MCAT Biology
1/229
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
230 Terms
Classifications:
Eukaryotes
Prokaryotes
Eukaryotes:
Protists, Fungi, Plants, Animals
Cell wall present in fungi and plants
Nucleus
Multiple linear chromosomes
Ribosomal subunits 40S and 60S
Membrane-bound organelles
Steroids in plasma membrane
Mitosis/meiosis
Prokaryotes:
Bacteria, Archaea
Cell wall present
No nucleus; nucleoid
Predominately circular DNA (plasmid)
Ribosomal subunits 30S and 50S
No membrane-bound organelles
No steroids in membranes
Binary fission
Endosymbiotic Theory
The organelles of eukaryotic cells evolved through symbiosis or prokaryotes
There are similarities between mitochondria, chloroplasts, and prokaryotic cells:
Multiply through binary fission
Contain circular DNA
Have transport proteins called “porins”
Approximate Size and Density of Macromolecules
Nucleus: Diameter (5-10 um) ; Density (1.4-1.7 g/cm3)
Mitochondria: Diameter (1-2 um) ; Density (1.1 g/cm3)
Ribosome: Diameter (0.02 um) ; Density (1.6 g/cm3)
Chloroplasts: Diameter (5-10 um) ; Density (1.6 g/cm3)
Archaea and Bacteria Similar Traits
Similar shape
Same ribosomal density
Multiply through binary fission
Phospholipids
Most have cell walls but some do not
Found in animals, soil, oceans
Archaea Only
Genetically more similar to eukaryotes than bacteria are
Some varieties can survive very high temperatures
Membrane lipids have ether bonds (more resistant than ester)
Example: methanogens are archaeal cells that produce methane
Bacteria Only
Membrane lipids have ester bonds
Some varieties are pathogenic
Cell walls contain peptidoglycan
Example: cholera, E.coli

B. The ribosomes would be further from the surface than the mitochondria
Smaller and denser items will get pushed further down
Ribosomes are smaller and denser than mitochondria
Remember: ribosomes are not organelles
Fluid Mosaic Model
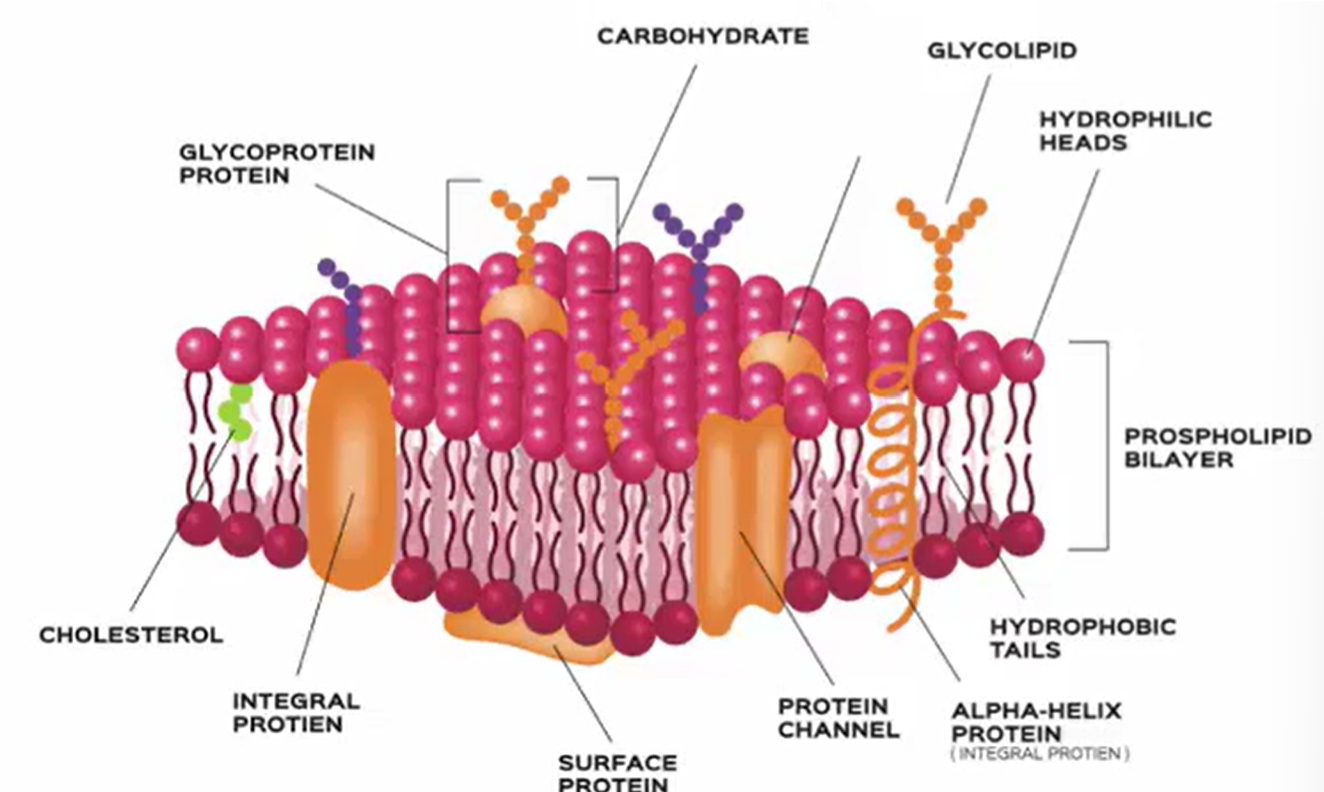
Phospholipid Bilayer
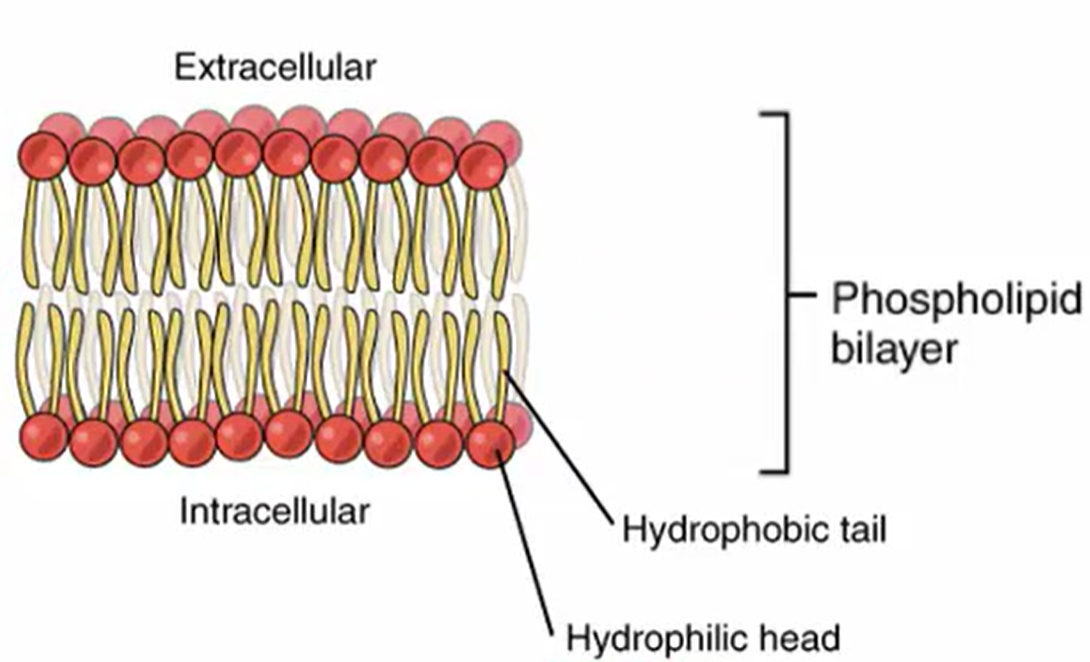
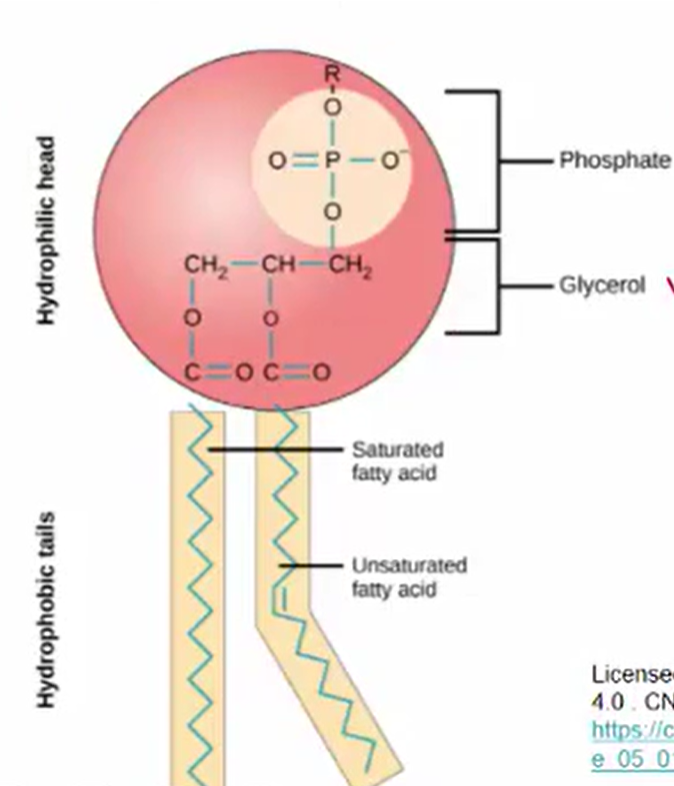
Amphipathic
Something that possess both hydrophilic (water-dissolving, polar) regions and hydrophobic (water resistant, nonpolar) regions
Structure of Phospholipid
Types of Proteins in Membrane
Peripheral
Surface proteins
Some glycoproteins
Integral
Completely embedded
Transmembrane
Transport proteins (i.e., channel proteins)
Some glycoproteins
Membrane receptors
Peripheral vs Integral Abundance
Many more peripheral proteins than integral proteins
Embedded within the Lipid Bilayer
Integral proteins can either be partly or fully embedded within the lipid bilayer
Proteins that are entirely embedded in the lipid layers are hydrophobic
Transmembrane proteins
A type of integral protein that can cross the membrane
Some act as transport proteins for ions and molecules
When an integral protein crosses the lipid bilayer, it…
Becomes shaped like an alpha-helix
Glycoproteins and Glycolipids
On the extracellular surface of membranes, proteins and lipids often have short carbohydrate chains extending out
Form hydrogen bonds with the water surrounding the cells which reinforce the cell’s structure
Glycoproteins can be integral or peripheral
Membrane Receptors
Type of integral protein that binds to molecules outside of the cell and transfer messages from outside the cell to inside the cell
Can be:
Ligand-gated ion channel receptors
Enzyme-coupled
G-protein coupled
Lipid Raft Theory
Lipid rafts are dense regions of the plasma membrane that are heavy in cholesterols and serve as protein signaling platforms
They can move across regions of the cell membrane as a unit
They can also be broken down into smaller rafts
Controversy of Lipid Rafts
They can only be observed indirectly, so the existence of lipid rafts is controversial
Passive Transport Types
Simple Diffusion
Facilitated Diffusion
Osmosis
Filtration
Active Transport Types
Primary vs Secondary
Exocytosis
Endocytosis
Pinocytosis (cell-drinking)
Receptor-Mediated
Phagocytosis
Passive Transport Definition
Molecules move in and out of the cell according to the concentration gradient
Relies on KINETIC energy
No ATP is needed
Simple Diffusion
Something small
Going towards gradient
Gases
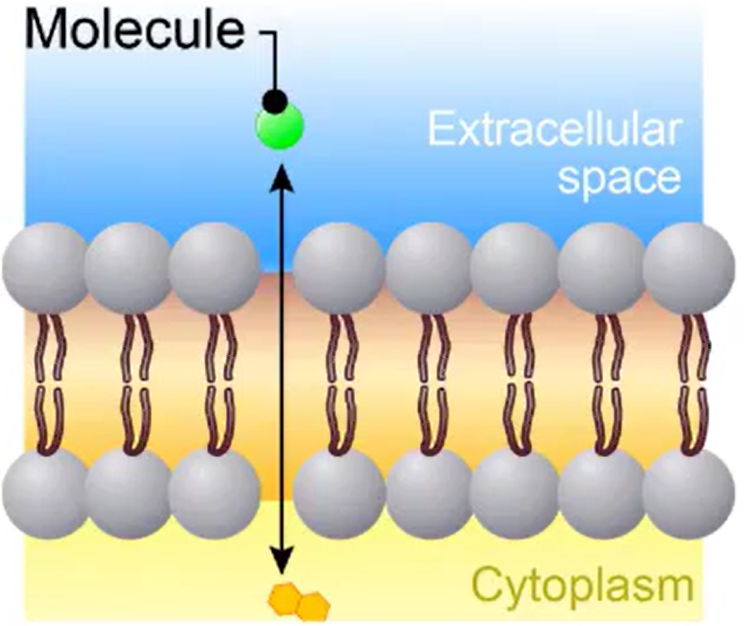
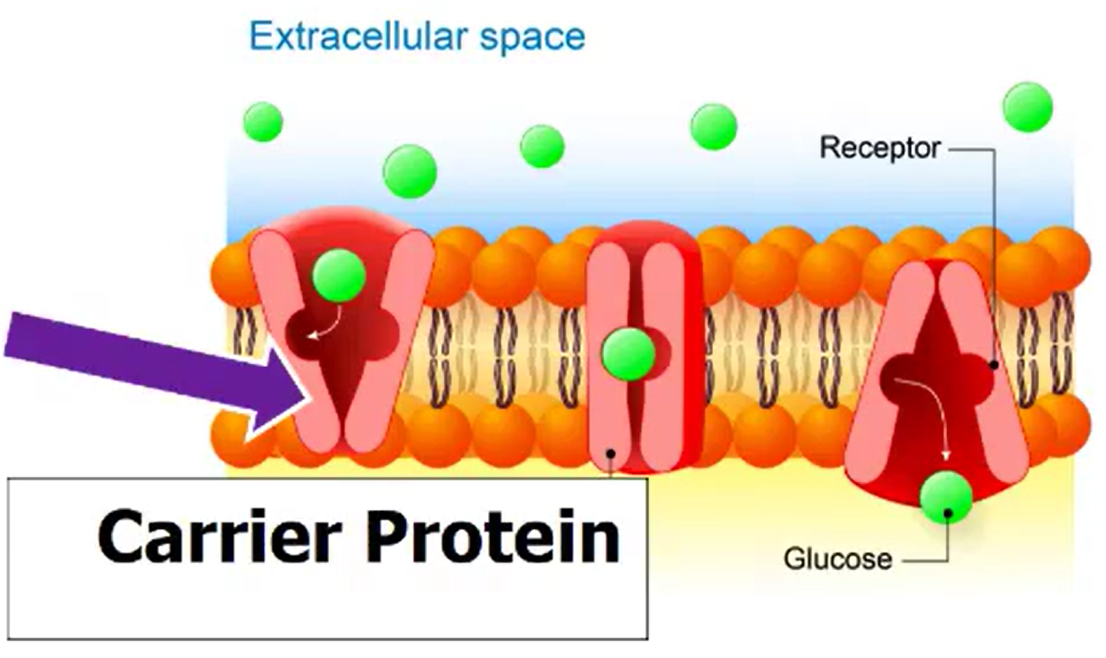
Facilitated Diffusion
Requires transport proteins
Polar, hydrophilic molecules
Larger molecules (glucose)
Carrier Proteins: molecules bind to proteins
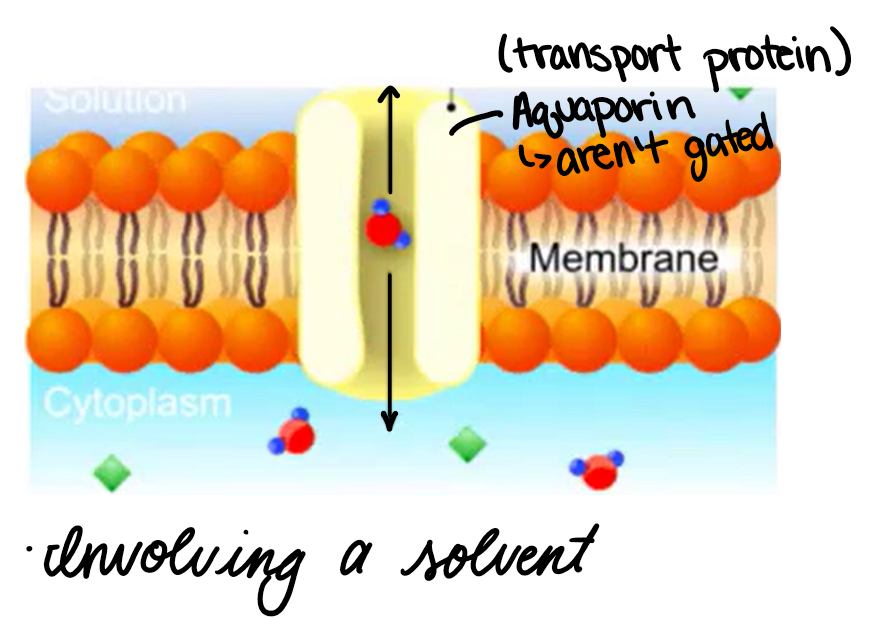
Osmosis
Involving a solvent
Water across a semi-permeable membrane
Direction of flow depends on the concentration of solute within cell vs external solute
Filtration
Water molecules and small solutes are pushed through a selectivity permeable membrane due to hydrostatic pressure
Usually a consequence of cardiovascular activity
Osmolarity
Solute concentration per volume solvent
Osmolality
Solute concentration per mass solvent
Too high → ADH
Tonicity
Measure of osmotic pressure gradient between two solutions
It is only influenced by solutes that can’t cross the membrane
Isotonicity
Concentration is the same in and out of cell
Hypertonicity
Greater concentration outside the cell
Shrivel = Crenation
Hypotonicity
Greater concentration inside cell
Bloat
Passive Transport Summary:


A. Lyse
The cell will bloat since it is in a hypotonic solution
No change in volume and dividing does not cause bloating
Active Transport Definition
Molecules move against the concentration gradient
Cellular energy (ATP)
Primary Active Transport
Makes DIRECT use of ATP to push molecules against the concentration gradient
Sodium-potassium pump
Secondary Active Transport
Uses INDIRECT forms of ATP to push molecules against the concentration gradient
Electrochemical Potential Driven
Sodium-Potassium Pump
3 Na+ OUT
2 K+ IN
ATP → ADP
Energy from conversion drives the pump
Electrochemical Potential Driven
Coupling of power from the primary active transport
Countertransporter: Na+ H+
More sodium than usual
Makes extracellular Na+ more driven to go back in (more stronger with pump)
Indirectly driven by Na+/K+ pump
Na+ coming in causes H+ to go out
Countertransporter (aka Antiport)
2 molecules are transported in opposite directions at the same time
Ex. Na+/H+ and Na+/Ca2+
Co-transporters (aka Symport)
2 or more molecules transported in same direction at the same time
Ex. Na+/glucose and SGLT: sodium-dependent glucose transporters
SGLT body absorbs filtered glucose
SGLT 2 inhibitor for diabetes 2
Exocytosis
Ways in which larger molecules are transported from the cell to the extracellular environment
Proteins, hormones, antibodies, glycogen
Happens via vesicles, which allow watery substances across fatty bilayer
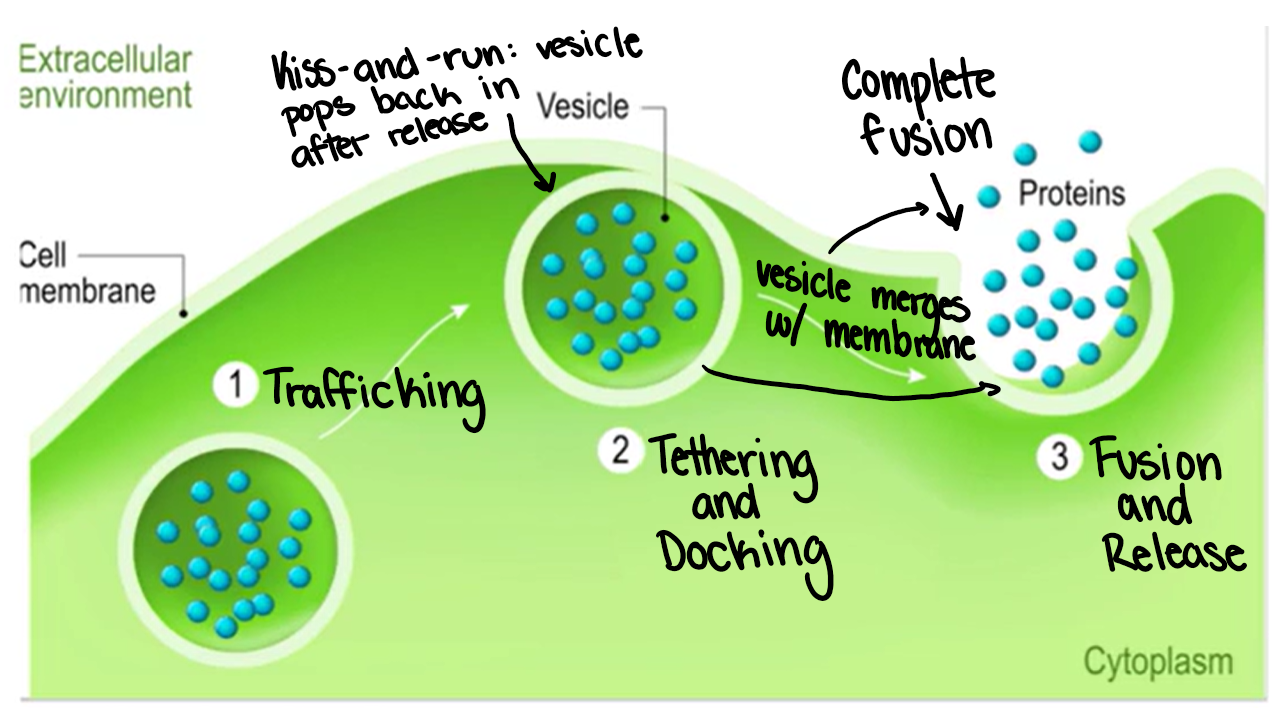
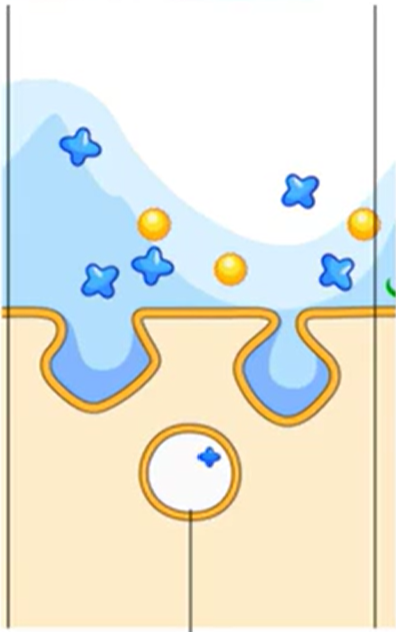
Endocytosis
Bringing large molecules into the cell
Uses a vesicle
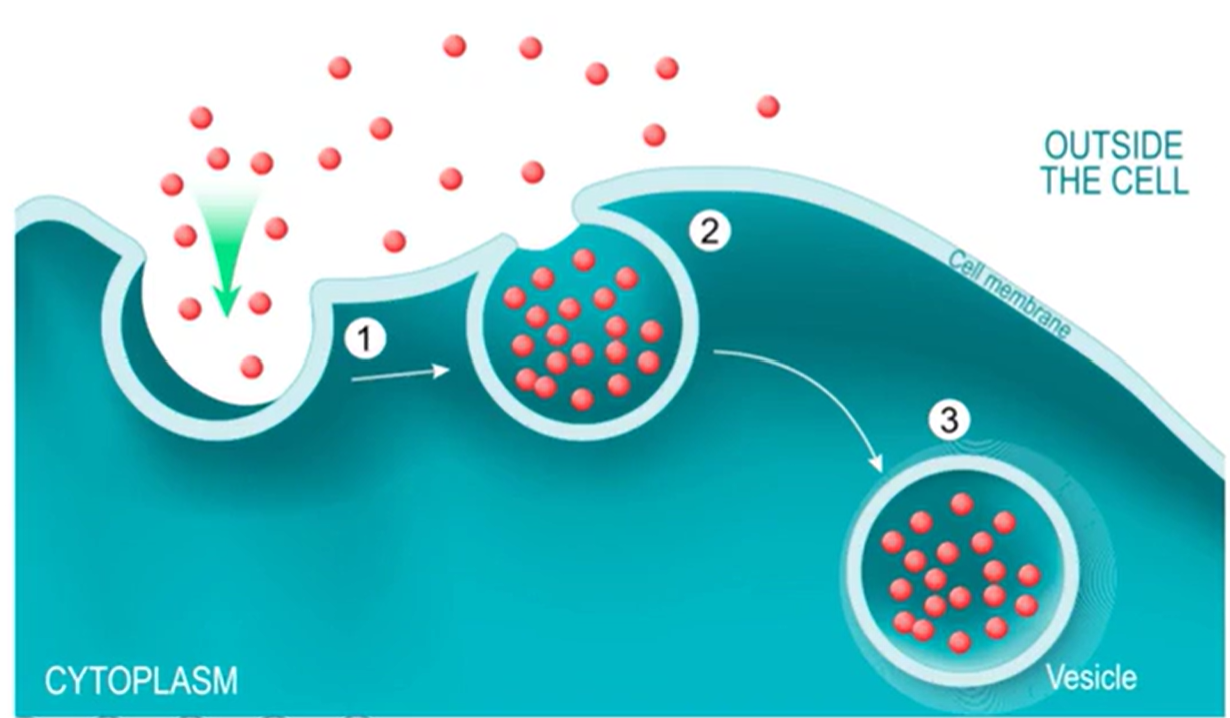
Pinocytosis
Cell takes in molecules across the water in big quantities from the extracellular environment
Membrane invaginates, then pinches off and brings in particles through the newly formed vesicles
Lysosomes come and bind to vesicles
Produce hydrolytic enzyme mixtures, so they can cut through water
Also break down whatever is floating in water
Efficient way to grab a lot of things all at once and haul them into the cell
Non-specific
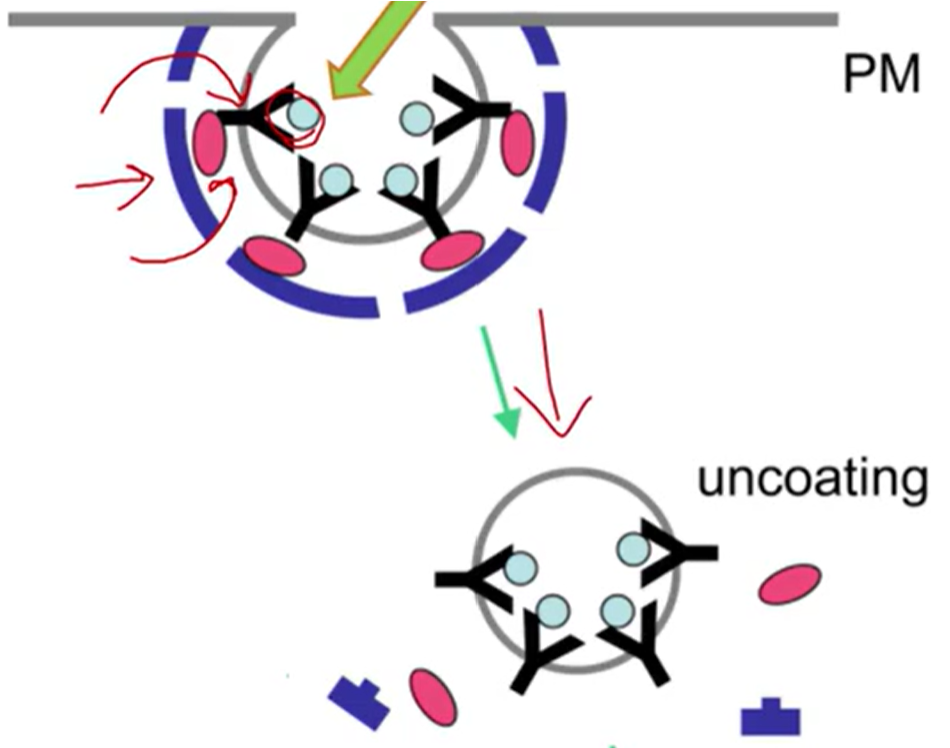
Receptor-Mediated
Clathrin-dependent endocytosis
Extremely selective
Works through receptors clathrin and adaptor proteins
Adaptor proteins: govern communication and signaling
By linking up in certain configurations
Form coordinated communication across the cell
Some carry material between organelles within the cell
AP-2: bind with clathrin
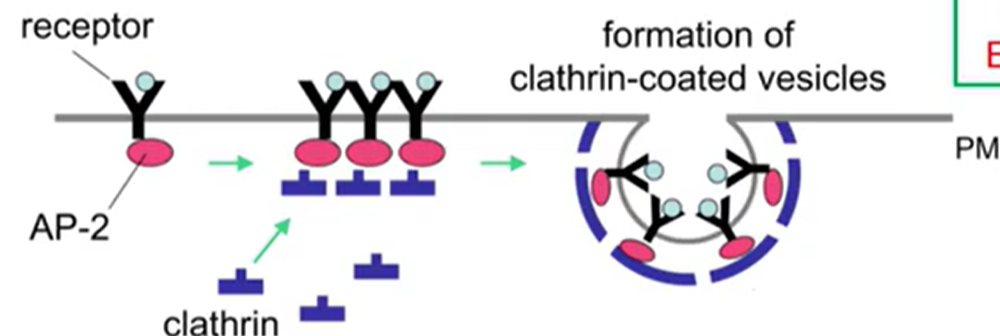
Clathrin Triskelion
One leg binds to adaptor protein, the other 2 bind to more clathrin
Ligand
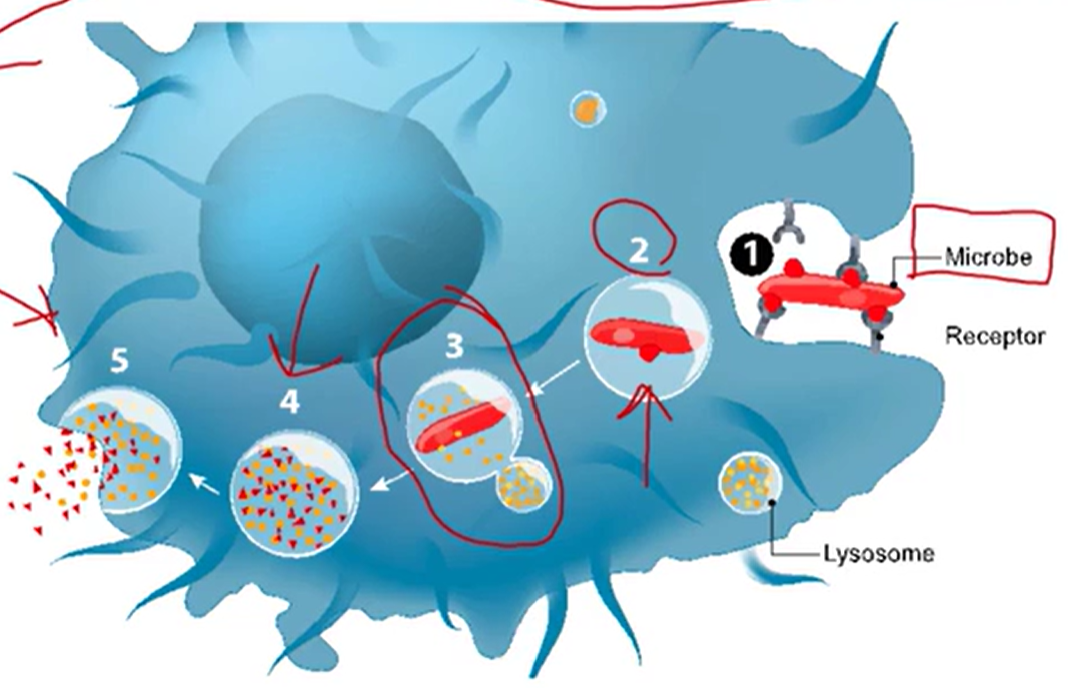
Phagocytosis
Cell-eating
Cell is moving around to find targets to eat
cell = phagocyte
Target large items they want to ingest
Frequently carried out by immune cells, which detect foreign pathogens
Where binding is occurring
Shape changes and target is internalized
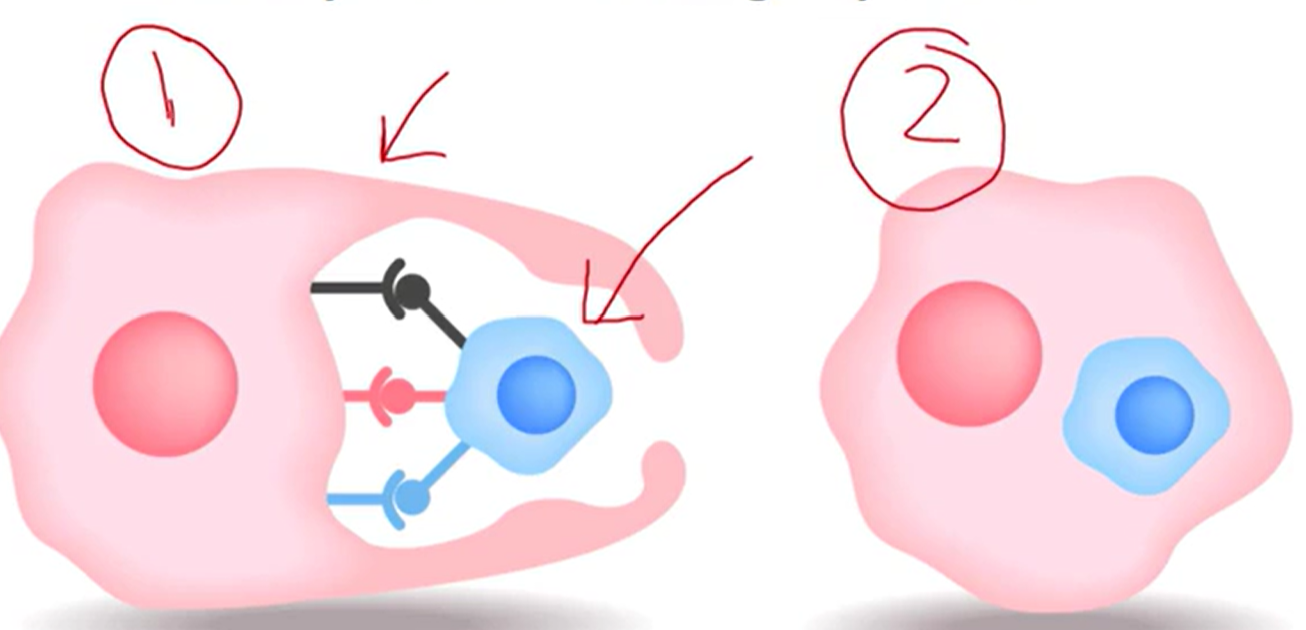
Process of Phagocytosis
Binding and absorption
Phagosome formation
Phagosome and lysosome to form a phagolysosome
Digestion
Release of microbial products
Exocytosis
Viruses
Comprised of:
Protein Coat = capsid protein
Single or double strand DNA or RNA
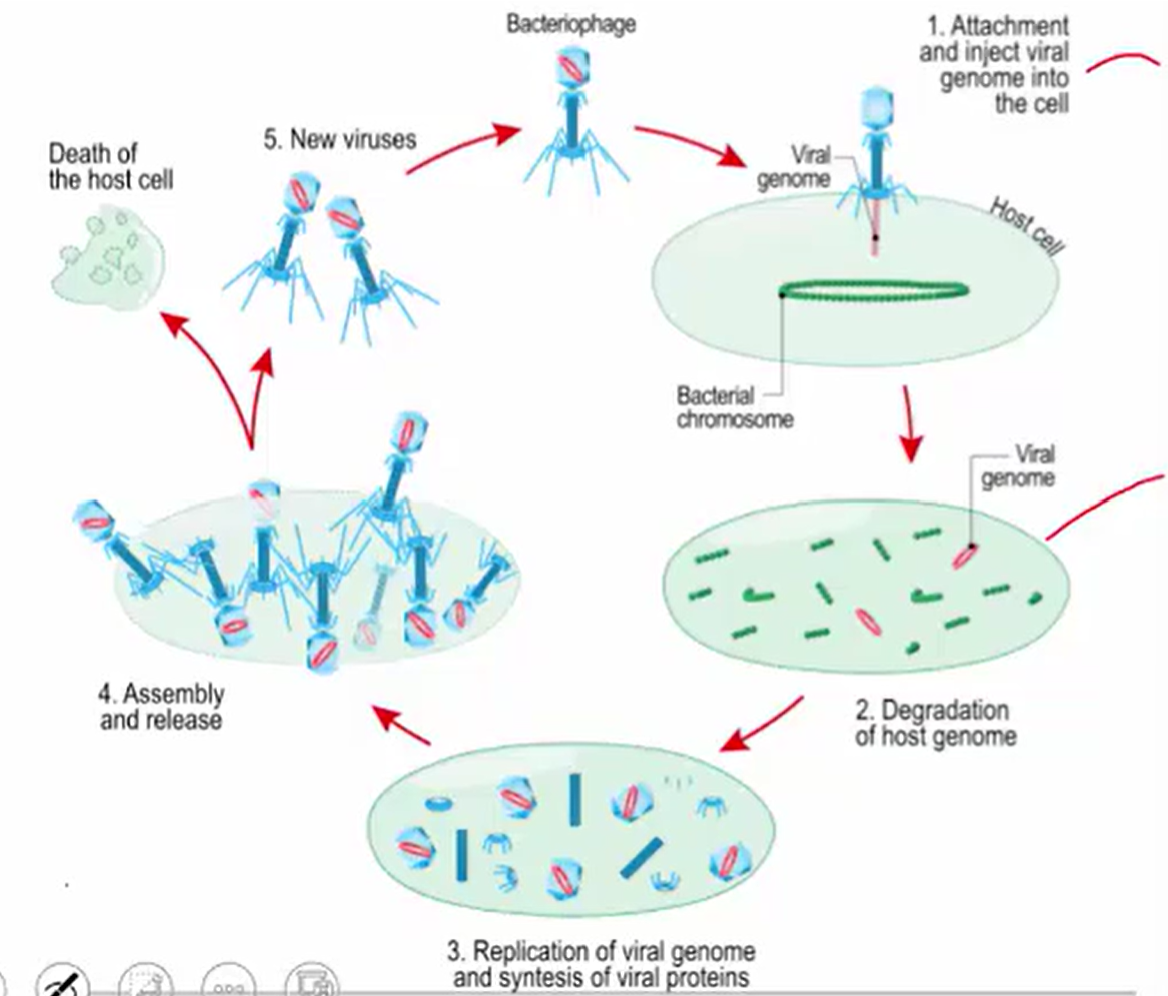
Bacteriophages
Viruses that exclusively infect bacteria via lytic or lysogenic processes
Lytic Cycle
Virus Enters Cell
Virus Replicates
Host Cell Lyses (destructs)
Release of Viruses
New Viruses Infect Other Cells
Lysogenic Cycle
Viral Nucleic Acid Incorporated into Host Cell Chromosome
Nucleic Acid is Replicated along with DNA
No Lysis. Host Cell Survives
If DNA Exits Cell, it can Enter Lytic Pathways and Infect
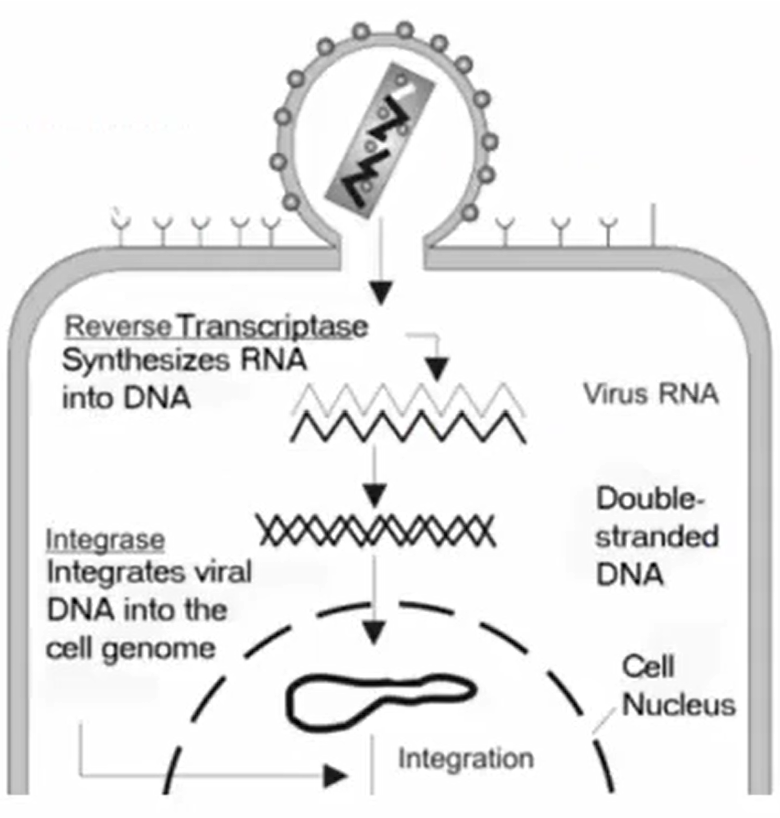
Retrovirus
Single-stranded RNA uses reverse transcriptase to make DNA
DNA-Containing Viruses
DNA viruses that replicate in host cell nucleus use host cell’s DNA polymerase
DNA viruses that replicate in cytoplasm need their own DNA and RNA polymerases
RNA-containing viruses
Replicate in host cell’s cytoplasm
Central Dogma
DNA → (transcription) mRNA → (translation) Protein
DNA Replication
DNA needs to make more of itself
Pyrimidine
CUT the pie
C: Cytosine
U: Uracil
T: Thymine
Purines
Pure as Gold (AG)
A: Adenine
G: Guanine
Nucleobase Bonding
G - C
A - T in DNA
A - U in RNA
G-C and H-bonds
The more G-C content, the more H-Bonds
Thus, higher melting or annealing temperatures
Nucleotide:
Sugar, Base, and Phosphate Group
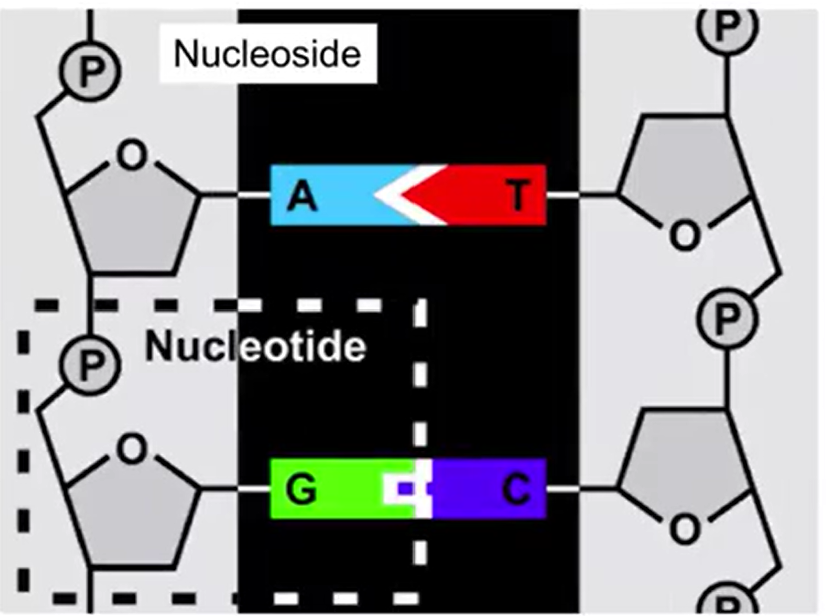
Nucleoside:
Sugar and Base only
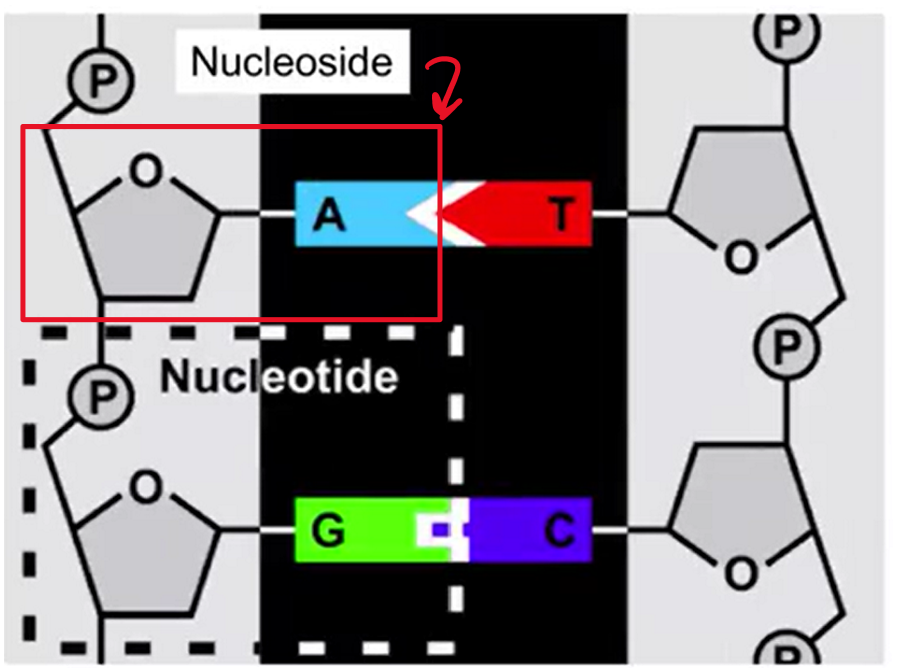
Sugar and Base Bond
N-Glycosidic Bond
Phosphate and Free 3’-OH bond
Phosphodiester bonds
Reverse Compliment / Complementary Strand
Complementary strand will be in 3’ to 5’ so make sure to make it 5’ to 3’ for final answer
C complements G
A complements T (in DNA) and U (in RNA)
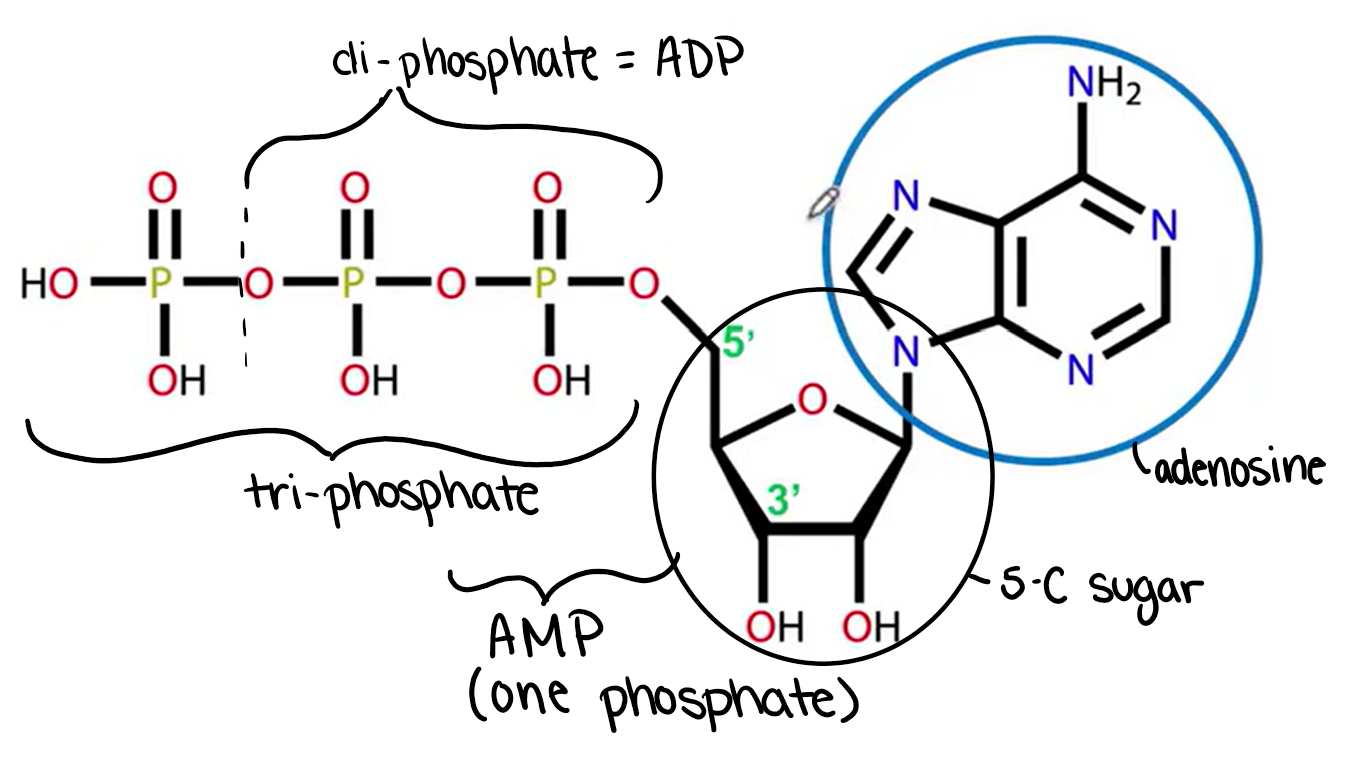
Structure of ATP
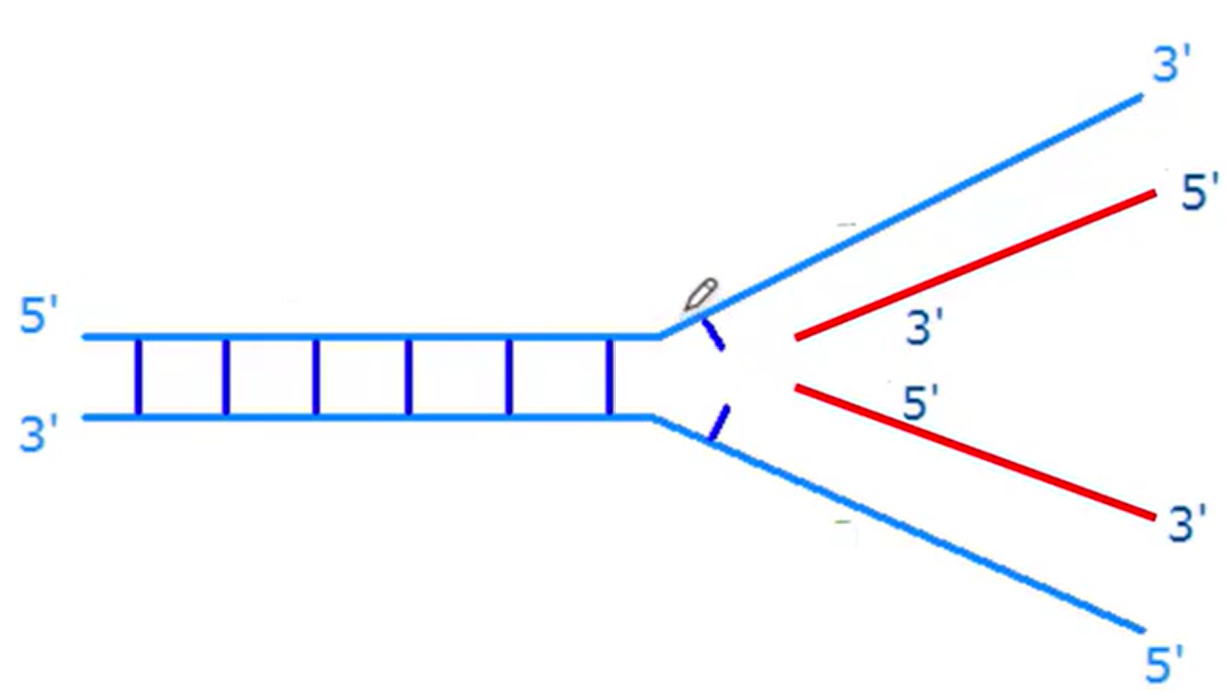
DNA is Semiconservative
Two strands running antiparallel
Half old and half new DNA once replicated
Helicase
Helps to unwind the DNA
Primase
A type of RNA polymerase
Polymerases
Enzymes that synthesis chains of nucleic acids
Polymerase III
Haloenzyme
Ligase
Enzyme that fuses Okazaki fragments
Okazaki Fragments
Each individual 3’ → 5’ sections
Single-Stranded Binding Proteins
To stabilize single-stranded DNA (ssDNA) during DNA replication and other cellular processes
Transcription
DNA → RNA
Direction: 5’ → 3’
RNA polymerase; sense vs anti-sense
Stopping
Post-transcriptional modifications (hnRNA → mRNA)

RNA Polymerases
rRNA → RNA pol I
mRNA → RNA pol II
tRNA → RNA pol III
RNA Processing Steps
Capping: Methyl G to 5’ end
Polyadenylation; adenosines to tail (3’ tail)
Editing: bases changed or deleted
Splicing: Intron removed, exons expressed in the final RNA
Translation
Codon: 3 letters; each codon stands for an amino acid
64 different combinations
Degeneracy, tRNA wobble
tRNA, rRNA, and protein synthesis
Direction of synthesis termination
Point mutation, frame shift mutation, nonsense mutation, missense mutation
Where is the information for eukaryotic cell survival and function stored?
In the nucleus as DNA
Transcription and translation allow the cell to…
Communicate general information as mRNA
used as a blueprint to build proteins
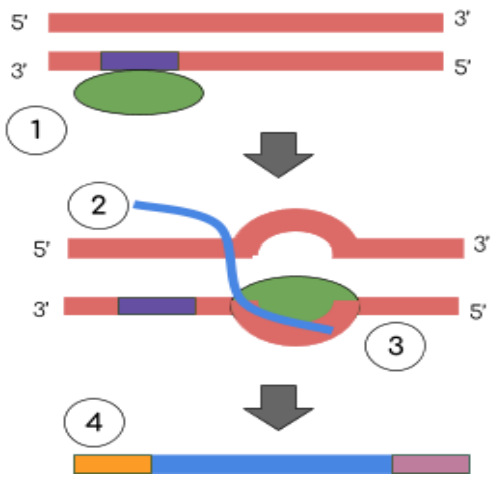
Transcription Steps
Initiation: RNA polymerase binds at the promoter region of a specific gene to open the DNA double helix
Elongation: Base-pairs are added in the 5’ to 3’ direction (U replaces T in RNA)
Termination: mRNA is released
Processing: prevent degradation: add 5’ cap and poly-A tail ; splicing: remove introns and join exons
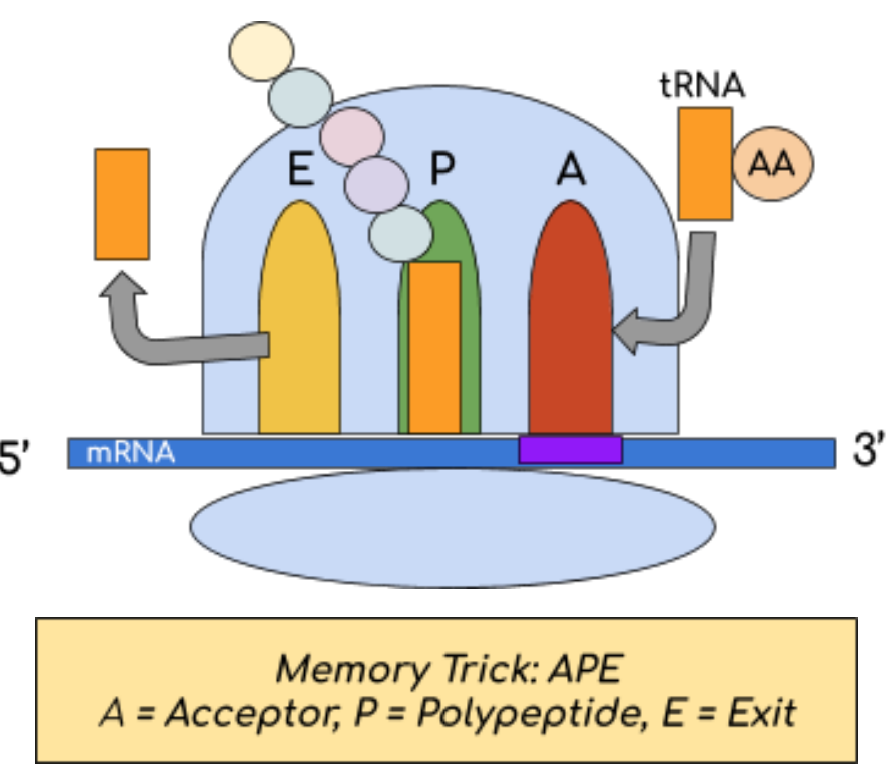
Translation Sites:
Ribosomes are composed of a large and small subunit that come together to form 3 sites:
A: a tRNA molecule carrying a specific amino acid binds to the corresponding mRNA codon
P: an amino acid is added to the growing polypeptide chain
E: tRNA is released from the ribosome
Catalysts
Increase rate of rxn (forward and reverse)
Enzymes and Activation Energy
Lowers activation energy
Enzymes do NOT change:
Equilibrium constant
Free energy (delta G)
Enthalpy (delta H)
Enzymes are ___ in reaction
Recycled
Appear in products + reactants
Enzymes are sensitive to…
pH and temperature
Purpose of Cofactors and Coenzymes
To improve enzymatic function
Apoenzymes
Holoenzymes
Prosthetic Groups
Cofactors:
In organic molecules, metal ions (Zn+2, Cu+2, Fe3+)
Coenzymes
Small, organic, vitamin derivatives (NAD+, FAD, coenzyme A)
Six Classification for Main Enzymes: LIL’ HOT
Ligase
Isomerase
Lyase
Hydrolase
Oxidoreductase
Transferase
Ligase
Join, use ATP
Isomerase
Rearrangement, isomers (constitutional and stereoisomers)
Lyase
Cleavage without H2O
Hydrolase
Cleavage with H2O