Sequencing and genomes
1/20
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
21 Terms
Sanger sequencing requirements
Developed in 1977 = still used today
DNTP:ddNTP = 100:1
Single primer used
Template = PCR product or plasmid
What are ddNTPs
→ chain terminating nucleotide bases
3’ OH group is replace by a H molecule, so once added, the DNA polymerase is unable to add another nucleotide onto the strand.
Blocks further polymerisation
Sanger sequencing 1977-1980s
A DNA template, primer, DNA polymerase, normal nucleotides (dNTPs), and a small amount of a fluorescently labeled ddNTP (A, T, C, or G) are mixed.
DNA polymerase adds nucleotides until it reaches the ddNTPs, where it stops
Produces lots of fragments of different lengths ending in fluorescent tags
Size separation by gel electrophoresis- 4 separate sequencing reactions, each with a different radioactive ddNTP
Photographic paper to detect radioactive DNA strands
manual process, takes time
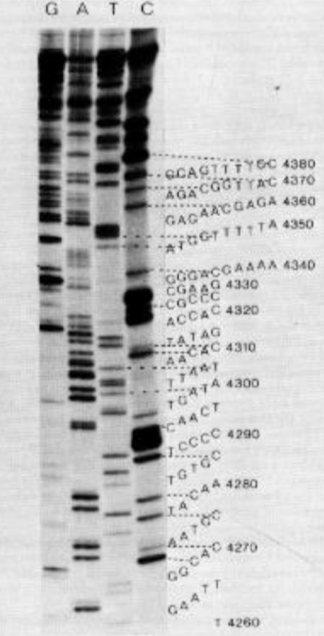
Sanger 19090s-present
Sequencing uses a mixture of flourescently labelled ddNTPs and normal dNTPs, each base has a different colour
Chain termination PCR produces mixture of DNA strands of different lengths
Size separation by gel electrophoresis in a capillary using a laser
automated

What come before sequencing always
PCR
Amplifies small bits of DNA into large numbers
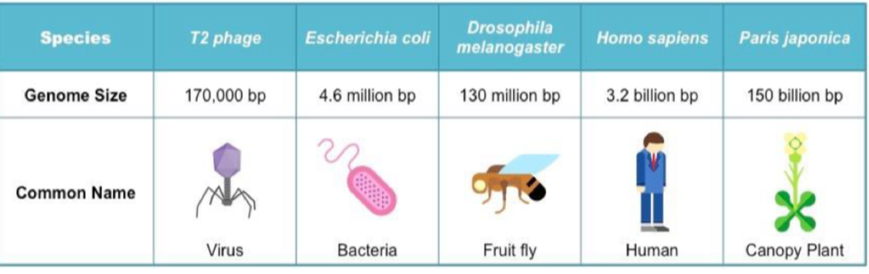
Human genome
→ complete set of genetic info present in a cell/organism
Genome size = total no. Of DNA base pairs in one copy of a haploid genome
Human genome = 3.2 billion bp
Human genome project timeline
1977- Sanger sequencing
1989- UK research council and USA jointly funded the project
1990- project begins, international team of researchers
2003- completed (almost) using Sanger sequencing
2023- fully completed genome
Cost = 3 billion pounds
Wellcome Trust Sanger Institute
HGP aims
identify and map all genes in the human DNA
Determine sequences
Store info in databases
Discover efficient technologies for data analysis
Sequence other genomes of medical importance e.g. mouse/yeast
Sequence content of human genomes
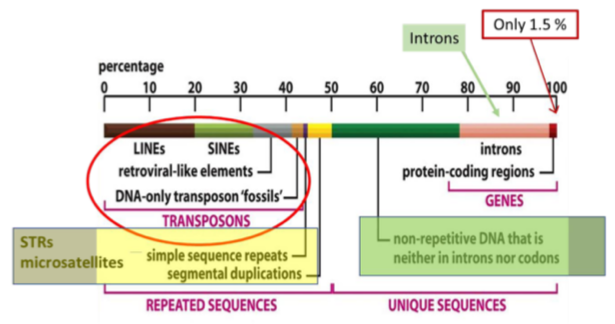
Transponsons:
LINEs = long interspersed nuclear elements
-Encode reverse transcriptase
SINEs = short…
-Don’t encode, steal it off LINEs
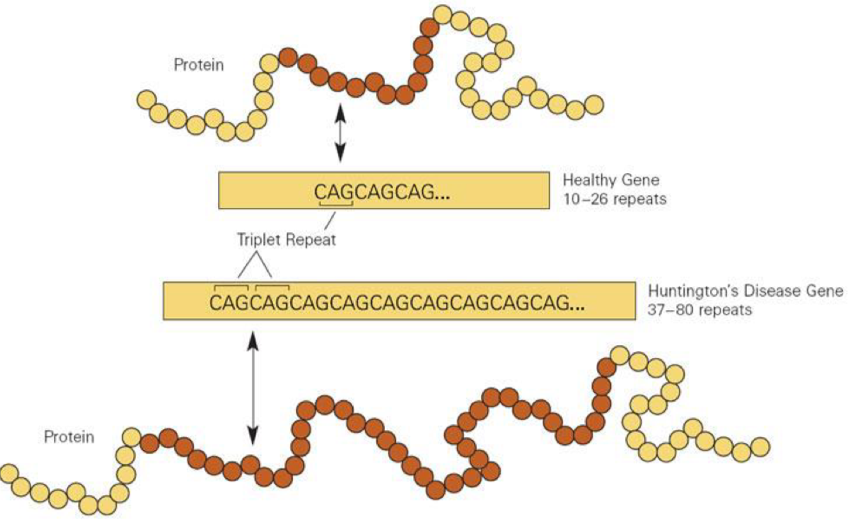
Micro satellites (STRs)
Repeats of 1-6 bp, typically repeated 5-50 times
Introns
non-coding DNA segments
Transponsons
→’jumping genes’→ try to make more of themselves
can replicate and insert into other parts of the genome
Discovered by Barbara McClintock
Two types:
reterotransponsons = transpose via mRNA, most mobile elements in eukaryotes
DNA Transponsons = no intermediate, ‘cut and paste’, most mobile elements in bacteria
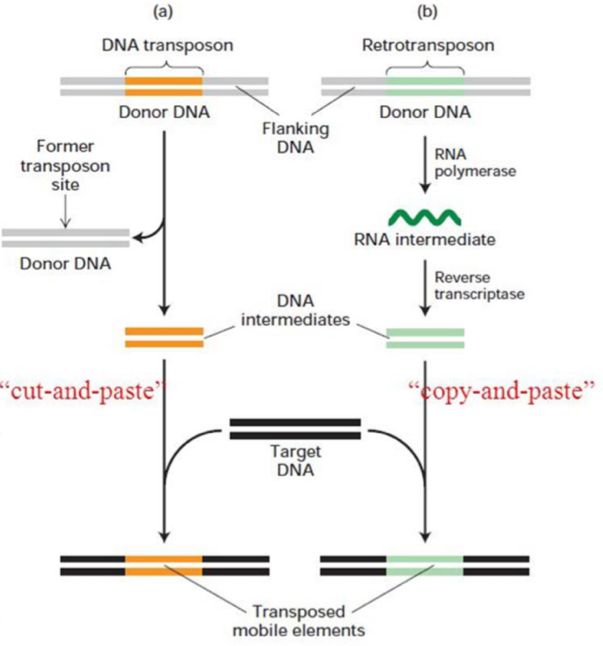
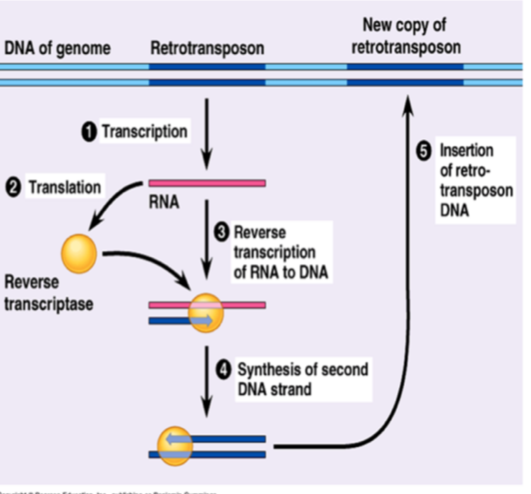
Reterotransponsons
→ encode reverse transcriptase
after transcription, the RNA is converted to DNA + inserted into another part of the genome (using mRNA intermediate)
LINEs encode reverse transcriptase, SINEs don’t
Alu is a SINE, 11% of human genome, uses LINEs for reverse transcriptase as don’t encode it
Micro/mini satellites
Micro satellites = STRs, 1-6bp repeats 5-50 times
Dinucleotide repeat = ATATAT
Trinucleotide = ATGATGATG
Mini satellites = longer repeats from 10-60bp, also repeated 5-50 times
HGP outcomes
approx. 22,300 protein coding genes
Similar to mouse, drosophila = 15,000
Many protein encoding genes in humans produce more than one type of protein via alternate splicing
Humans have more transcription factors and control elements
Prokaryotic genomes
Operons = cluster of genes with a single promoter
No introns
Non-coding, only 12% of the genome
Nonessential prokaryotic genes are commonly encoded on extrachromosomal plasmids
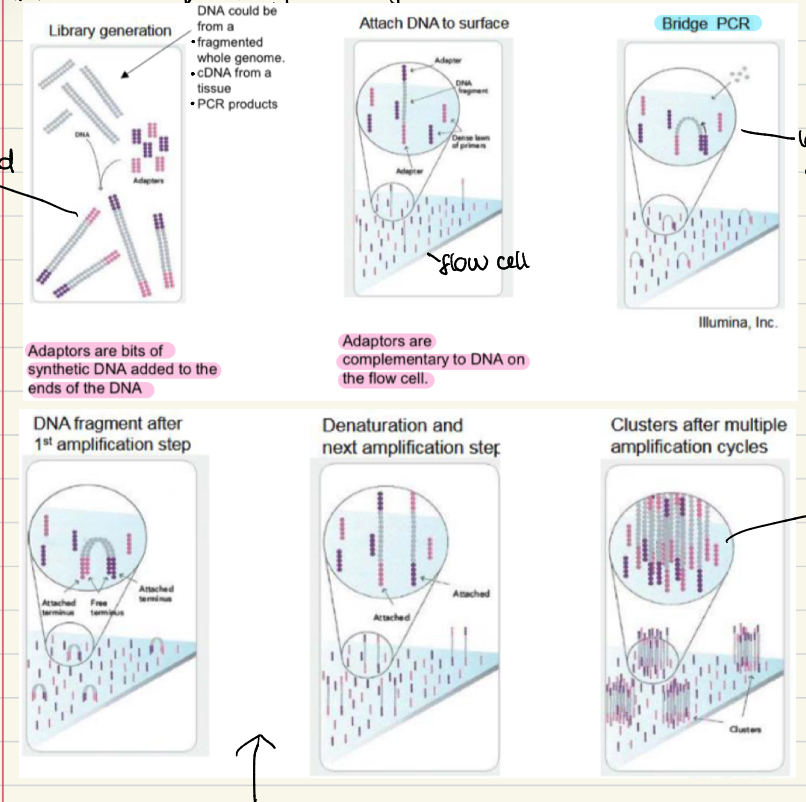
Illumina sequencing
Library generation- adaptors added with ligand (synthetic DNA added on the ends)
Attach DNA to surface of a flow cell
Bridge PCR- produces clusters after multiple cycles
Add 4 different (bases) reversible terminator nucleotides and image after each cycle
A picture is taken of the flow cell after each addition, using a powerful camera and microscope
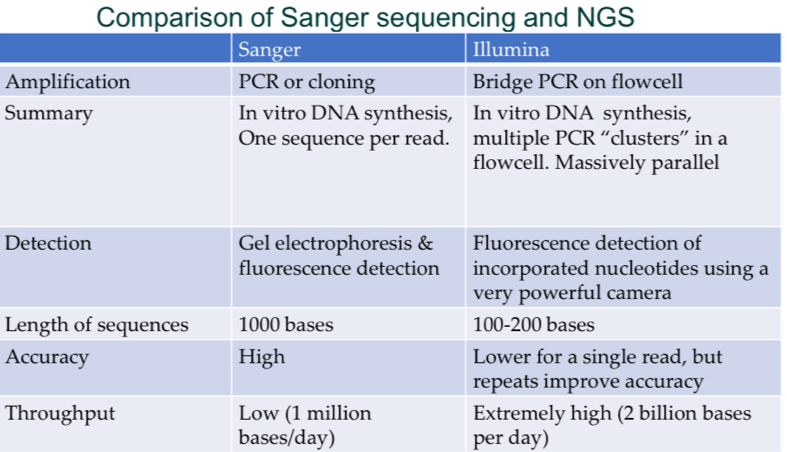
Comparison of Sanger vs illumina sequencing
Sanger
PCR or cloning amplification
Gel electrophoresis and fluorescence detection
1000 bp sequences
High accuracy
Low throughput (1 million bp/day)
Illumina
Bridge PCR on flow cell amplification
Fluorescence detection on powerful camera/microscope
100-200 bp sequences
Low accuracy for a single read but repeats improve
Extremely high throughput
3rd generation sequencing
long-read sequencing - up to 10,000bp
Urgently under active development
PACBIO - a large machine
Oxford nanopore tech, MinION, a handheld device
Completed human genome project in 2023
Chromosomal differences
Down’s syndrome = extra chromosome 21
Klinefelter = XXY, only symptoms later in life
Turner’s syndrome = lacks second X as a female
1,000 genomes project
launched in Jan 2008
International research effort to establish the most detailed catalogue of human genetic variation by 2020
2500 unrelated humans sequenced
Re sequencing with 2nd/3rd gen
→ to identify genetic variants
whole genome sequencing= sequence the genome from genomic DNA
Exogenous/transcriptome sequencing = expressed genes cDNA (mRNA coding)
Spotting differences→ align the short sequences to the reference genome to identify differences
Identify genetic variants whole= alleles, SNPs (single nucleotide differences) etc.
Common genetic variants
SNP: single nucleotide polymorphisms
Substitution of a single nucleotide at a specific position
Indels:
insertions and deletions, INDELs
Most genetic variants = no phenotypic effect
intergenic region = silent
Non-coding regions may be silent or affect gene expression e.g.promoters
Copy number variants (CNVs)
variations in which part of the genome is either deleted or repeated