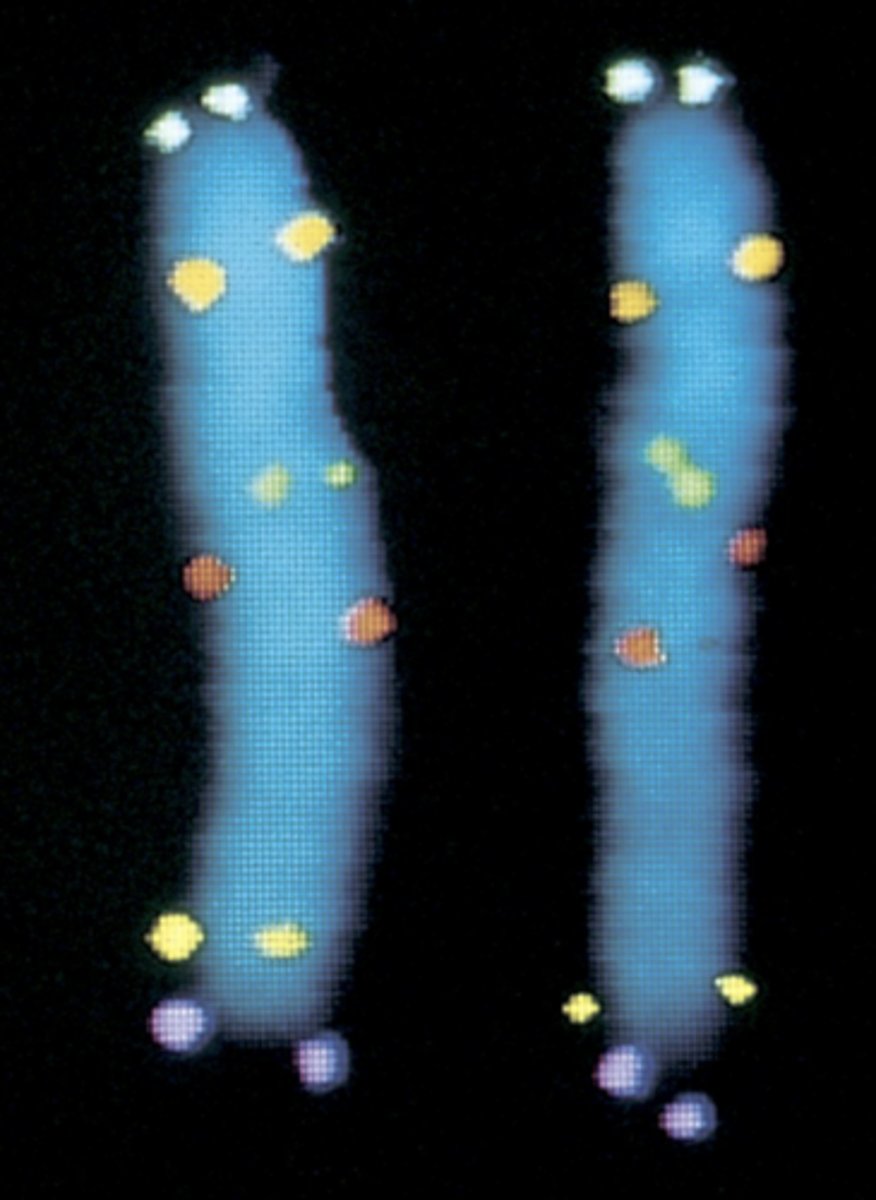
why are there 2 dots represented on each chromosome?
already went through S phase so each chromatid has 2x DNA
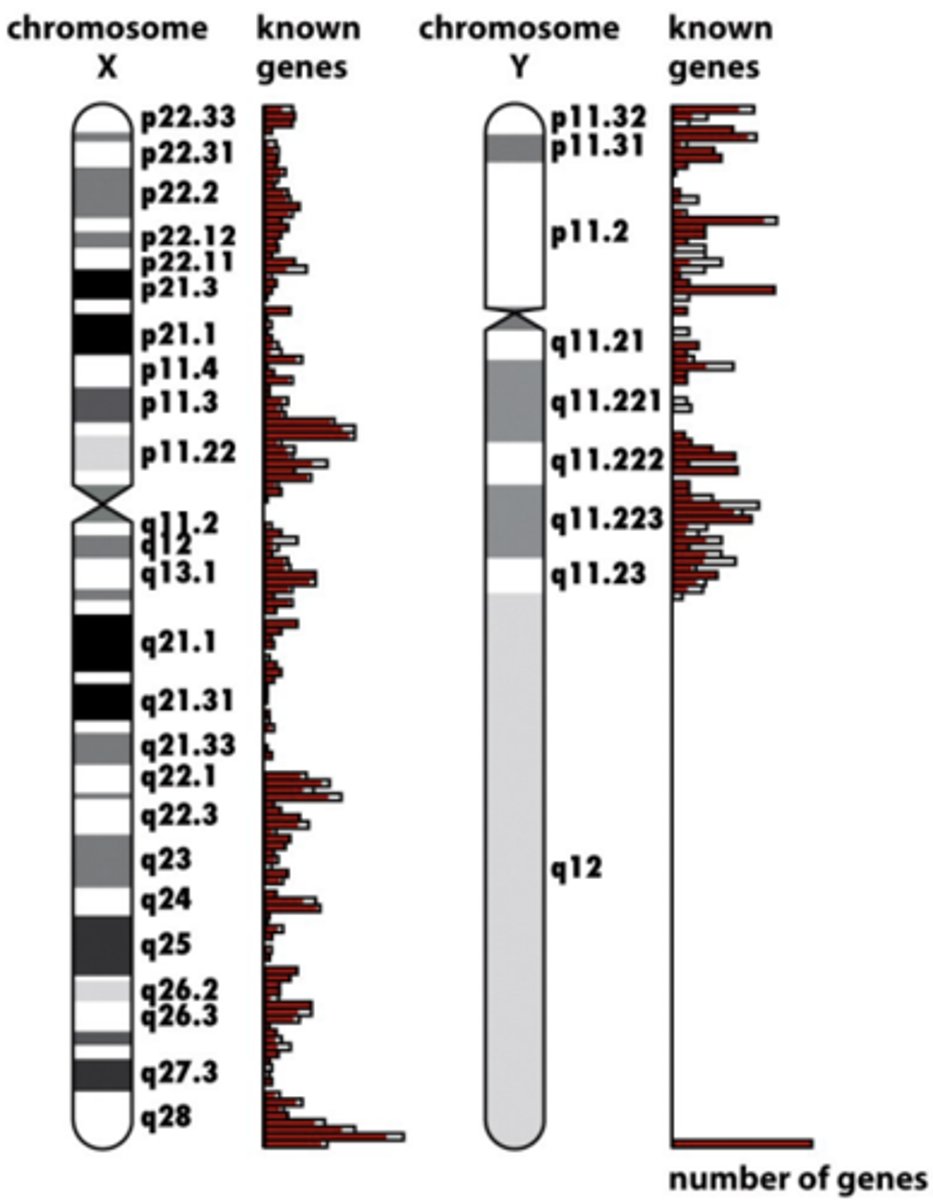
human genome project accomplished
mapped positions based on DNA sequences --> chromosome map
- 1st sequence of human genome
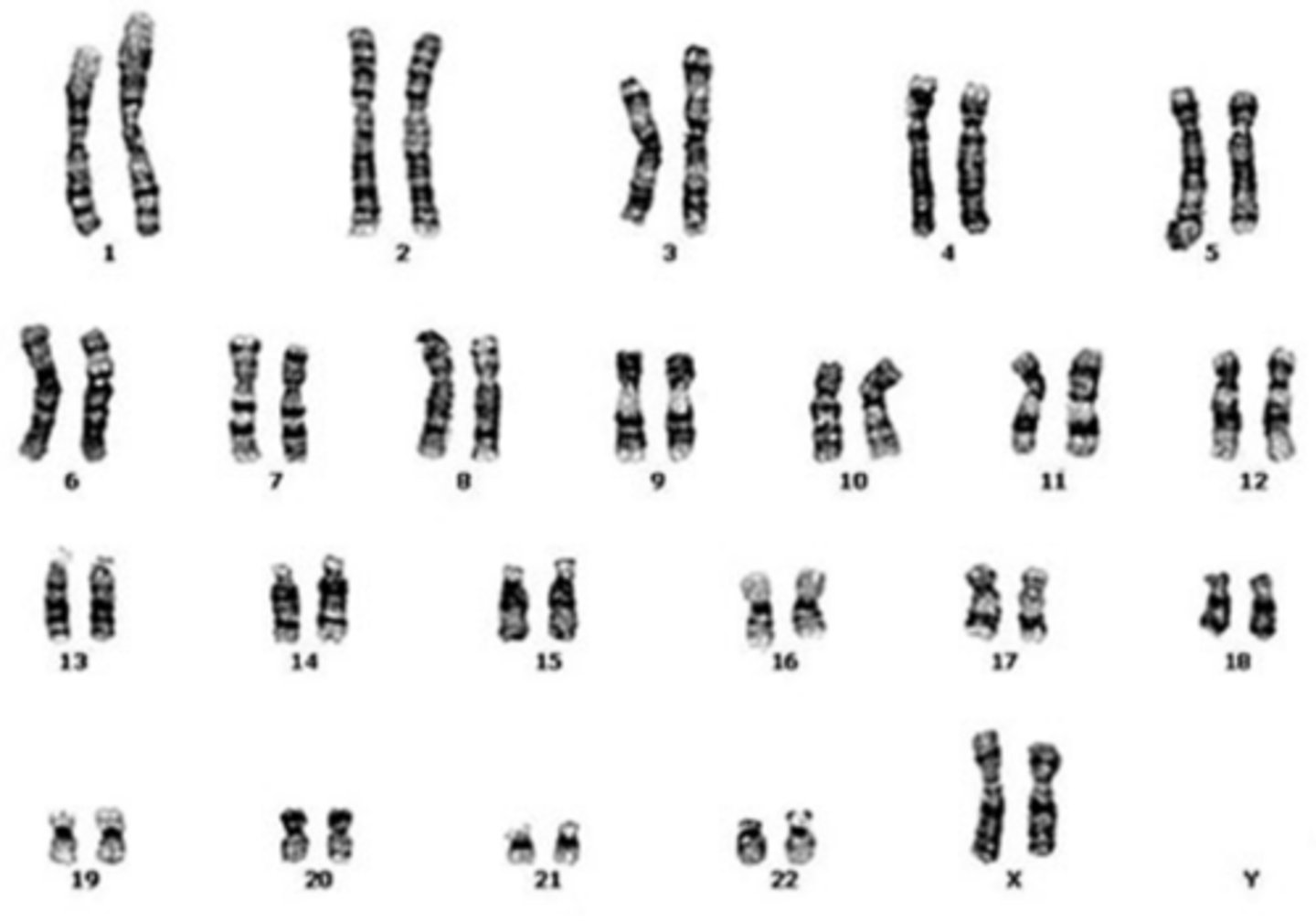
karyotype
a picture of all the chromosomes in a cell arranged in pairs
autosomes
all diploid, important in cancer development
2 categories of mammalian genes
1) housekeeping genes (10,000-15,0000)
2) tissue-specific genes (1000)
housekeeping genes
- 10,000-15,000 (more numerous)
- dedicated to maintaining fundamental biological functions (always on)
- common to all cell types
tissue-specific genes
- 1000 (less fewer)
- dedicated to production of proteins required by a specific differentiated cell
what creates phenotype?
proteins create it from genotype (nucleotide sequence)
types of proteins at work
- cytoskeleton (structural)
- extracellular matrix (structural)
- intermediary metabolism (biochem rxns)
- cell-cell signaling proteins and signal transduction proteins—central to cancer formation
cytoskeleton proteins
proteins involved in cellular scaffolding
types of cytoskeleton proteins
- intermediate filaments
- microfilaments
- microtubules
roles of cytoskeleton proteins
- cell shape
- motility
- cell division
- intracellular transport
*all matter in carcinogenesis and metastasis
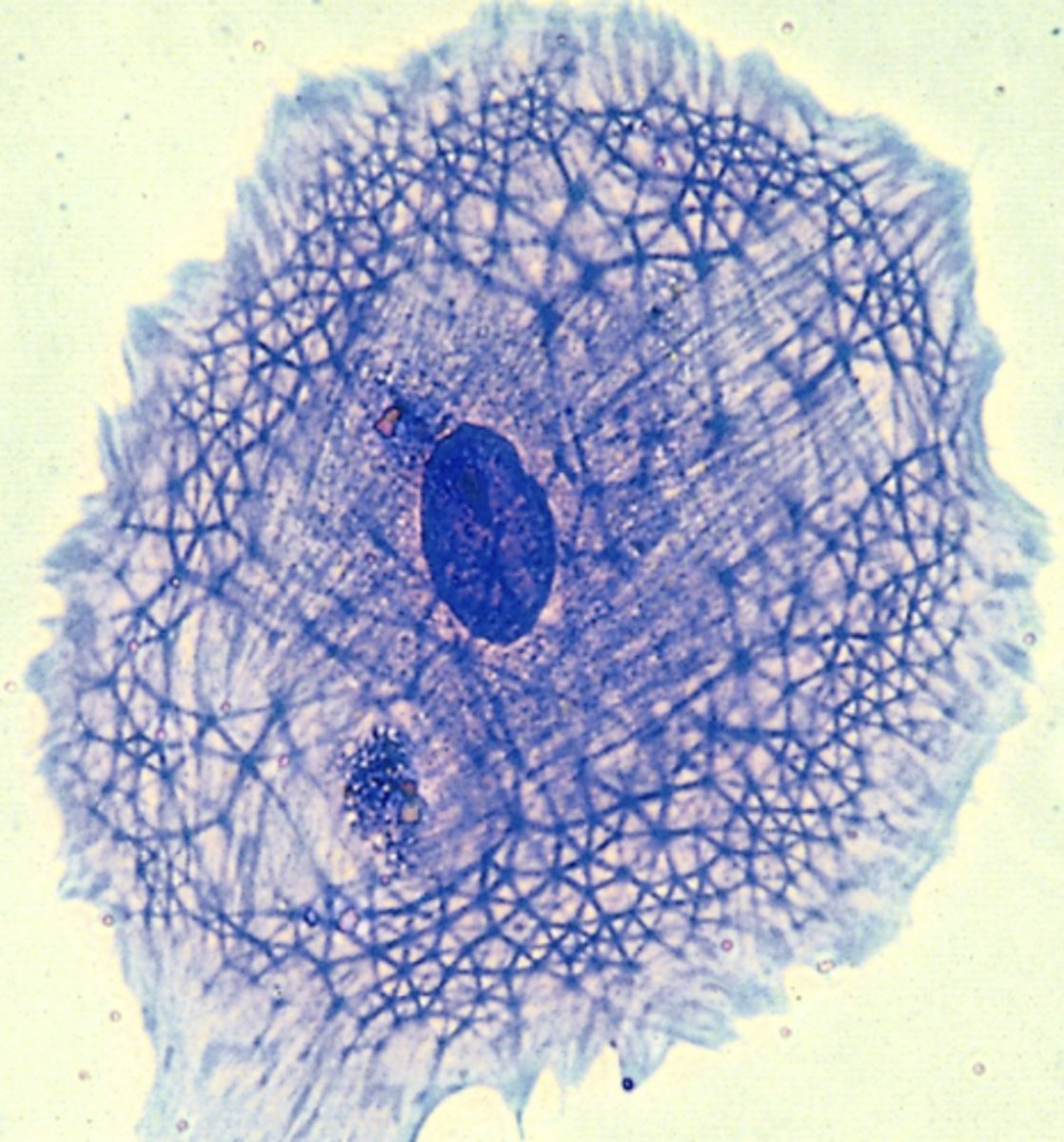
intermediate filaments
keratin, vimentin, laminin
- stationary; maintain cell shape within the cell
microfilaments
actin
- polymerize and depolymerize; movement and muscle contraction (w myosin)
microtubules
tubulin
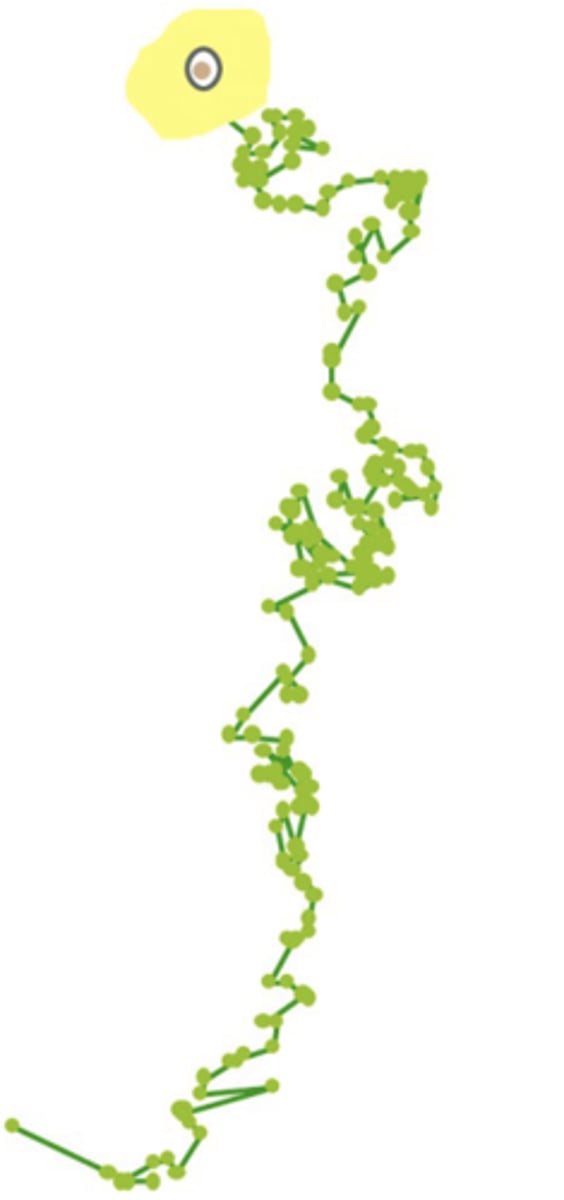
cell motility involves
cytoskeleton
human vascular endothelial cell movement plot
- experiment: growth factor is added to one end of culture dish to attract cells.
- each point represents an electronically plotted 10 min time interval.
**these cellular movements are critical to the formation of new blood vessels in cancer cells
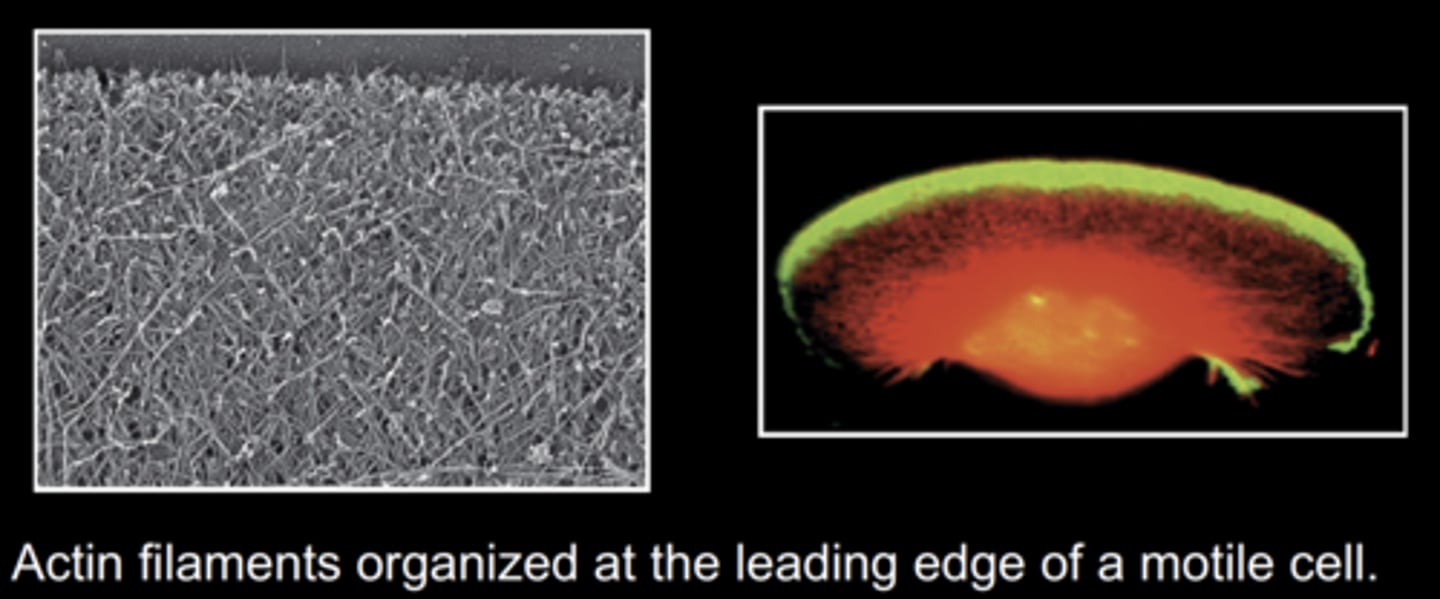
what allows cell motility?
actin filaments
extracellular matrix (ECM)
meshwork of collagen fibers, glycoproteins, hyaluronan, proteoglycans
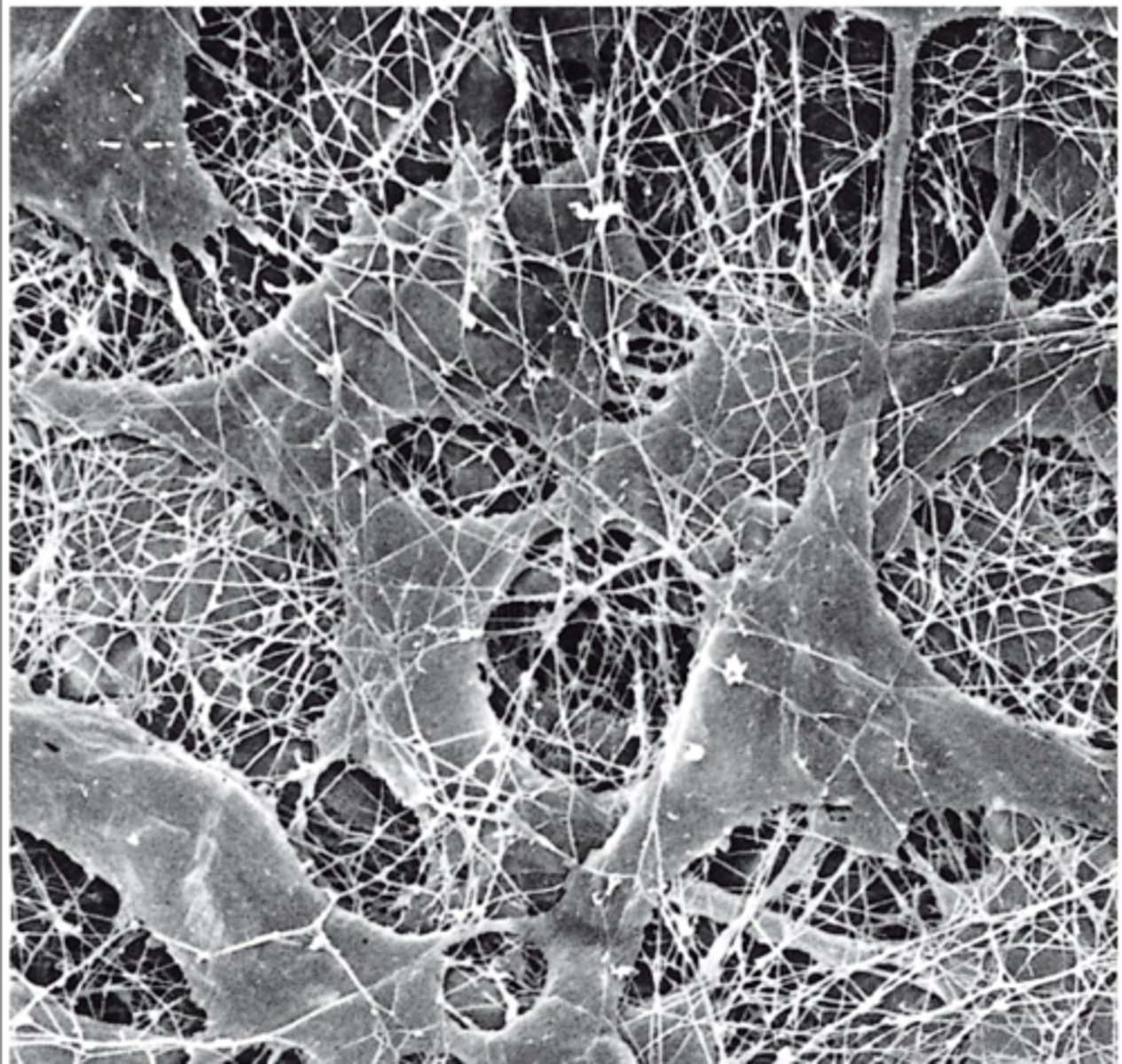
proteins of the extracellular matrix (ECM)
secreted by fibroblast cells
if every cell in a body has the same DNA, how do hundreds of cell types, each w distinct phenotypes, exists in the human body?
cellular differentiation and selective gene expression
nuclear equivalency
nucleus of 1 cell is enough to create a whole organism
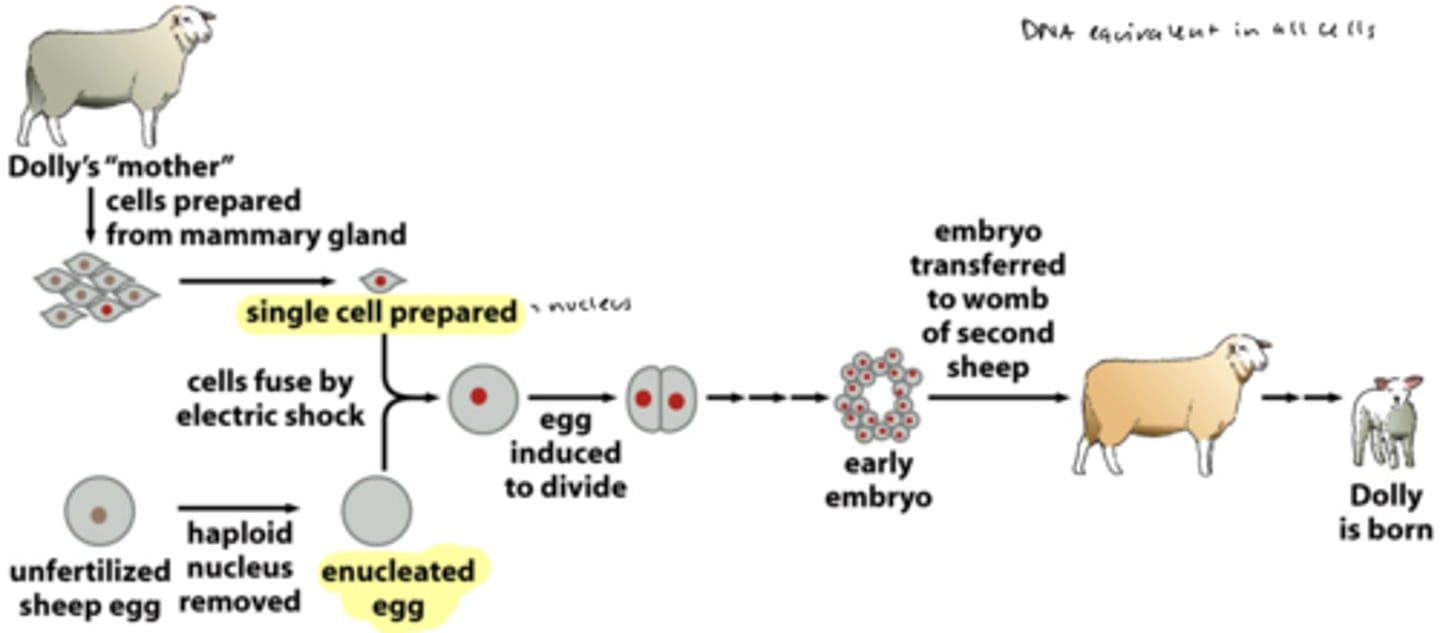
experiment to prove nuclear equivalency
cloning of Dolly from somatic cells
- cloned sheep from a somatic (diploid) cell from "mother" mammary gland = not by fertilization
gene regulation in cancer cells
the tight regulation is altered
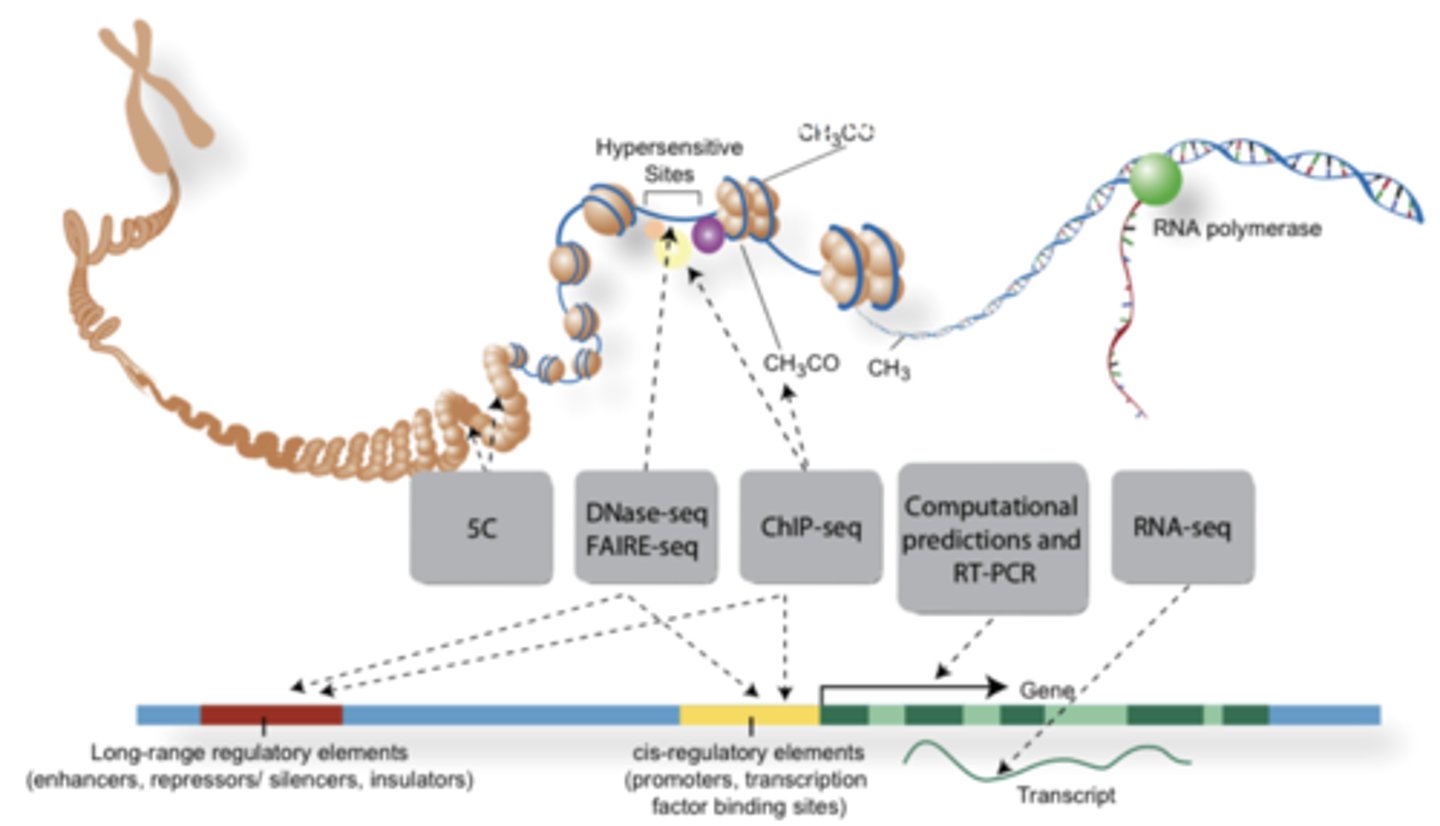
some modes of gene regulation at level of transcription
1) enhancer and silencer gene elements
2) transcription factors—combinations
3) alternative RNA splicing
4) change in chromatin state: methylation and acetylation of DNA and histones. (the histone code)
5) RNA interference
gene functional parts
- non-tx control regions (enhancers, promoters)
- transcribed sequences (become RNA)
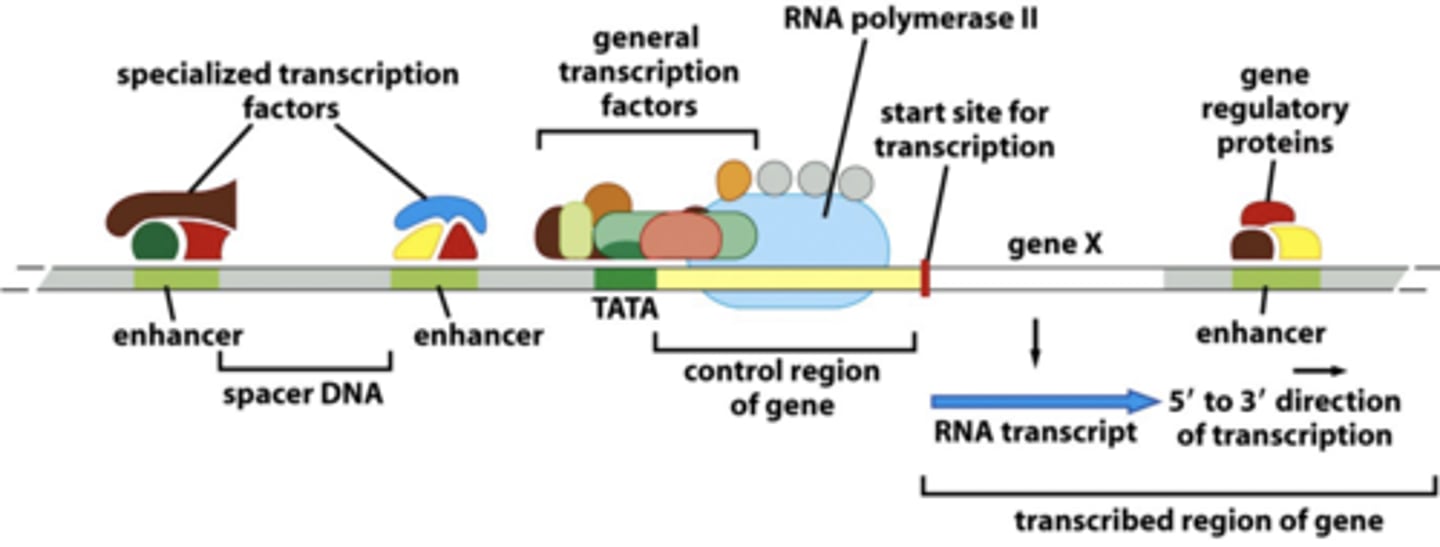
how do transcription factors control gene expression
by binding control regions and altering the DNA
eukaryotic enhancers
located several 1000 bps upstream and downstream of promoter
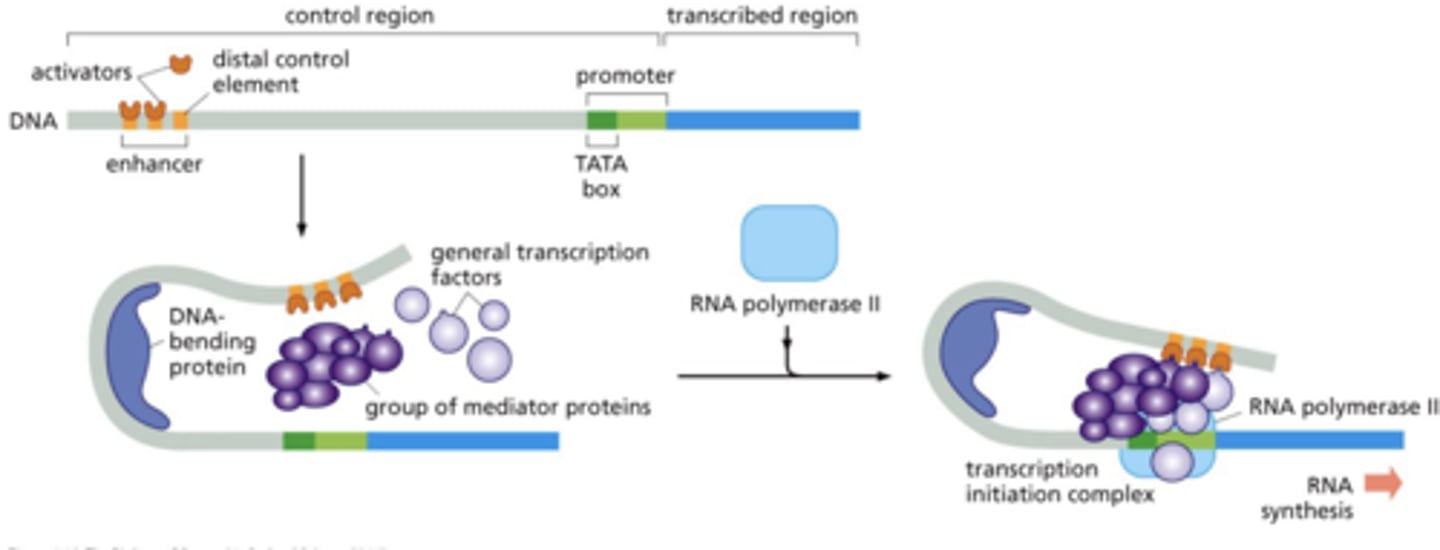
how do enhancers communicate with promoters?
DNA bends --> contact between initiation complex proteins and enhancer bound proteins
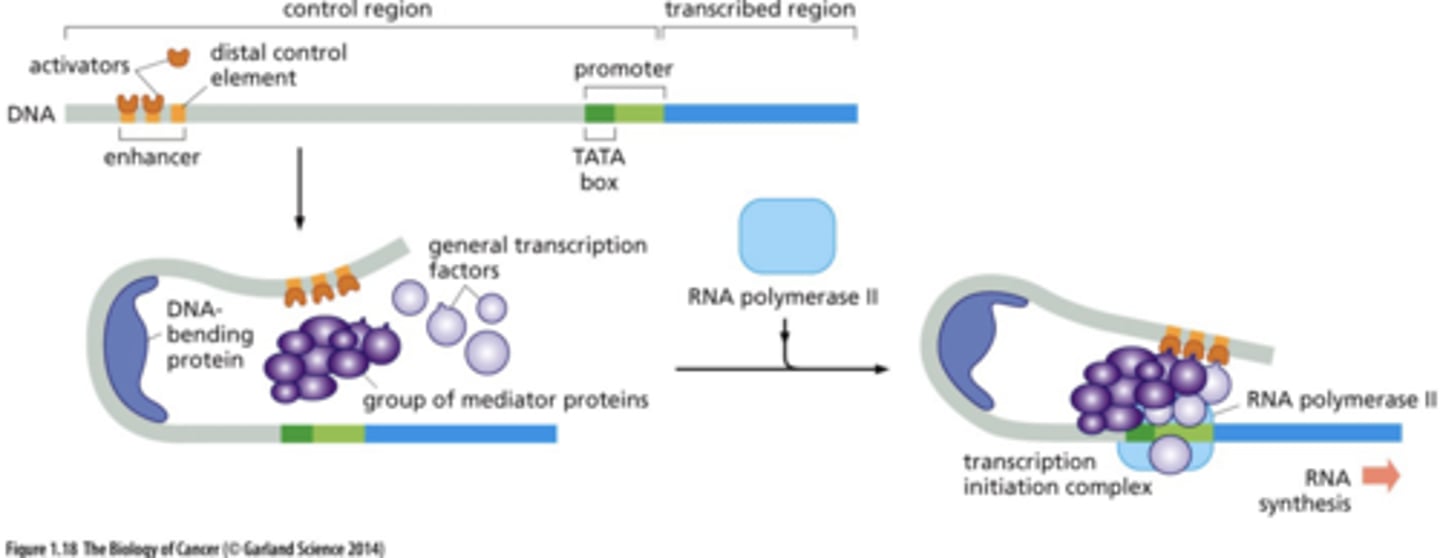
power of transcription factors
combinatorial action
pleiotropic action
combinatorial action
multiple transcription factors act in combination to create an expression program
pleiotropic action
a single type of transcription factor can elicit multiple changes within a cell by signaling a large cohort of responder genes.
what might happen if a pleiotropically acting tx factor malfunctions?
potential for launching a cancer program, where a mutated tx factor signals "gene on", affecting many responder genes
alternative RNA splicing
a single pre-mRNAs may be alternatively spliced to form distinct mRNAs and distinct proteins
- introns spliced out and exons ligated to form mature mRNA
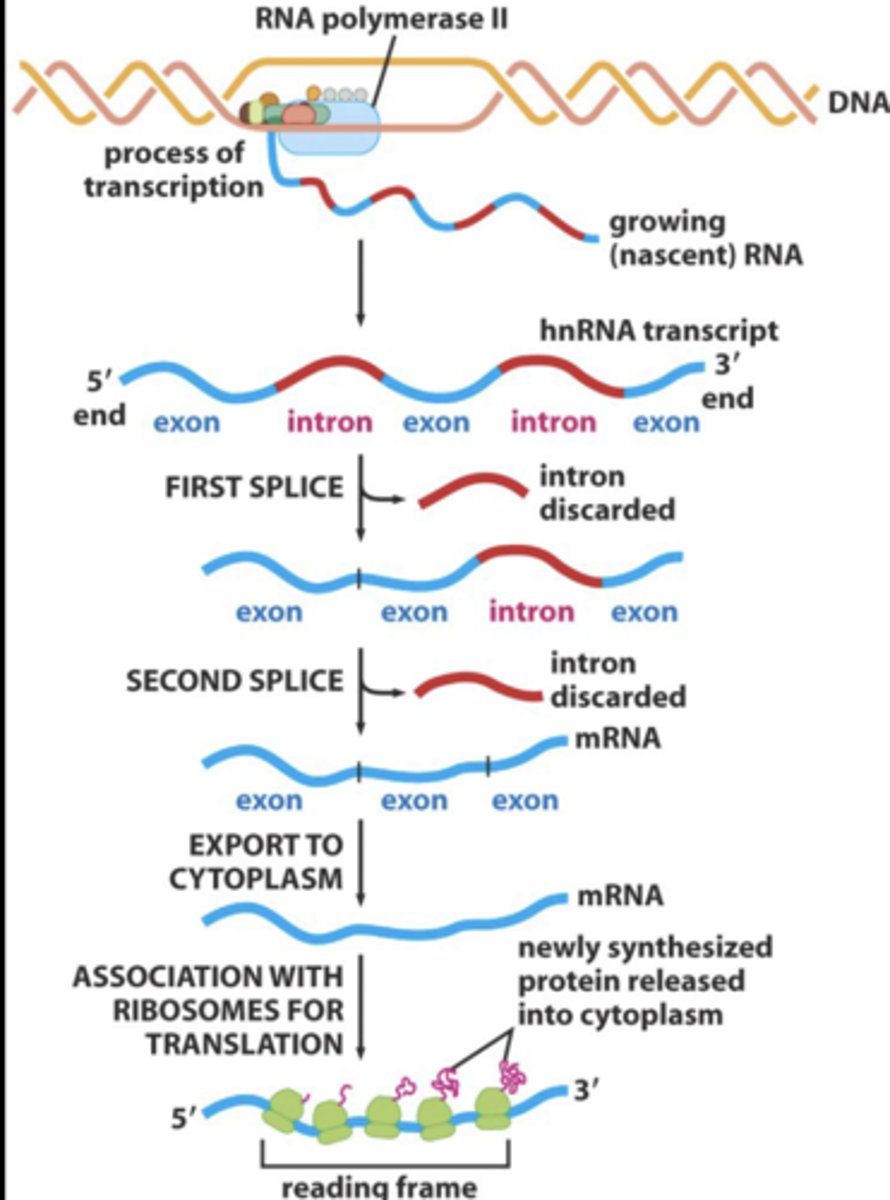
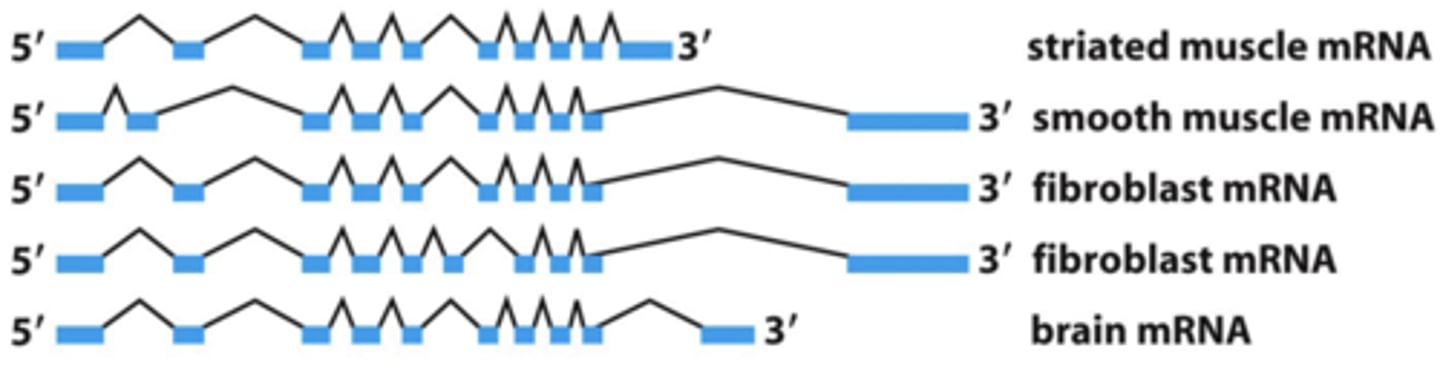
tissue specific alternative splicing of α-tropomyosin pre-mRNA
one RNA may encode several proteins.
- alternative splicing allows different proteins (like a family of proteins) to be expressed from the same gene, depending on cell type
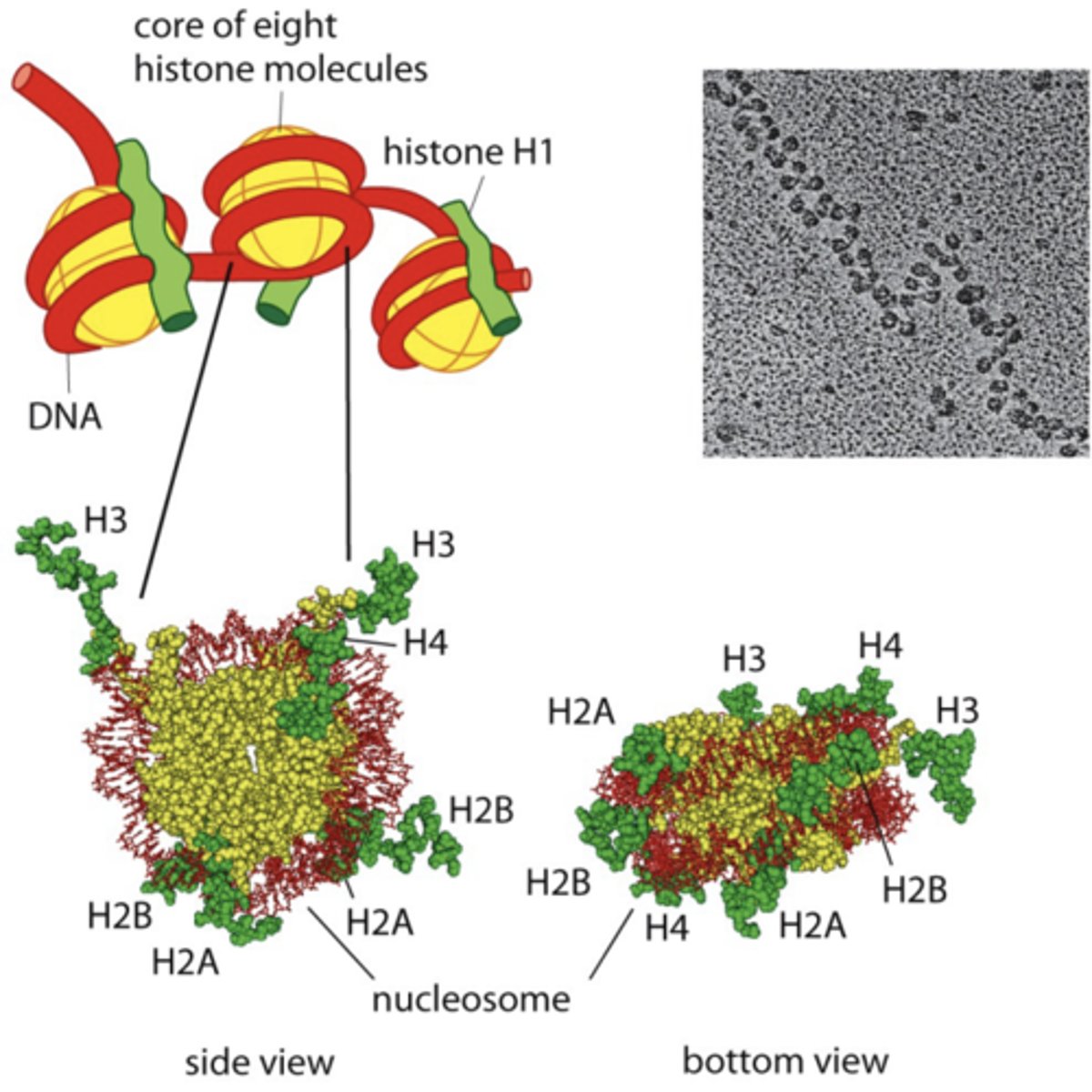
chromatin state affects gene expression
histones: positively charged octamer of proteins which (-) DNA wraps around --> packs DNA
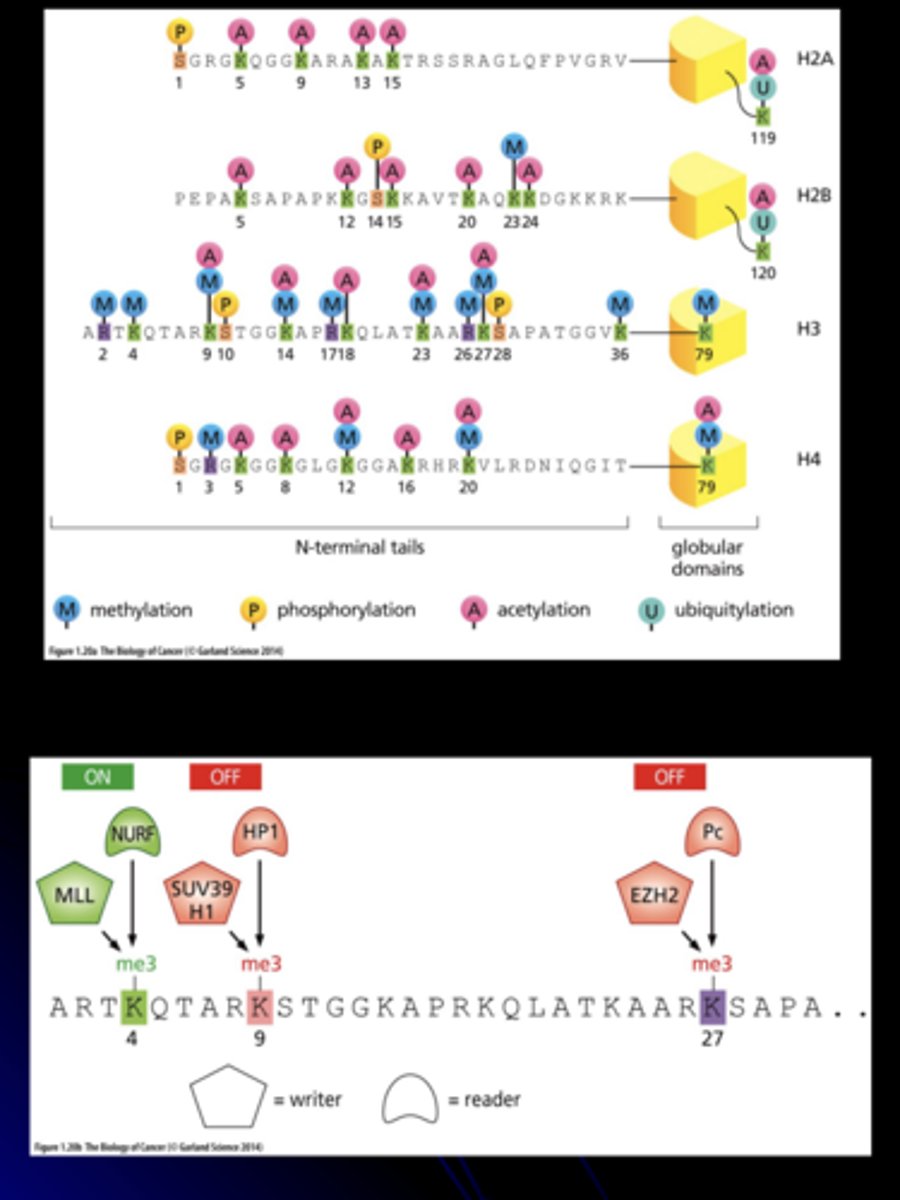
post-translational modification of histone tails
occurs via covalent attachment of methyl, acetyl, phosphate or ubiquitin groups
- "open" or "close" access of DNA to transcription factors by changing charge
RNA interference
control gene regulation
- short noncoding (functional) RNA sequences bind to specific target mRNAs to destroy the message.
- result: down regulation of gene product
example of RNA interference
microRNAs control level of mRNA in cytoplasm or efficiency of translating mRNAs
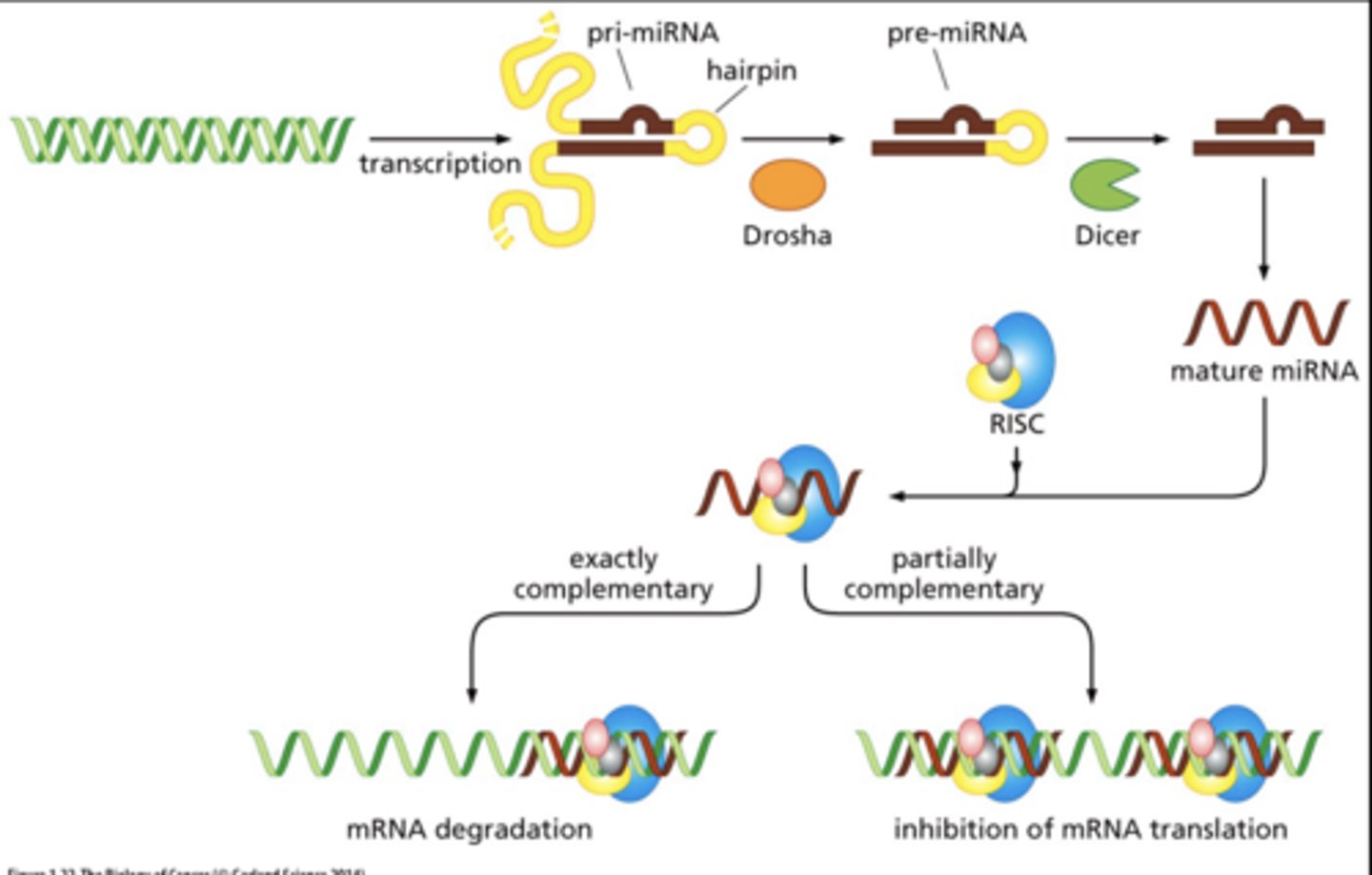
microRNAs and cancer
overexpression or loss of > dozen miRNA species has been associated with the formation of a variety of human cancers = "oncoMiRs"
chromosomal alterations in cancer cells
cancer cells accumulate various DNA mutations
- aberrant chromosomal number
- aberrant chromosomal structure:
• translocations (inversions, reciprocal)
• deletions
- amplification of chromosome
- extra copy of chromosome
- loss of entire chromosome
normal chromosomal complement
- giemsa stain binds phosphate groups on DNA, creates G-banding patterns.
- giemsa binds tighter to highly condensed DNA, creating a darker band.
- lighter bands=gene rich regions
- G-banding pattern is a chromosome identifier.
chromosome painting
- hybridize chromosomal specific, fluorescently labeled DNA probes to chromosomes
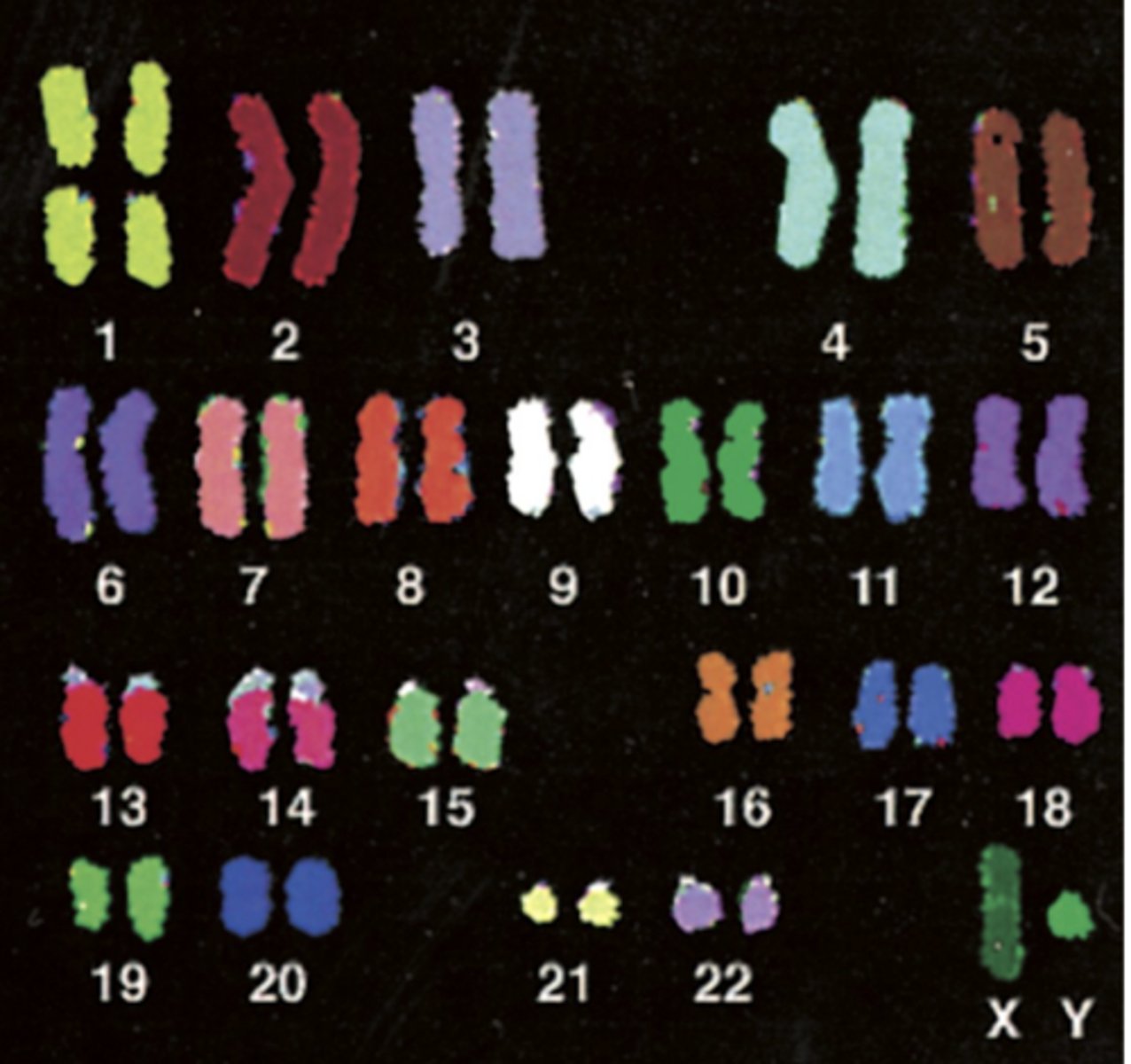
euploid
normal diploid karyotype = 22 autosomes and XX or XY
aneuploid karyotype
chromosomes present in:
- inappropriate numbers &/or
- structural abnormalities
present in >85% solid tumors
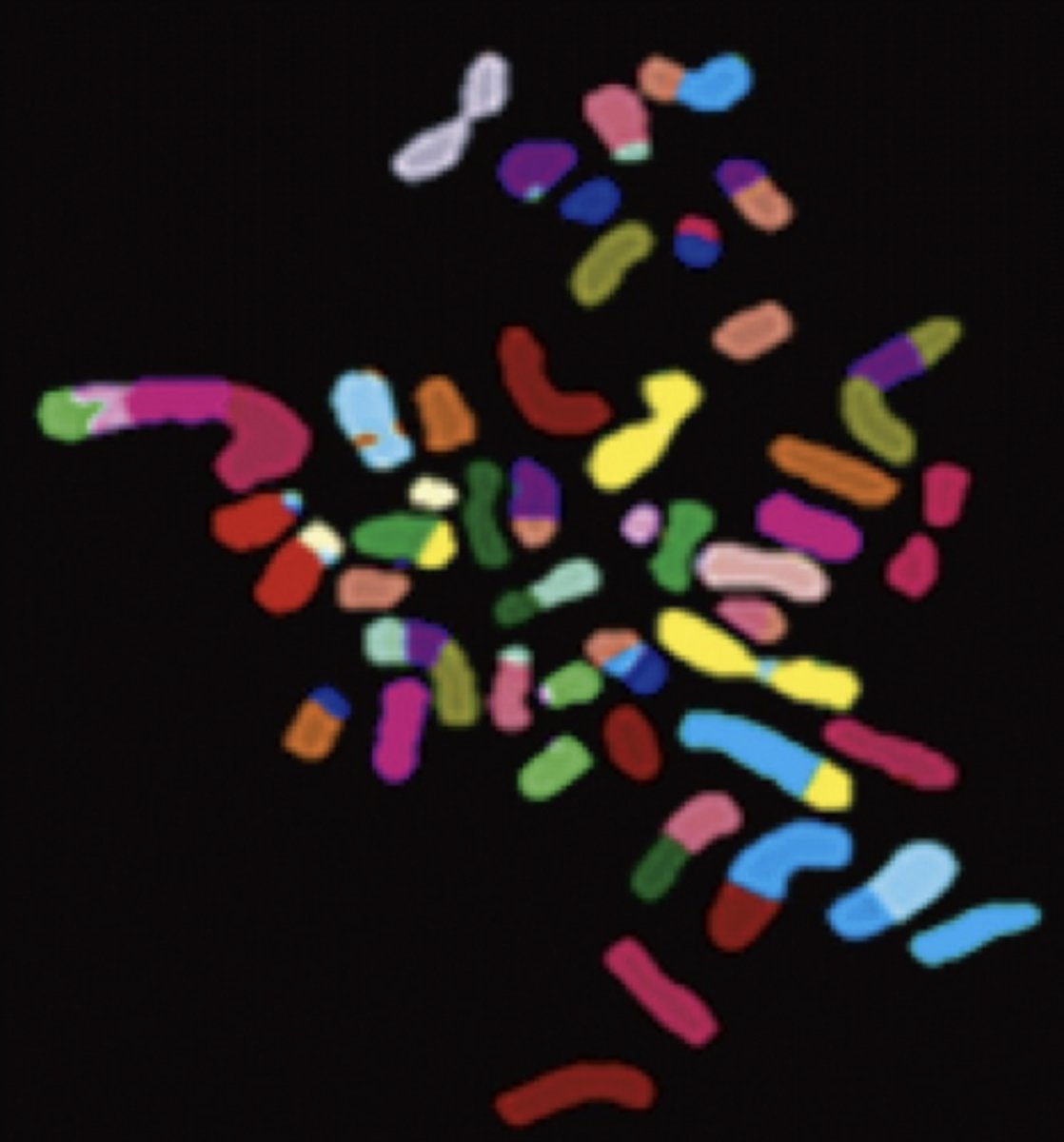
abnormal karyotype in cancer cells is characterized by
genomic instability
aneuploidy: how do incorrect chromosome #'s form in the cell?
normal cell in mitotic metaphase:
- chromosomes line up
- attach to microtubules of spindle (organized by centrosome)
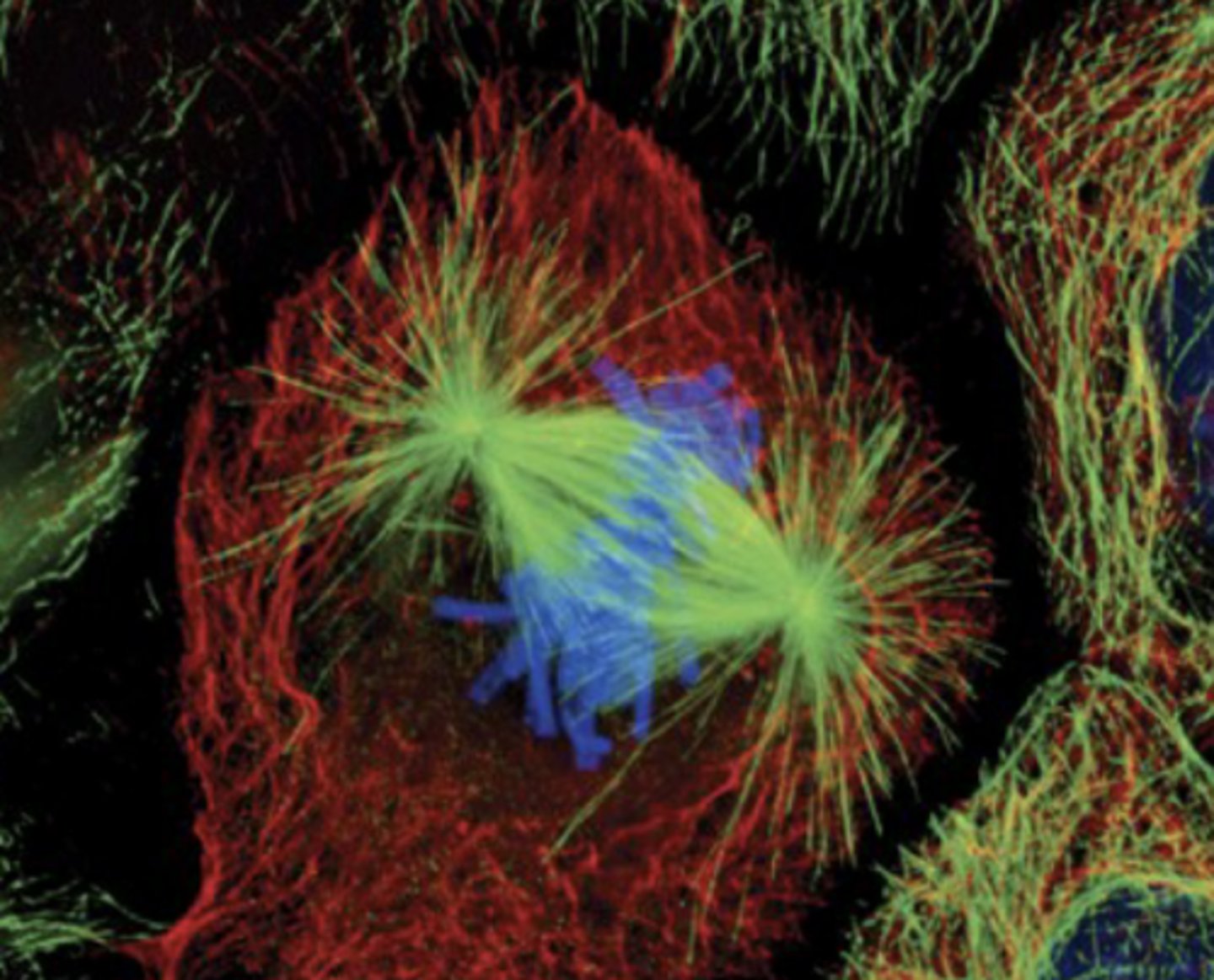
cancer cells exhibit
chromosomal instability (CIN)
- in vivo & in vitro
- consequential or causal?
what can lead to changes in chromosome #?
chromosome mis-segregation during mitosis
ex) non-disjunction
non-disjunction events
both sister chromatids of a chromosome pulled to one cell; other cell does not receive a copy
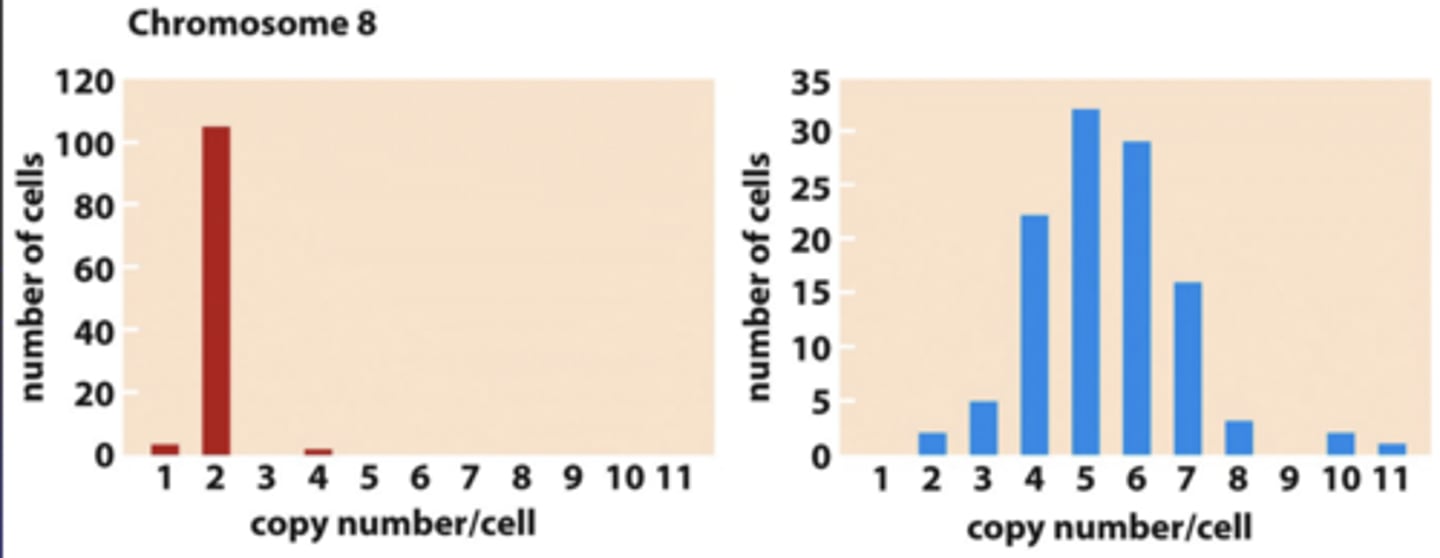
chromosome mis-segregation during mitosis can lead to
changes in chromosome #
- improperly assembled spindles
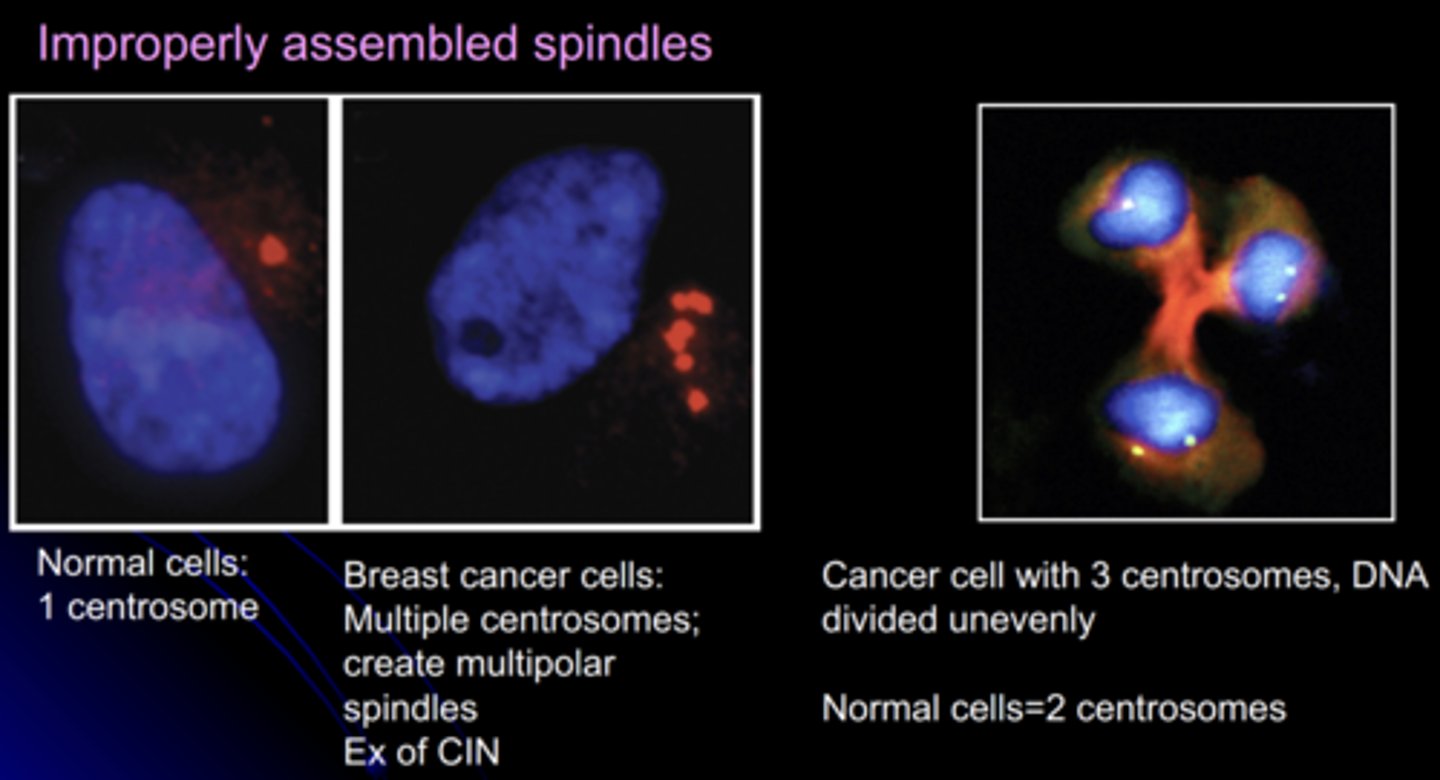
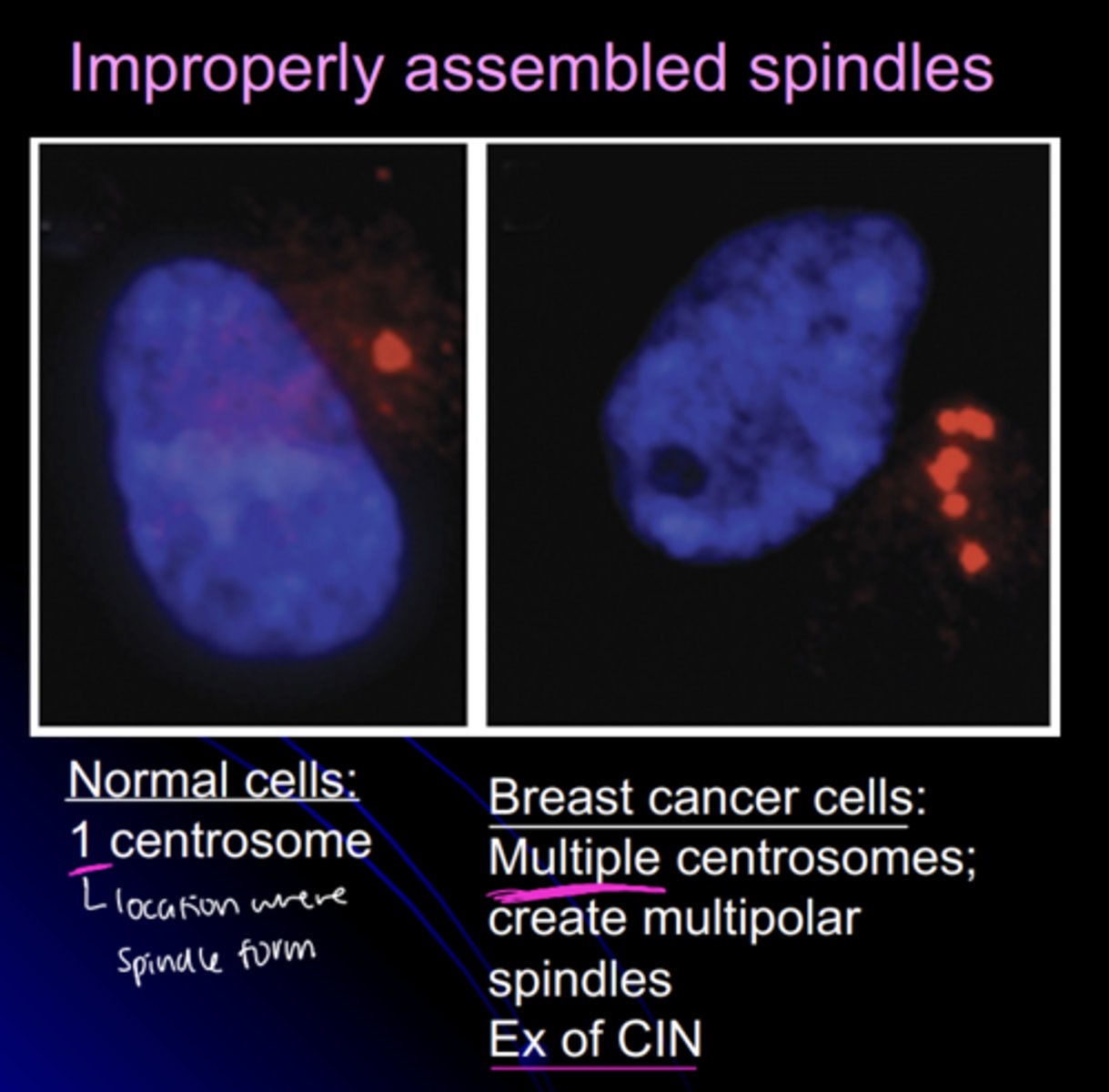
spindles in normal cells vs. breast cancer cells
normal cells:
- 1 centrosome
cancer cells:
- multiple centrosomes
- multipolar spindles
- example of CIN
translocations
fusion of two chromosomal segments that are not normally attached
types of translocations
- interstitial deletions
- reciprocal translocations
- inversions
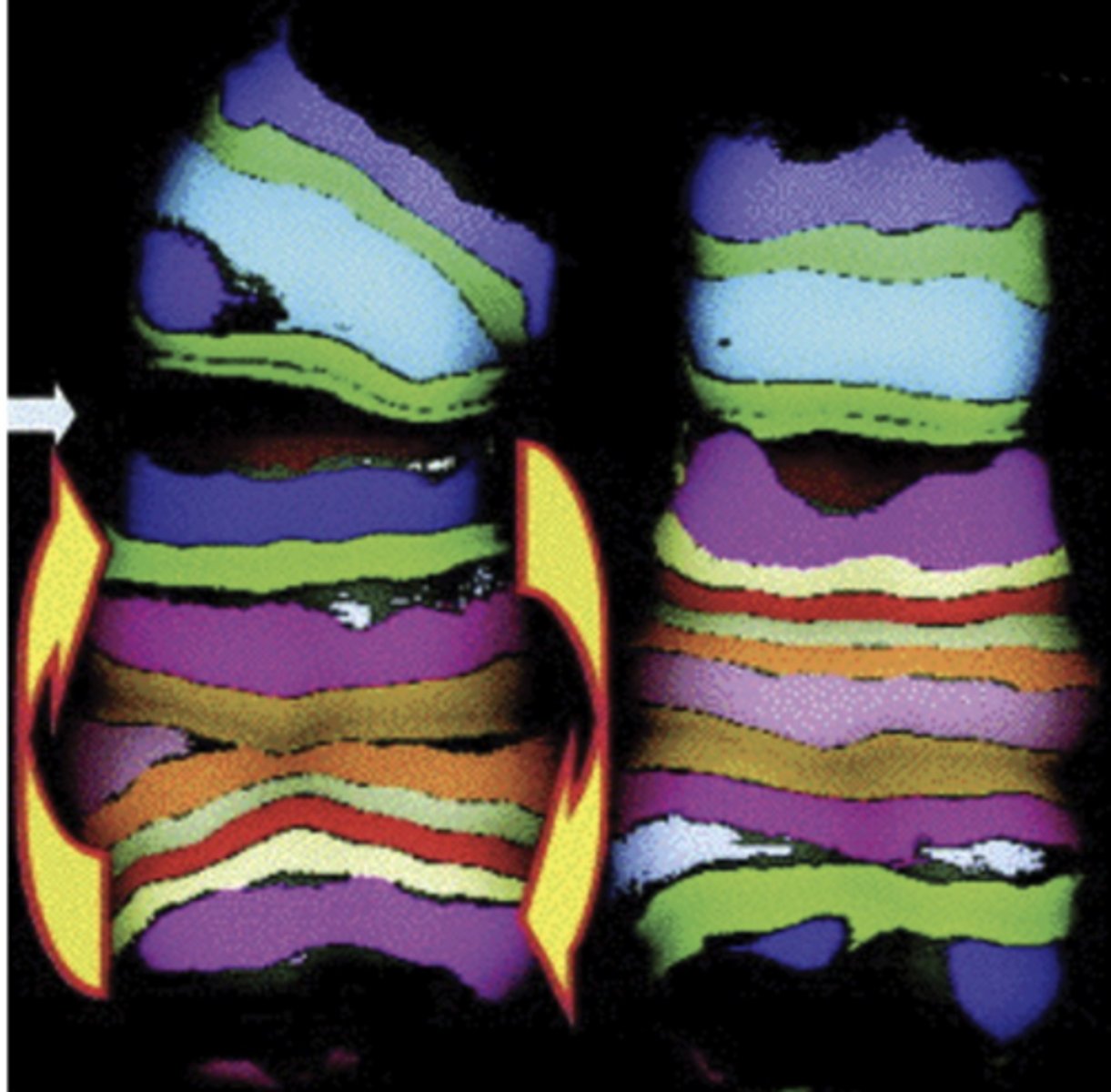
chromosomal inversion
when part of the chromosome becomes oriented in the reverse of its usual direction
- note specificity achieved: break location and inversion clearly identifiable
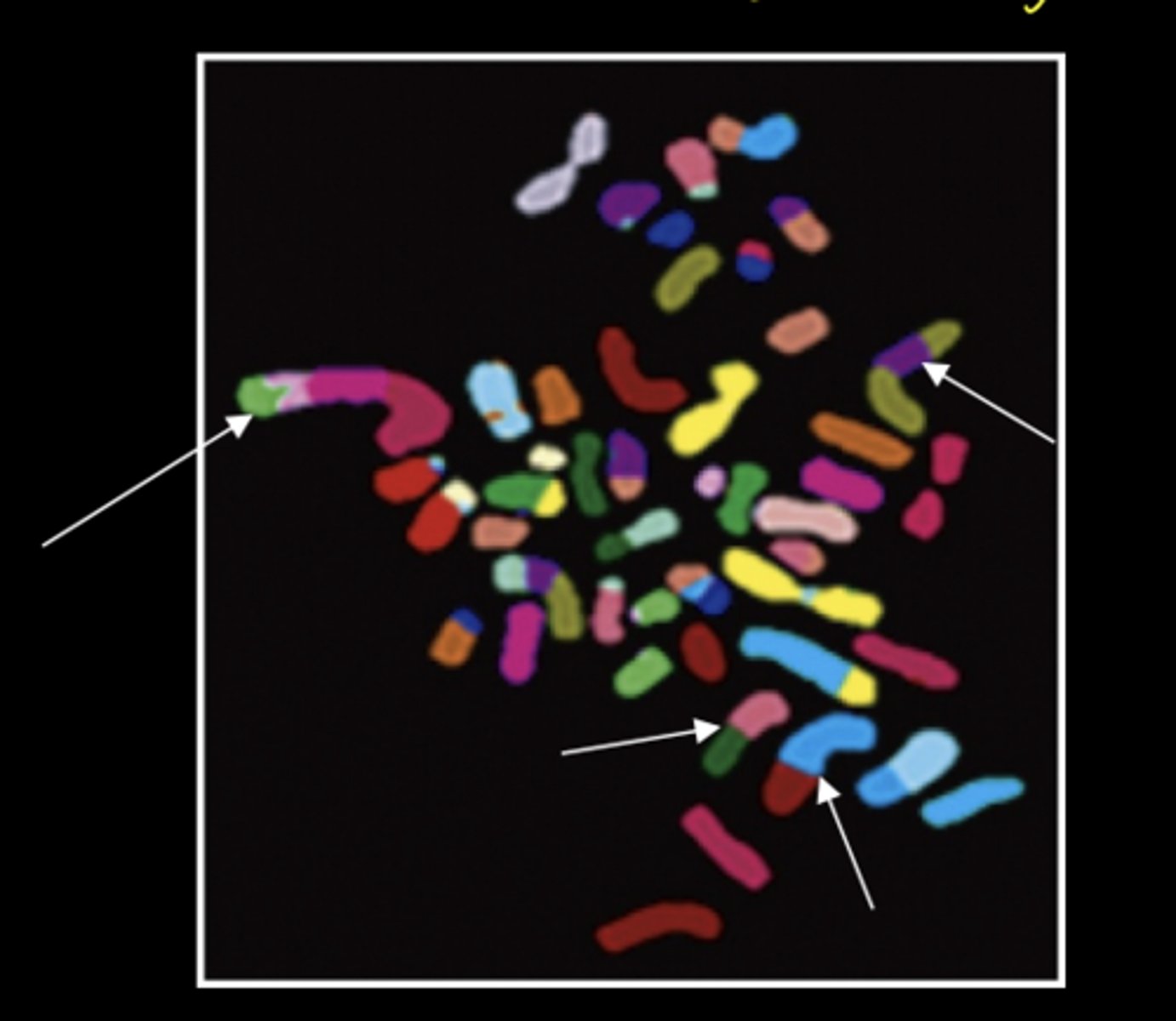
example of chromosomal inversion
FISH labeled intrachromosomal subregions reveals chromosome 5 inversion in a plutonium worker's cells
- power of FISH in determining aneuploidy
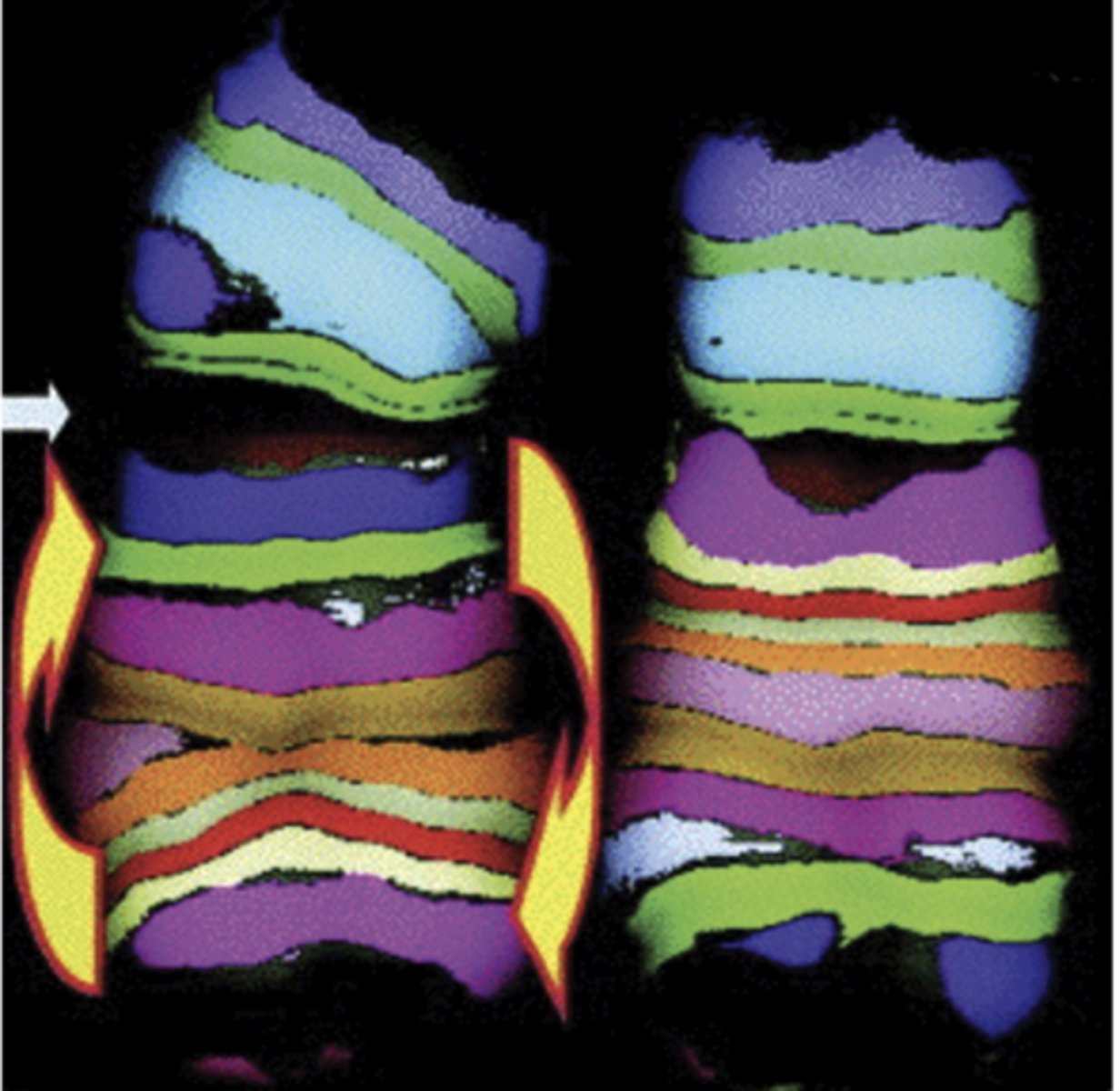
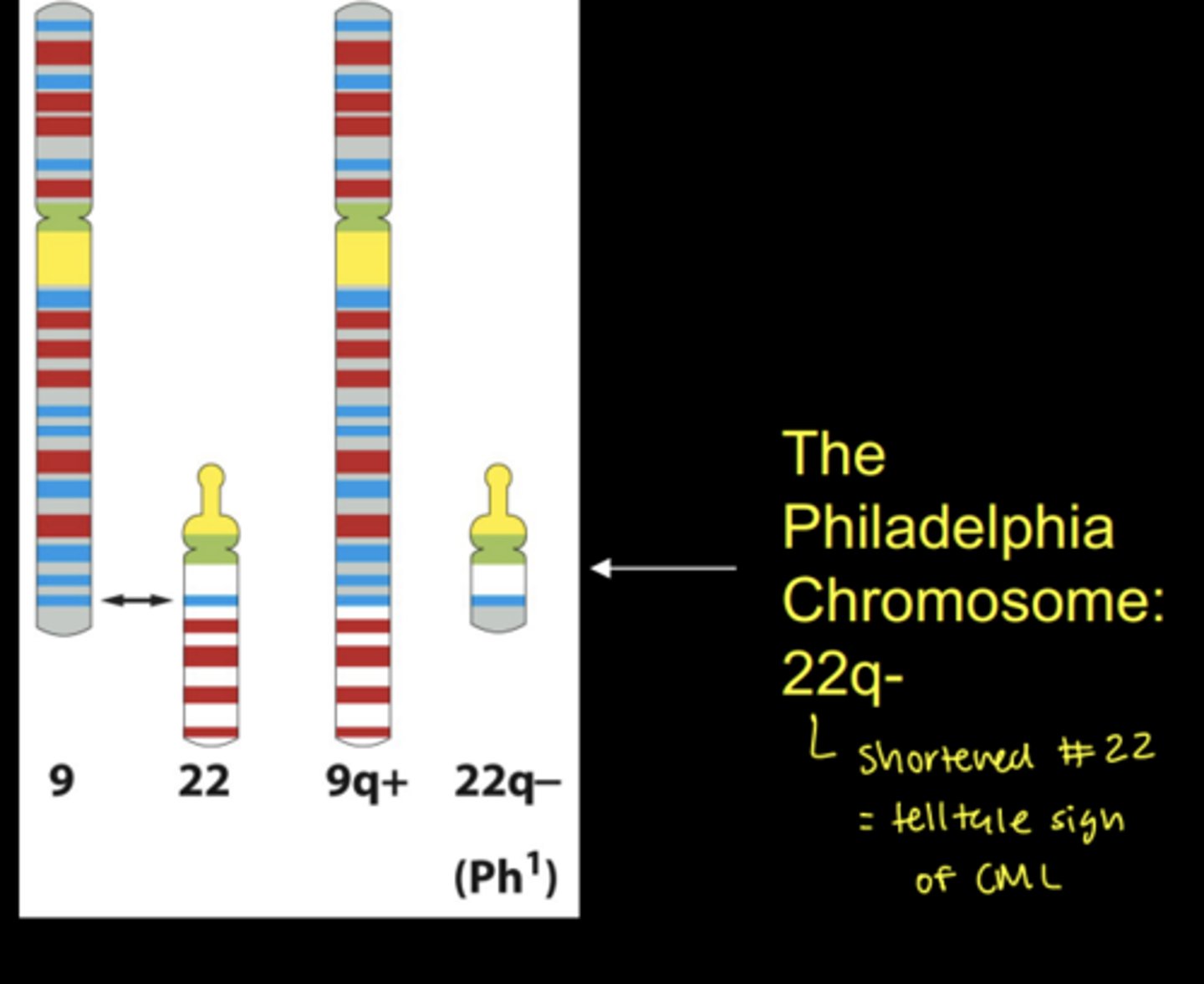
reciprocal translocations
chromosomal segments exchanged between nonhomologous chromosomes
- ex) exchange between 9 and 22 forms CML, chronic myelongenous leukemia
interstitial deletions
gene segment between arrows deleted and the flanking ends rejoined
- cancer ex. loss of inhibitory gene
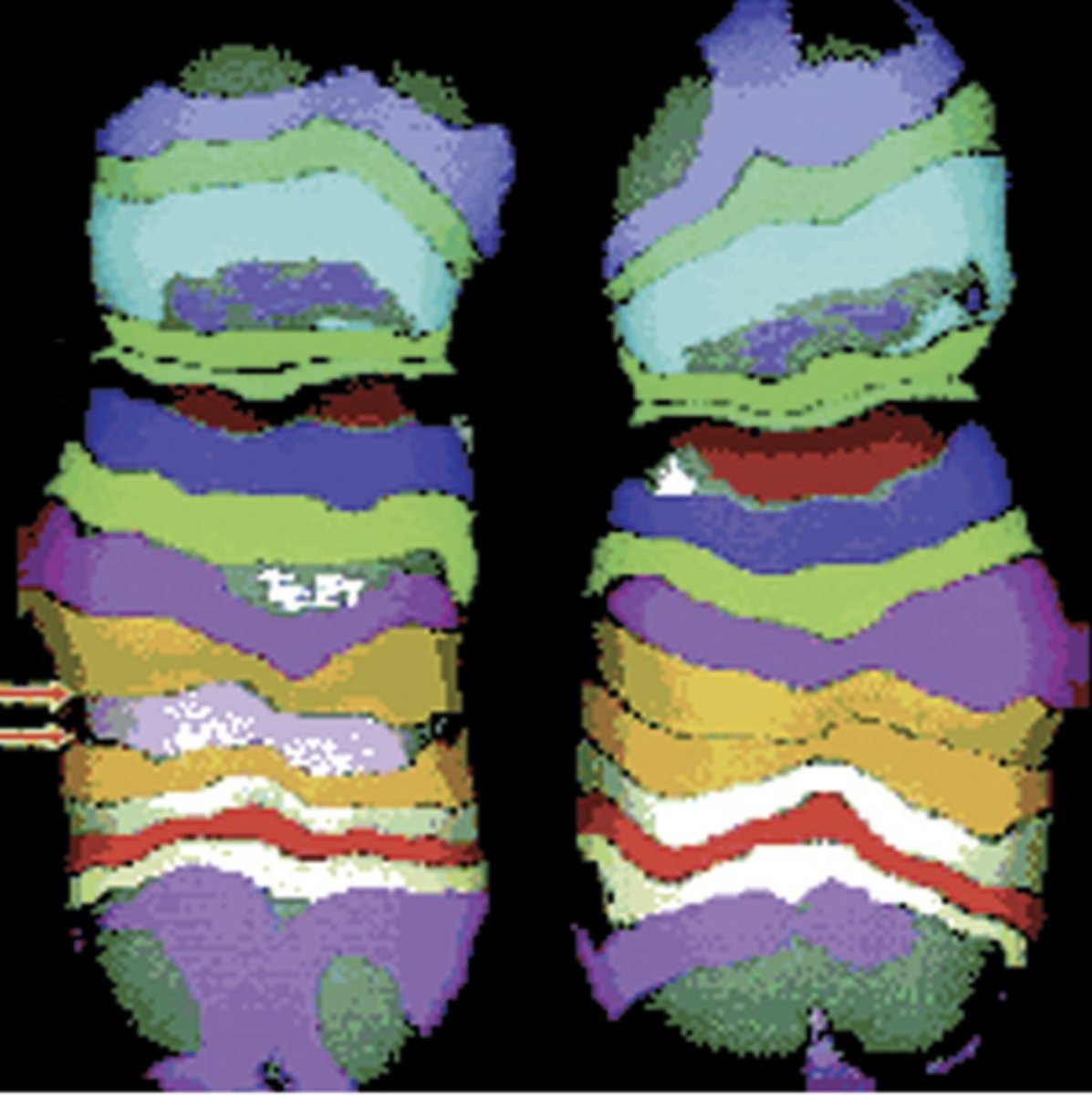
what would not count as an interstitial deletion?
a total loss of a chromosomal segment since no segment rejoins to itself.
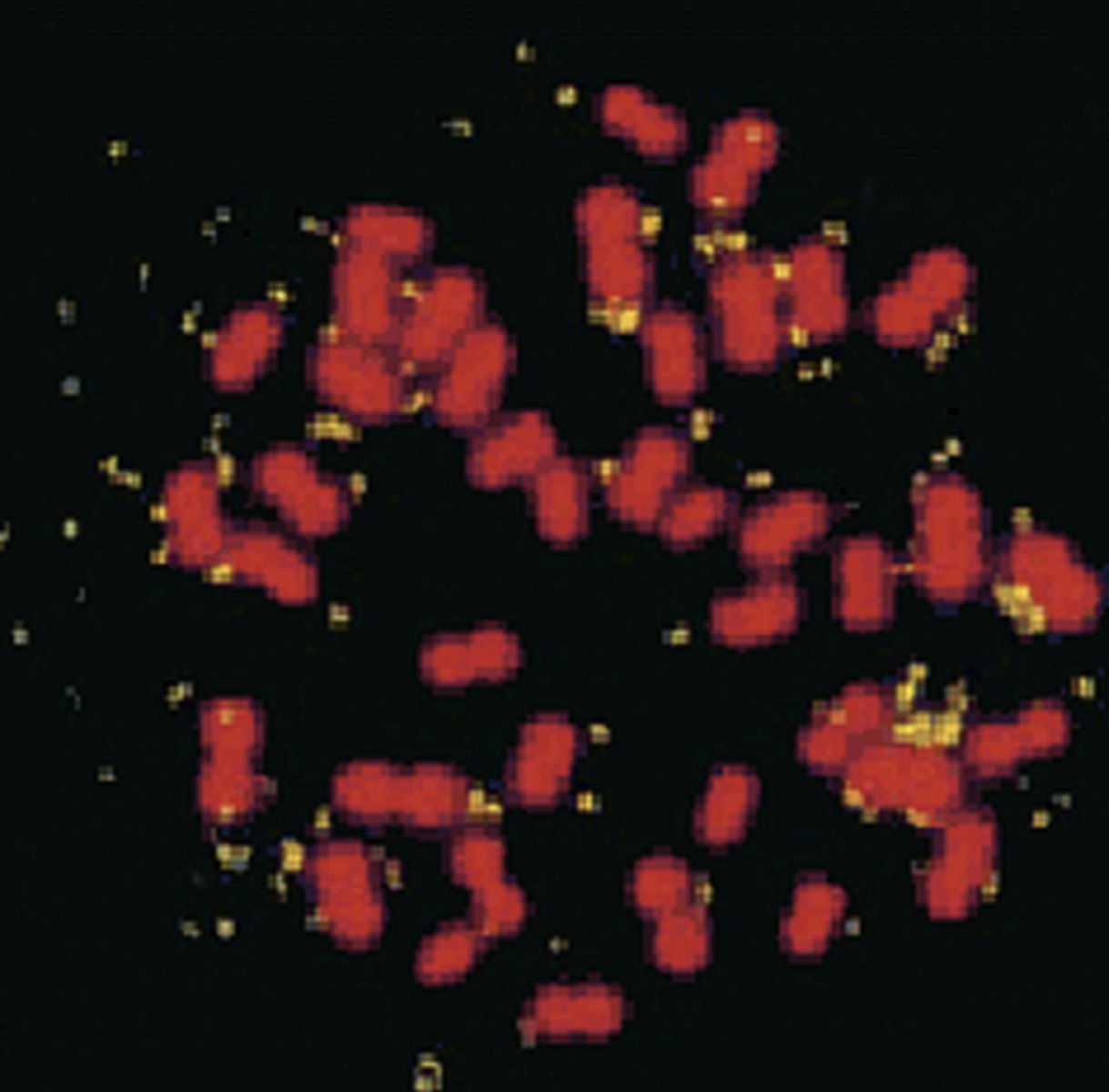
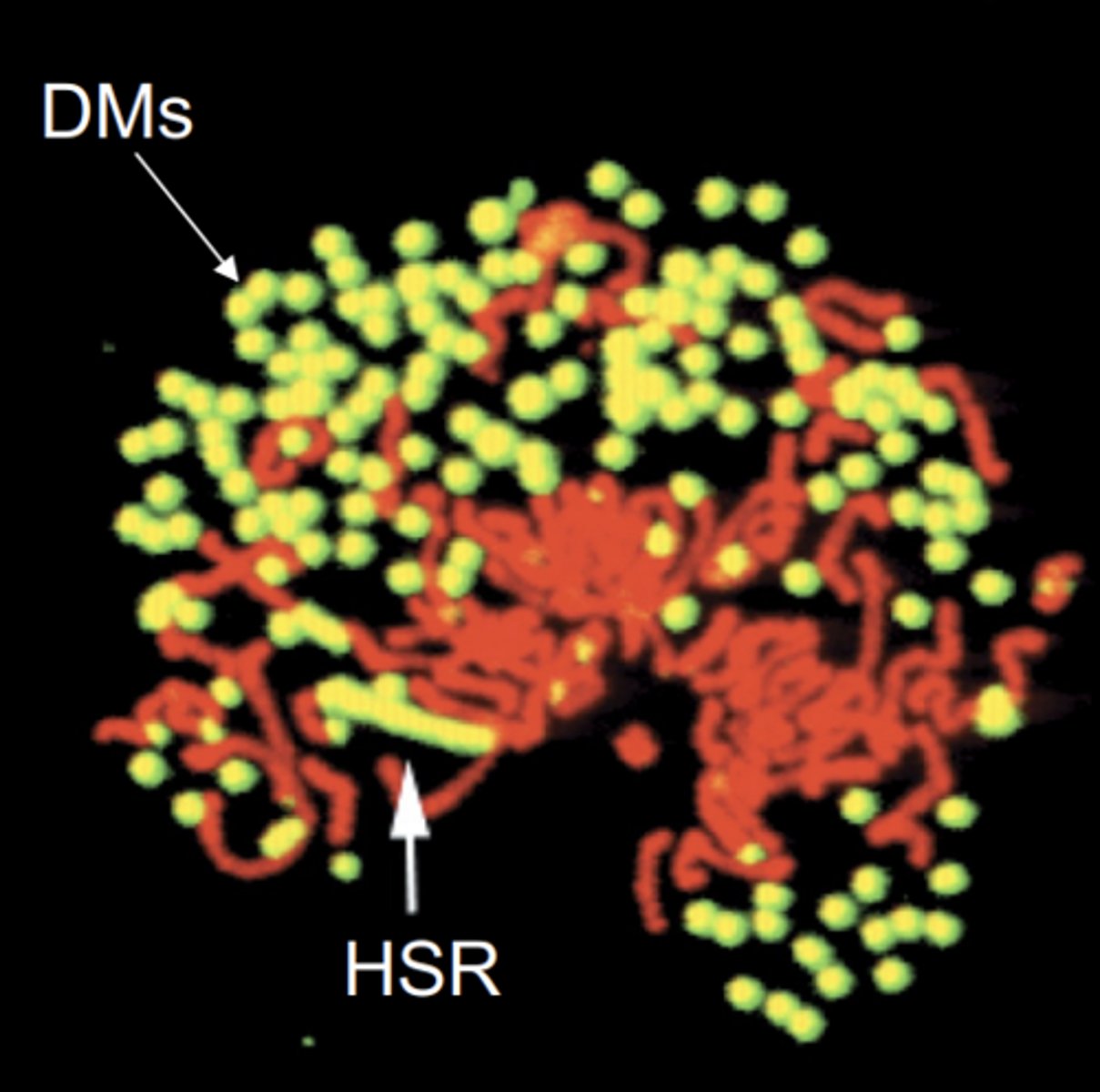
amplifications
increase in gene copy number
2 types of amplifications
1) HSR: homogeneously staining region
2) DMs: double minutes
HSR amplification
repeated rounds of chromosomal reduplication result in an elongated chromosome with head to tail repeats of a particular segment
ex) giemsa stain reveals banding of a typical streched appearance, as well as highly abnormal karyotype
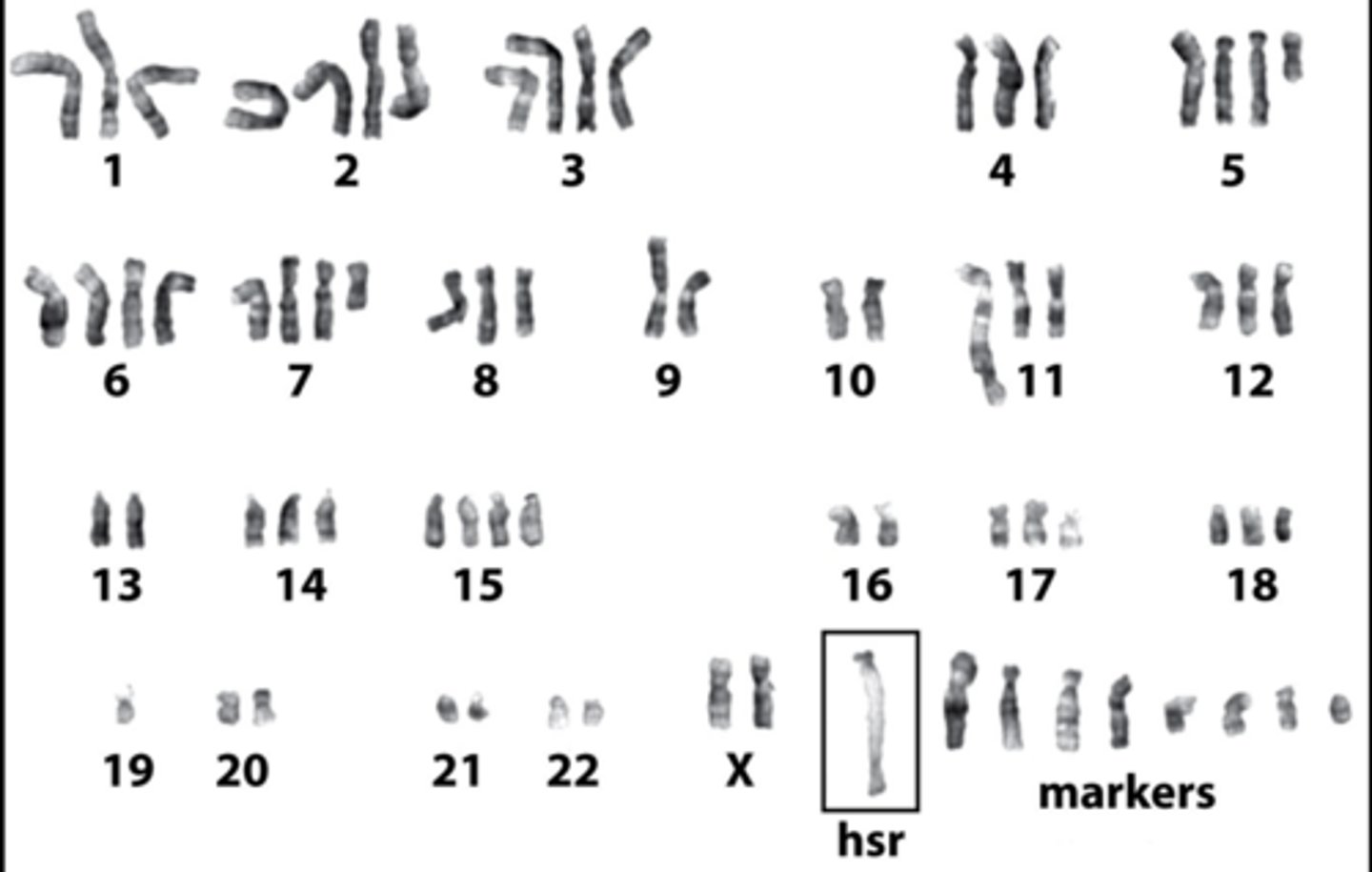
double minutes
extrachromosomal, autonomously replicating segments that were originally part of the chromosome
ex) breast cancer cells with amplified HER2/neu oncogene borne on DMs—causes a great increase in the dosage of that gene
2 types of cancer genes
oncogenes and tumor suppressor genes
oncogenes
genes that have the potential to cause cancer once mutated; can enhance growth
tumor suppressor genes
a gene whose protein product inhibits cell division --> when mutated causes cancer
besides oncogenes and tumor surpressors, cancer cells also have __________ mutations
passenger
passenger mutation
mutation in a cancer that does not contribute to tumorgenesis
can a tumor have HSR and DMs in the same cell?
yes, COLO320 tumor cells
paranchymal cells
functional cell type within a tissue that performs the specific tasks associated with that tissue's role
- tumor cells may obstruct or replace them, disrupting normal tissue function
somatic vs. germline mutations
germline mutations are only 5-10% and can be passed down/inherited
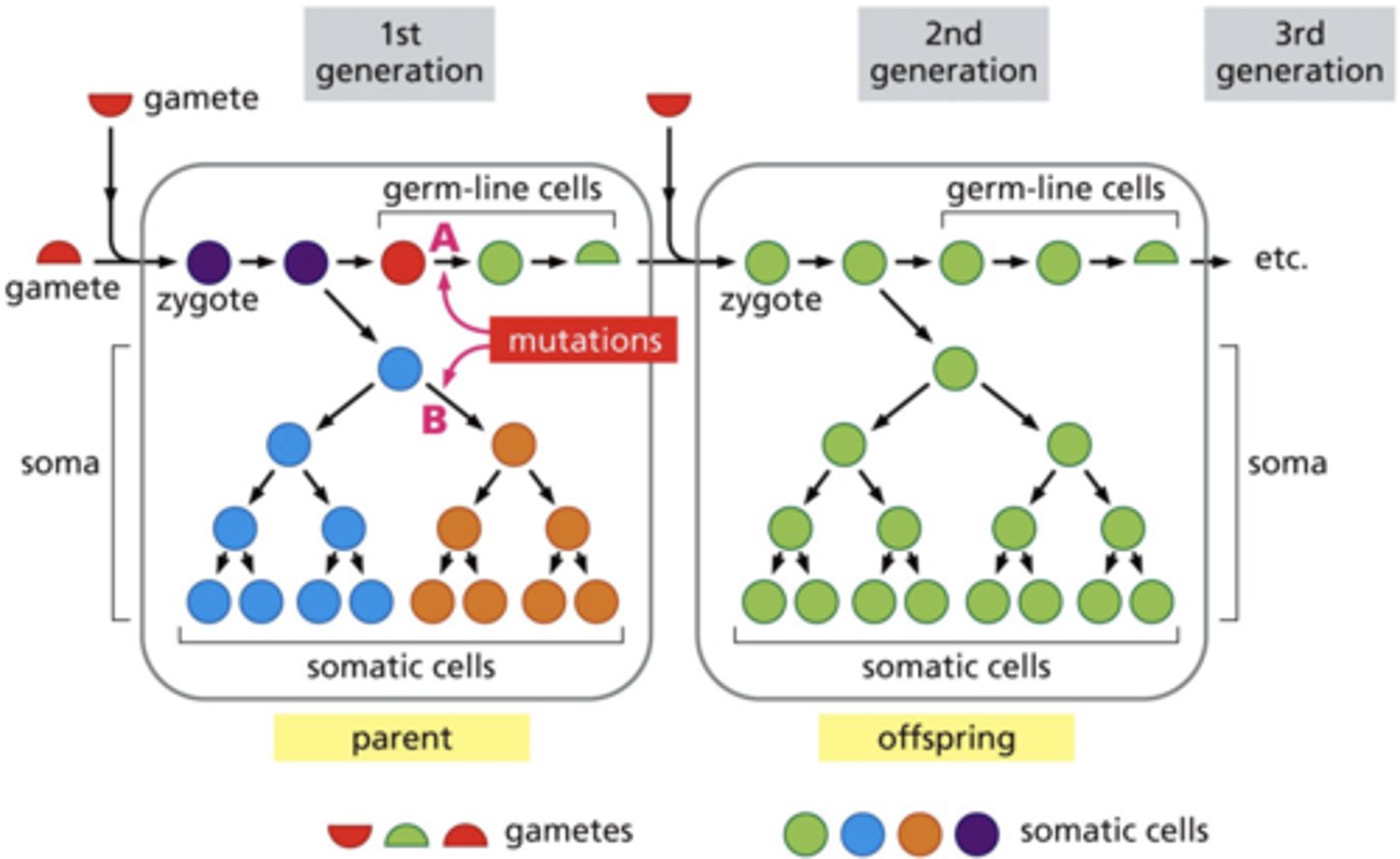
somatic mutations
- not inherited by offspring
- form a clonal population from a single progenitor
germline mutations
- occur in sperm, eggs or their precursor cells
- affect offspring
how can mutations occur with cellular repair mechanisms in place?
while cellular repair mechanisms in place to maintain genomic integrity, they are not infallible
early detection methods
screening for cancers
driver mutations
protein biomarkers
effective cancer screening tests (FDA approved testing)
- mammography (breast cancer)
- colonoscopy (colorectal cancer)
- PAP Smear (cervical cancer)
- PSA test (prostate cancer, efficacy?)
genomics and oncology in cancer testing
using DNA sequencing and RNA transcripts to determine the structure and function of genomes
genomics and oncology testing methods
1) karyotype & FISH
2) microarrays
3) GWAS single nucleotide polymorphism screen
4) NGS next generation sequencing – paired comparison of patient’s normal tissue and tumor tissue
5) genomic assays for circulating tumor cells DNA and protein biomarkers (CANCERSEEK)
2) microarrays reveal
selective gene expression
gene expression microarray analysis
allows evaluation of thousands of gene expression patterns in a given cell type
- small red bars = high gene exp
- small green bars = low gene exp
- vertical axis= exp levels of 1800 genes in this array
- horizontal axis = mRNAs from 142 different human tumors
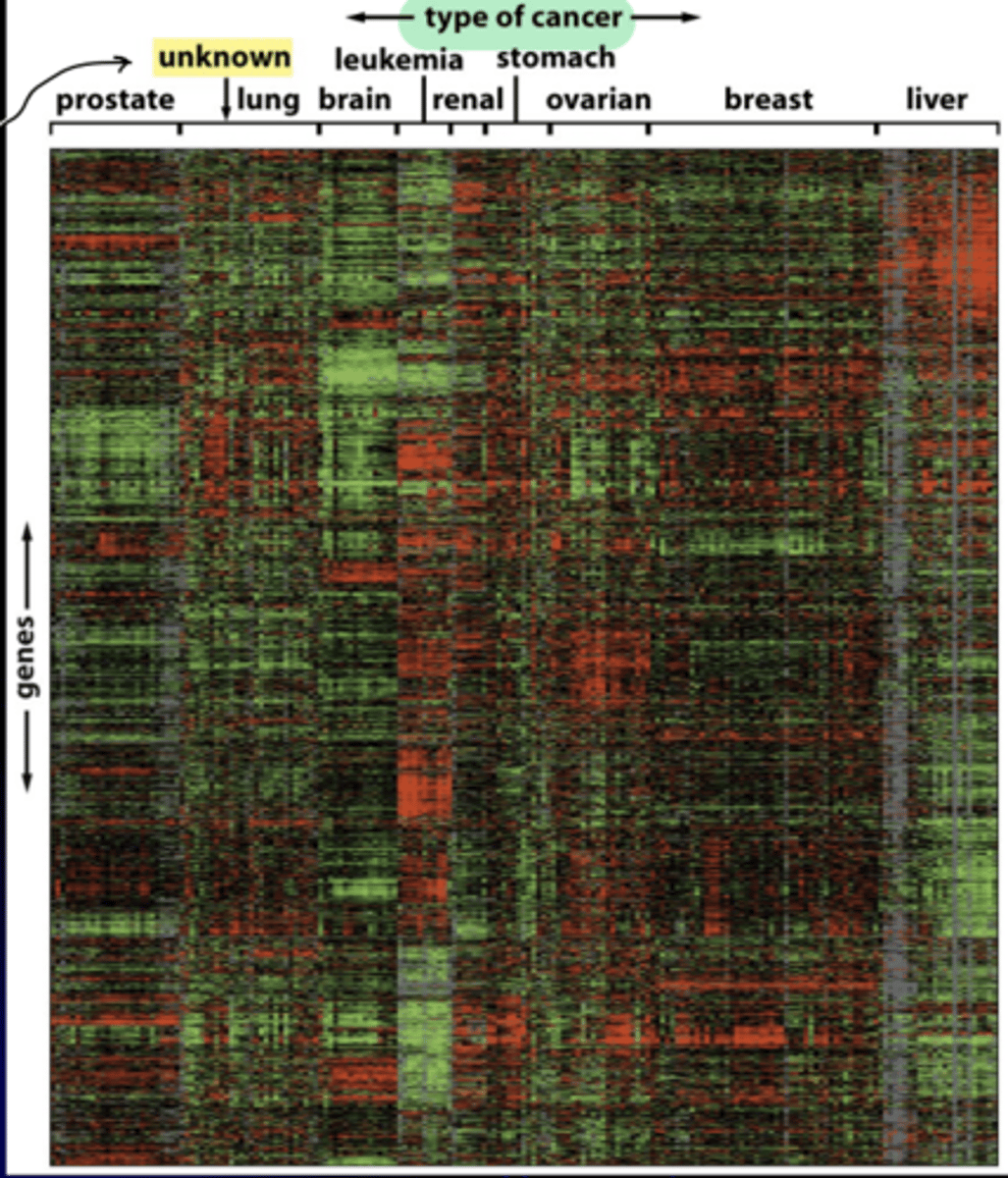
oncotyping
like normal tissues, each class of tumor has its own characteristic spectrum of gene expression
- allows identification of tumor of unknown origin and individualized treatment
ex) tumor of unknown origin was lung cancer
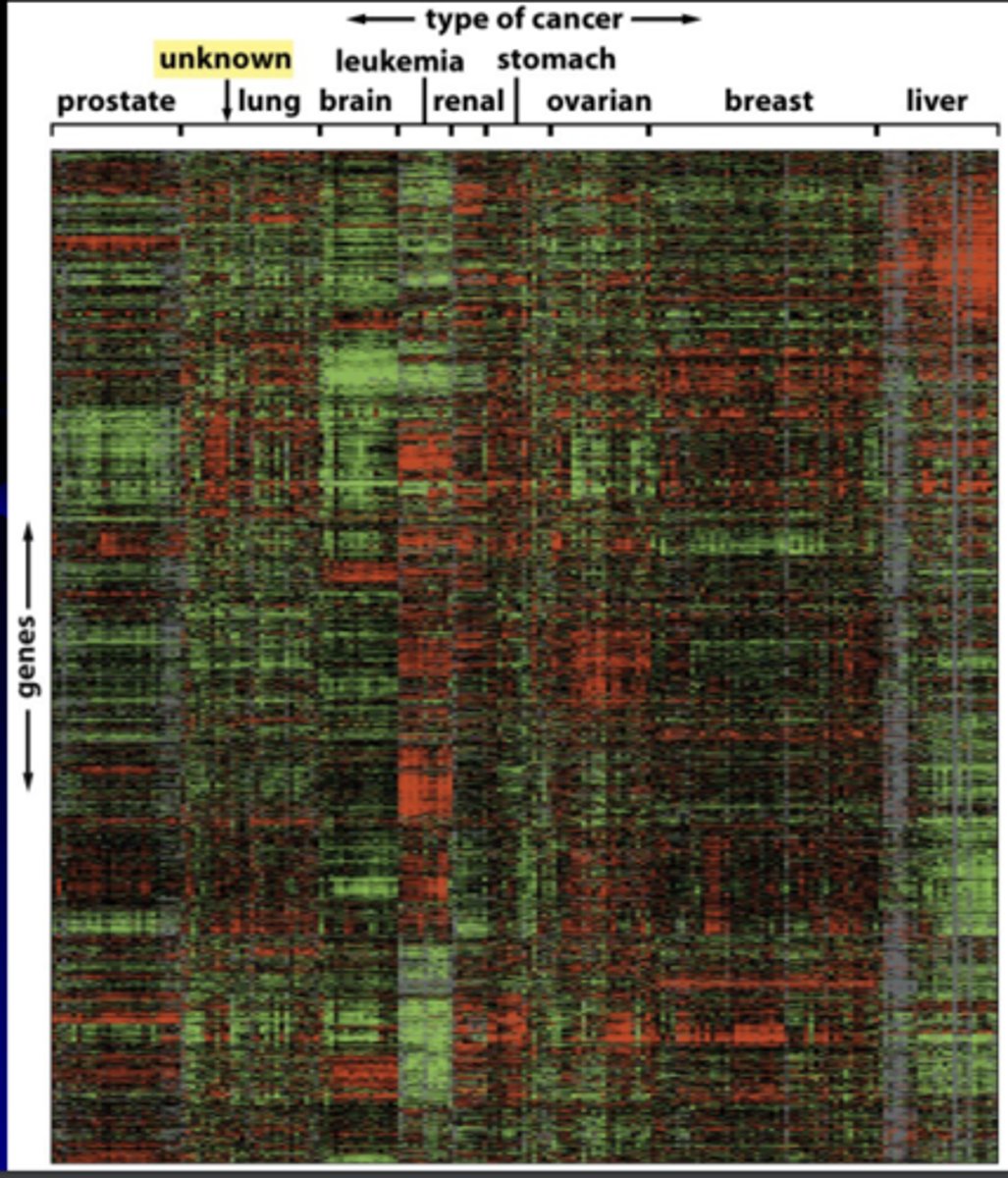
3) GWAS: genome-wide association studies
genetic epidemiology
- phenotype first studies
- examine a group with particular disease -> screen 100,000s of loci simultaneously for single nucleotide polymorphisms (SNPs)
- NCI: PLCO =prostate, lung, colorectal, ovarian GWAS
pros of GWAS
- find common gene trends among patients
- identification of at-risk population (screening for SNP biomarkers)
- allows for potential causal relationships
- tailoring of treatment options (gene-drug interactions)
cons of GWAS
- may not be predictive of disease
- cannot assess rare genetic variants
- associations represent small effect size
- causality and patient risk not clear
- genes and other risk factors not known (environment, diet)
4) NGS next generation sequencing
classify cancers according to genomic characteristics, potential to guide therapy
- paired sample testing reveals genetic differences between normal and malignant tumor tissue
types of NGS
- WGS whole genome sequencing
- WES whole exome sequencing
- RNA-seq
WGS (whole genome sequencing)
- sequence entire genome in one day, $1000 cost
- detects point mutations, indels, copy number variation, rearrangements and translocations