ch 15- intracellular compartments and protein transport
1/81
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
82 Terms
Prokaryotic cells regarding intracellular compartments
single membrane-bound compartment — includes the contents of the entire cell surrounded by the plasma membrane
membrane closed organelles
any organelle in eukaryotic cells that is surrounded by a lipid bilayer
ex: ER, golgi, and lysosomes
each have specialized functions
cytosol
Organelles are surrounded by cytosol, which is enclosed by the plasma membrane
contains many metabolic pathways
protein synthesis
the cytoskeleton (The cytoskeleton is a network of protein filaments (microtubules, actin filaments, and intermediate filaments) that extends throughout the cytosol.The cytosol is the fluid environment that surrounds and supports the cytoskeleton.)
Nucleus
contains the main genome
DNA + RNA synthesis
most prominent organelle, it is surrounded by a double membrane called the nuclear envelope & communicates with the cytosol via nuclear pores that perforate the envelope
ER
JOB:
synthesis of most lipids
synthesis of proteins for distribution to many organelles and to the plasma membrane
Details:
The outer nuclear membrane is continuous w/ the membrane of the ER- system of interconnected membranous sacs and tubes that extend throughout most of the cell
ER= the major site of synthesis of new membranes in the cell
have ribosomes attached to the cytosolic surface - rough ER
ribosomes- actively synthesizing proteins that are inserted into the ER membrane or delivered into the ER interior —> LUMEN
smooth ER… lacks ribosomes
site of steroid hormone synthesis in some endocrine cells of adrenal glands
site where organic molecules like alchohol are detoxified in liver cells
in eukaryotic cells the smooth ER sequesters Ca2+ from the cytosol
the release and reuptake of Ca2+ from the ER is involved in muscle contraction and other responses to extracellular signals
The outer nuclear membrane is continuous w/
he membrane of the ER- system of interconnected membranous sacs and tubes that extend throughout most of the cell
What is the difference between the rough ER and the smooth ER?
golgi apparatus
JOB
modification
sorting
packaging of proteins and lipids for either secretion or delivery to another organelle
details
usually situated near the nucleus
received proteins and lipids from the ER and modifies them —> dispatches them to other places in the cell
Lysosomes
intracellular degradation
small digestive enzymes degrade organelles that are worn out or damaged + macromolecules + particles taken into the cell by endocytosis
On their way to the lysosomes, endocytosed materials first pass through a series of compartments called
endosomes
Endosomes
sorting of endocytosed material + recycle some of them back to the plasma membrane
Mitochondria
site of ATP synthesis by oxidative phosphorylation
surrounded by a double membrane
contains internal membranes that are highly specialized for ATP production
Chloroplast
ATP synthesis and carbon fixation by photosynthesis
surrounded by a double membrane
contains internal membranes that are highly specialized for ATP production
Peroxisomes
oxidative breakdown of toxic molecules
small organelles that contain enzymes that break down lipids and destroy toxic molecules producing hydrogen molecules
cytoskeleton
attachment
provides tracks for moving the organelles around and for directing the traffic of vesicles between one organelle and another— the movements are driven by motor proteins that use the NRG of ATP hydrolysis to propel the organelles and vesicles along the filaments
evolution of membrane enclosed organelles
to grow in size and volume the ancient archaeal cell enlarged its plasma membrane — how? — by forming protrusions that allowed it to exchange metabolites with its environment + interact with other cells including aerobic bacteria. at some point the protrusions pushed out further and further and segments of the plasma membrane started to tuck in — over time the invaginations pinched off forming double layered enelope that encloses the nuclear compartment
some of the membrane infoldings split off to produce the endomembrane system (ER, golgi, peroxisomes, endosomes, and lysosmes)
ancestral archaeon + endosymbiosis of ancestral bacteria
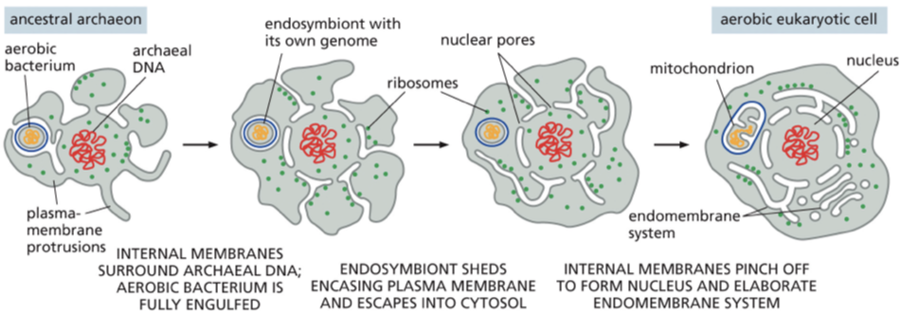
mitochondria and chloroplast endosymbiont theory
both of these organelles evolved from bacteria that were engulfed by ancestral archaeal cells
mitochondria and chloroplast arose separately from the components of the endomembrane system — remain isolated from the vesicular traffic that connects the interiors of the other membrane enclosed organelles
SA to volume ratio + plasma membrane
archaea + bacteria
present day eukaryotic cells
archaea + bacteria: small size — high surface area to volume ratio — their plasma membrane can sustain all of the cells vital functions
present day eukaryotic cells: by contrast, have volumes that are 1000 to 10,000 times greater. Such a large cell has a small surface-to-volume ratio and presumably could not survive with a plasma membrane as its only membrane.
in a typical human secretory cell, which membrane has the largest surface area
rough ER
The rough ER is folded up to form an extensive maze of interconnected spaces. This organelle can, in some cases, compose about half of the total membrane present in the cell.
How do the interiors of the ER, Golgi apparatus, endosomes, and lysosomes exchange contents with each other?
by small vesicles that bud off of one organelle and fuse with another
before a eukaryotic cell divides what needs to happen to the membrane enclosed organelles
it must duplicate the membrane enclosed organelles
does organelle growth require lipids?
Organelle growth requires a supply of new lipids to make more membrane and a supply proteins—both membrane proteins and the soluble proteins that will be in the interior (or lumen) of the organelle. Even in cells that are not dividing, proteins are being produced continually
Proteins are delivered to the mitochondria, chloroplast, and the interior of the nucleus from the?
proteins are directly delivered from the cytosol
Proteins and lipids are delivered to the golgi, lysosomes, endosomes and the inner nuclear membrane by?
indirectly via the ER
although some of these proteins are retained in the ER, most are transported by vesicles to the
Golgi apparatus and then onward to the plasma membrane or to other organelles.
the synthesis of all proteins
where does synthesis begin
where does the protein go after being synthesized
sorting signals
almost every protein the cell begins on ribosomes (make proteins) in the cytosol
depends on its amino acid sequence — can contain a sorting signal that directs the protein to the organelle that needs it — proteins that lack a sorting signal just stay in the cytosol
different sorting signals direct proteins into the nucleus, mitochondria, chloroplast, peroxisomes, and the ER
3 mechanisms of Protein transport
•Via nuclear pore complexes
•Transmembrane transport
•Vesicular transport
proteins are moving from
cytosol to nucleus— transported via nuclear pores
the nuclear pores penetrate both the inner and outer nuclear membranes
pores= are selective gates that actively transport specific macromolecules + free diffusion of smaller macromolecules
proteins remain folded
cytosol into the ER mitochondria (or chloroplast) — transported via protein translocators (located in the membrane).
the transported protein = unfolded to move it across the hydrophobic interior of the membrane
bacteria have similar protein translocators in the plasma membranes— export proteins from the cytosol to the cell exterior
moving from the ER — moving from one compartment of the endomembrane system to another— transported via transport vesicles (pinch off from the membrane of one compartment then fuse w/ the membrane of the second compartment)
deliver soluble cargo proteins + the proteins/ lipids that were a part of the vesicle membrane
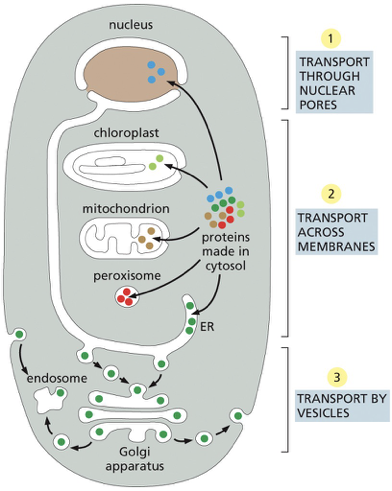
Signal sequences
amino acid sequence that directs a protein to a specific location in the cell
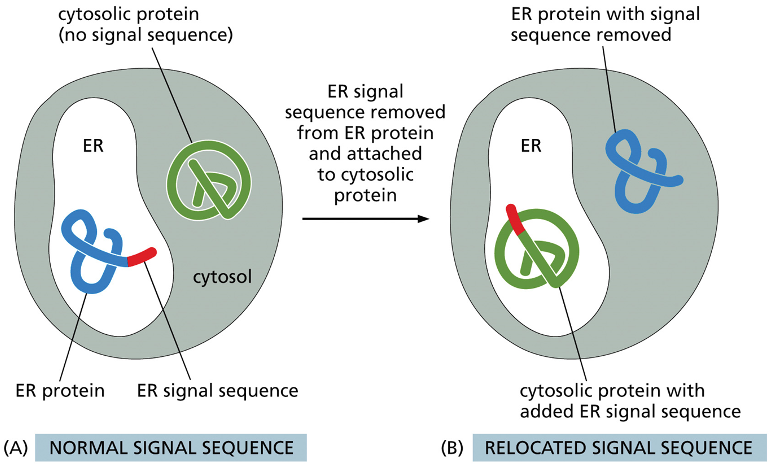
describe what is happening here
in diagram A the blue protein is and ER protein that has a ER signal sequence so it is in the ER, the green line is the cytosolic protein it doesnt have a signal sequence so it stays in the cytosol
the ER signal sequence is removed from the ER protein and is attached to the cytosolic protein… now the ER protein is in the cytosol without a signal sequence and the green one is in the ER with the signal sequence
now they arent in their proper locations
Signal sequences that specify the same destination
same function but the proteins can vary in
physical properties (hydrophobicity)
placement of charged amino acids (important for the function of the signals)
once the signal sequence has arrived to its destination what happens?
often removed
some matured proteins keep their signal sequence
proteins that are goin to the nucleus tend to hold onto their localization signal— why— because the nuclear membranes break down during cell divisons — nuclear proteins need to “save their ticket” to return to the nucleus when those membranes are reassembled to form the nuclei of the daughter cells.
Signals can be primary aa sequence or
3-dimentional structure formed by aa’s of the protein
Proteins enter the nucleus through
nuclear pores
Nuclear envelope
Inner vs outer nuclear membrane
encloses the nuclear DNA + defines the nuclear compartment
2 concentric membranes (sharing the same circle)
inner nuclear membrane: has proteins that act as binding sites for the chromosomes + that provide anchorage for the nuclear lamina (woven meshwork of protein filaments that lines the inner face of this membrane and provides structural support for the nuclear envelope)
outer membrane: composition resembles the ER membrane. Outer membrane is continuous with the ER
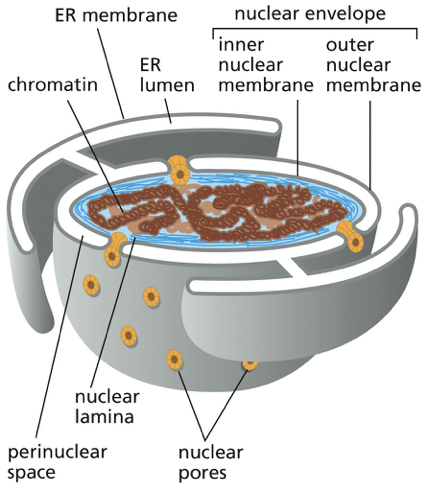
the outer nuclear membrane is continuous with ?
the ER membrane
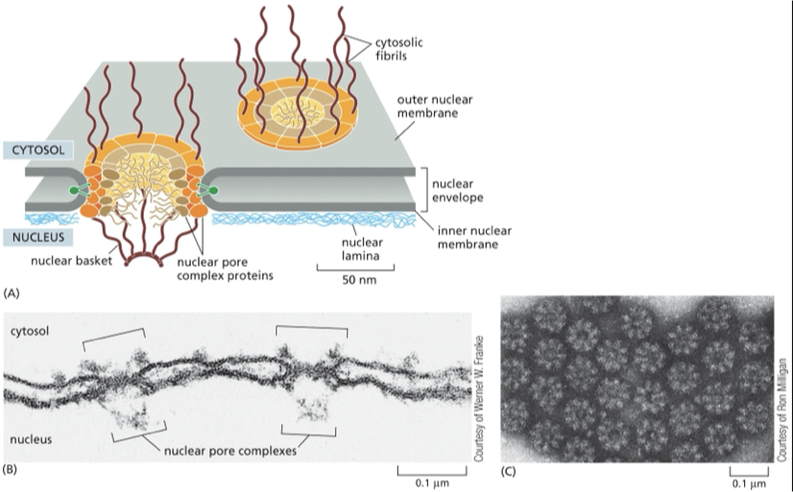
nuclear pores
channel that large molecules can move between the nucleus and the cytoplasm
all eukaryotic cells nuclear envelope is perforated by nuclear pores
large + elaborate structure— composed of complex of 30 different proteins
Nuclear pore proteins have unstructured, disordered regions that form a soft, tangled meshwork (like a kelp forest) in the center of the channel. This mesh blocks large molecules but allows small, water-soluble molecules to pass freely and non-selectively between the nucleus and cytosol.
The nuclear pore complex
• giant complex (50-100 polypeptides)
•diameter of complex: 120 nm
•diameter of opening: 25 nm
passive diffusion through the nuclear pores vs active transport
discuss the difference
what is the nuclear localization signal and nuclear import receptor
Passive diffusion:
small molecules
small proteins <50kDa (no energy required)
Active transport
Required for large proteins and RNAs
RNA molecules are synthesized in the nucleus. Ribosomal units are assembles in the nucleus and they must be imported from the cytosol.
Newly made proteins that are going to the nucleus need to be imported from the cytosol… how do the larger molecules gain entry?
Nuclear Localization signal [aka NLS] (signal sequence that directs a protein from the cytosol into the nucleus)
7 amino acid stretch, typically have several (+) charged amino acids (Lysines and arginines) in the middle of the polypeptide
nuclear import receptors [aka importins] : the NLS proteins that are trying to get from the cytoplasm to the nucleus are recognized by the import receptors
recptors guide the newly synthesized protein to a nuclear pore by interacting with the fibrils that extend from the pore like a tentacle. Those tentacles extend from the pore to the cytosol
After locating a pore, the receptors with their cargo jostle their way through the gel-like meshwork formed from the unstructured regions of the nuclear pore proteins until nuclear entry triggers cargo release.
After cargo delivery, the receptors return to the cytosol via nuclear pores for reuse
![<p><span><strong><span>Passive diffusion:</span></strong></span></p><ul><li><p><span><span> small molecules</span></span></p></li><li><p><span><span>small proteins <50kDa (no energy required)</span></span></p></li></ul><p><strong>Active transport</strong></p><ul><li><p>Required for large proteins and RNAs</p><ul><li><p>RNA molecules are synthesized in the nucleus. Ribosomal units are assembles in the nucleus and they must be imported from the cytosol.</p></li></ul></li></ul><p>Newly made proteins that are going to the nucleus need to be imported from the cytosol… how do the larger molecules gain entry?</p><ul><li><p><strong>Nuclear Localization signal</strong> [aka NLS] (signal sequence that directs a protein from the cytosol into the nucleus) </p><ul><li><p>7 amino acid stretch, typically have several (+) charged amino acids (Lysines and arginines) in the middle of the polypeptide</p></li></ul></li><li><p><strong>nuclear import receptors [aka importins] :</strong> the NLS proteins that are trying to get from the cytoplasm to the nucleus are recognized by the import receptors </p><ul><li><p>recptors guide the newly synthesized protein to a nuclear pore by interacting with the fibrils that extend from the pore like a tentacle. Those tentacles extend from the pore to the cytosol </p></li><li><p>After locating a pore, the receptors with their cargo jostle their way through the gel-like meshwork formed from the unstructured regions of the nuclear pore proteins until nuclear entry triggers cargo release.</p></li><li><p> After cargo delivery, the receptors return to the cytosol via nuclear pores for reuse</p></li></ul></li></ul><p></p>](https://knowt-user-attachments.s3.amazonaws.com/973c38c4-eb33-48c5-879a-e67792cab3f7.png)
what prevents the nuclear import proteins from entering the nucleus empty handed and then returning from the nucleus with nuclear proteins
movement is guided by the hydrolysis of a nucleoside (in this case its GTP)
the hydrolysis is controlled by a single sub unit of GTPase Ran
GTP hydrolysis drives nuclear transport in the appropriate direction
Ran
Ran GTP
Ran GDP
Ran-GAP
Ran- GEF
Two forms on RAN
molecule has GTP
molecule has GDP
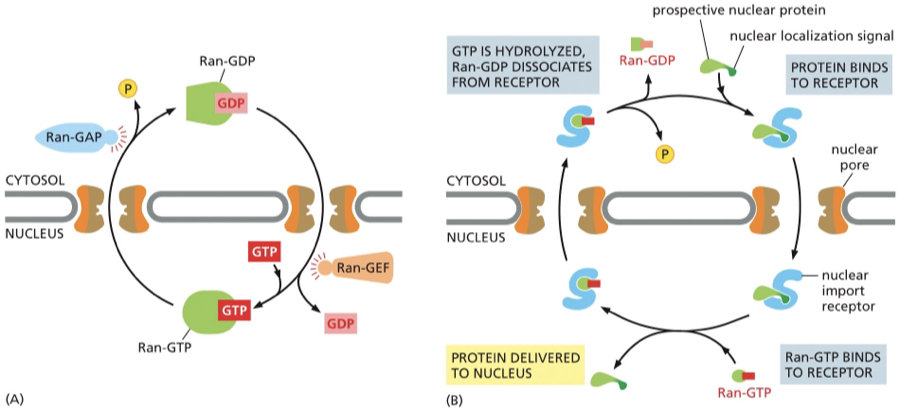
Figure 1 shows
Ran GTP is present in high concentration in the nucleus
Ran GDP= in the cytosol
Ran is converted from one form to the other with the help of accessory proteins that are found in different locations.
Ran-GAP (GTPase- activating protein) =triggers GTP hydrolysis (removing a phosphate) — Ran- GAP is found exclusively in the cytosol —- converts Ran-GTP to Ran- GDP
Ran- GEF (guanine nucleotide exchange factor)= t (importiin — found exclusively in the nucleus
Because these two proteins are in different locations, you get a gradient:
High Ran-GTP inside the nucleus (thanks to RanGEF)
High Ran-GDP in the cytosol (thanks to RanGAP)
That difference in concentration drives the direction of nuclear transport — it tells import and export receptors where to bind or release their cargo.
Figure 2:
Nuclear import receptor (importin) in the cytosol picks up a prospective nuclear protein by binding to the NLS of the nuclear protein and enters the nucleus
Encounters Ran-GTP — Ran GTP binds to the import receptor — causes a release the nuclear protein — cargo is released in the nucleus. (importin can only release cargo inside of the nucleus if it interacts with Ran-GTP)
the receptor still carrying Ran-GTP is transported back through the pore to the cytosol (Importin bound to Ran-GTP will exit the nucleus)
in the cytosol the interaction with RanGAP hydrolyzes its bound GTP —GDP
Ran GDP falls off the import receptor — which is then free to bind to another protein goin to the nucleus
GDP has less affinity for the import receptor— dissociates— leaving receptor free to pick up another protein that needs to go to the nucleus
Ran GDP cant bind importin and release it back into the cytoplasm
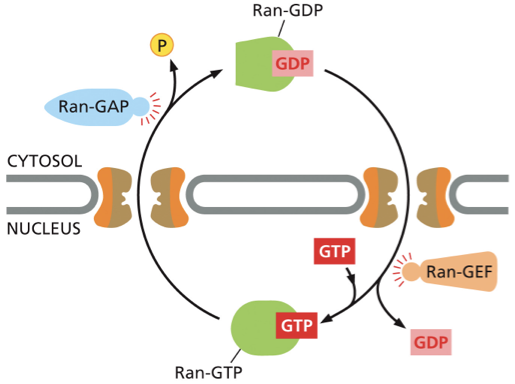
Ran-GAP & Ran-GEF— ppt notes
Ran GAP (Ran GTPase activating protein)
interacts with Ran and activates Ran’s GTPase activity
located on the cytoplasmic side
triggers GTP hydrolysis (removing a phosphate) —- converts Ran-GTP to Ran- GDP
Ran-GEF (Ran GTPase Guanine nucleotide exchange factor)
interacts with Ran and induces Ran to release GDP (the accessory protein that causes Ran-GDP to release its GDP and take up GTP)
naturally high levels of GTP result in binding of GTP
located inside of the nucleus
cytosol to nucleus— transported via nuclear pores
what feature distinsguishes this type of nuclear transport mechanism from the other mechanisms that transport proteins into most other organelles
proteins are transported into the nucleus in their fully folded conformation and ribosomal components as assembled particles
this is different than the mitochondria and chloroplast where the proteins have to be unfolded to cross membranes
The __________is the most extensive membrane system in a eukaryotic cell
ER
Importance of ER regarding proteins
ER serves as an entry point for proteins that are destined for other organelles and the ER itself
proteins that need to go to the golgi, endosome, and lysosome all enter the ER from the cytosol — once inside of the ER lumen or embedded in the ER membrane — the individual proteins will not reeneter the cytosol during their journey forward — instead they are ferried by transport vesicles from organelle to organelle w/ in the endomembrane system/ plasma membrane
What are the 2 kinds of proteins that are transferred from the cytosol to the ER
water soluble proteins - that are translocated across the ER emmbrane and released into the ER lumen
destined for secretion (release at the cell surface) OR for the lumen of an organelle of the endomembrane system (either gonna leave the cell or stay inside of another organelle)
prospective transmembrane proteins — partly translocated across the ER membrane and become embedded in it
destined to stay in the membrane of the organelles or in the plasma membrane
The proteins that are headed to the ER are initially directed to the ER by?
an ER signal sequence— a segment of 8 or more hydrophobic amino acids — involved in process of translocation across the membrane
Rough ER
region of the ER associated with ribosomes and involved in the synthesis of secreted and membrane bound proteins
membrane bound ribosomes vs free ribosomes
membrane- bound ribosomes
are attached to the cytosolic side of the ER membrane or the outer nuclear membrane
are making proteins that are being translocated into the ER
Free ribosomes
unattached to any membrane
making all the other proteins encoded by the nuclear DNA
if the synthesized protein is destined for the nucleus chloroplast, mitochondria, or stay in the cytosol translation occurs in the free ribosomes
All of these ribosomes, whether membrane-bound or free, are part of a common pool
membrane bound ribosomes and free ribosomes are structurally and functionally identical they differ only in the proteins they are making at a given time
• if the synthesized protein is destined for the ER and Golgi as a transmembrane, lysosomal, or secreted product: translation occurs in ?
ribosomes that are associated with the rough endoplasmic reticulum (rough ER), and the newly synthesized protein is translocated into the lumen of the ER
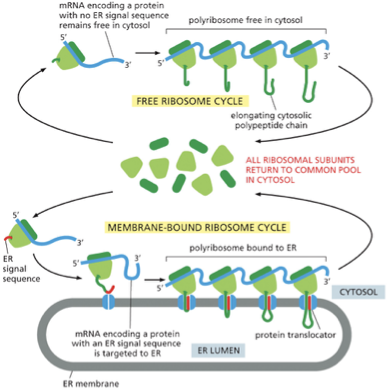
describe what is goin on in this figure:
A common pool of ribosomes is used to synthesize all the proteins encoded by the nuclear genome
ribosomes that are translating proteins with no ER signal sequence remain free in the cytosol
ribosomes that are translating proteins containing ER signal sequence (red) on the growing polypeptide chain will be directed to the ER membrane
polyribosome: many ribosomes bind to each mRNA molecule
At the end of each round of protein synthesis, the ribosomal subunits are released and rejoin the common pool
the elongation of each polypeptide provides the thrust needed to push the growing chain through the ER membrane
Soluble proteins made on the ER are released into the________
ER lumen
2 protein components that help guide ER signal sequences to the ER membrane
Signal Recognition particle (SRP)
cytosol
binds to both the ribosome and the ER signal sequence as it emerges from the ribosome
SRP receptor
embedded in the ER membrane recognizes SRP
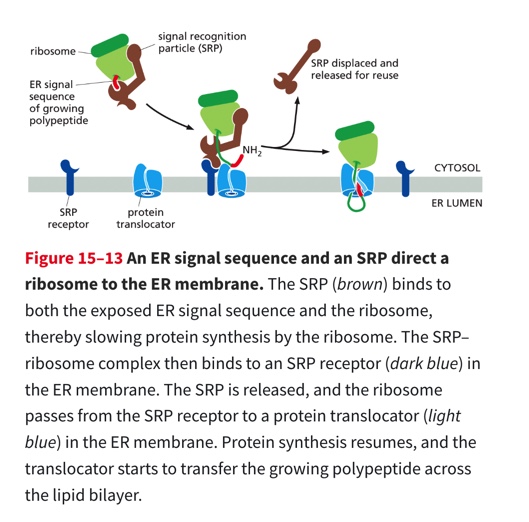
Process
Secreted proteins contain a signal sequence at their amino terminus (8 or more hydrophobic amino acid sequences)
the SPR binds to a ribosome that has a ER signal sequence (this slows protein synthesis)
SPR is then bound to the SPR receptor — Once bound SPR is released & the receptor passes the ribosome to a protein translocator in the ER membrane + protein synthesis recommences
polypeptide is threaded across the ER membrane through a channel in the translocator
SPR relocated the ribosome, mRNA, and protein to the ER to continue translation across the ER membrane
The SRP and SRP receptor function as molecular matchmakers: they bring together ribosomes that are making proteins that have the ER signal sequence with protein translocators w/ in the ER membrane
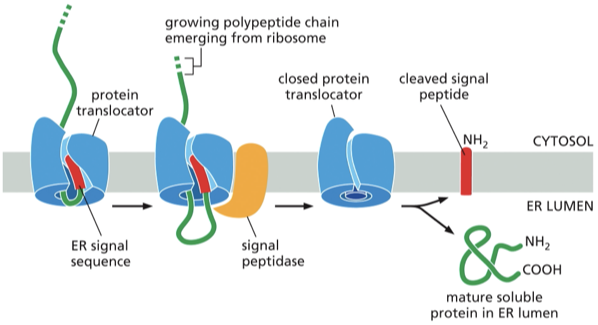
Describe the process of a soluble protein crossing the ER membrane and entering the lumen
the signal sequence ( @ the N terminus aka the end that was synthesized first) function= to open the protein translocator
the signal sequence at that opened the protein translocator will remain bound to the translocator, while the rest of the polypeptide chain is threaded through the membrane to form a big loop
for soluble proteins: the signal sequence is removed by a transmembrane signal peptidase (has an active site facing the lumenal side of the ER membrane.)
what happens to the cleaved signal?
it is released from the protein translocator into the lipid bilayer and than degrades rapidly
Once the C- terminus of the soluble protein has passed through teh translocator the protein is released into the ER lumen
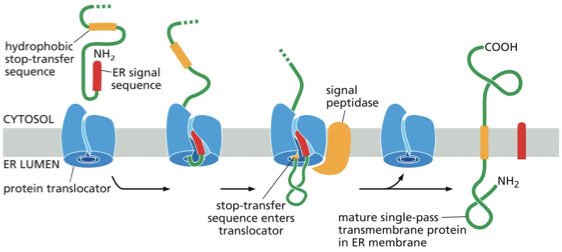
Not all proteins made by ER-bound ribosomes are released into the ER lumen, discuss transmembrane proteins
Transmembrane proteins: remain embedded in the ER membrane
•Transmembrane proteins have additional signals resulting in release into the membrane
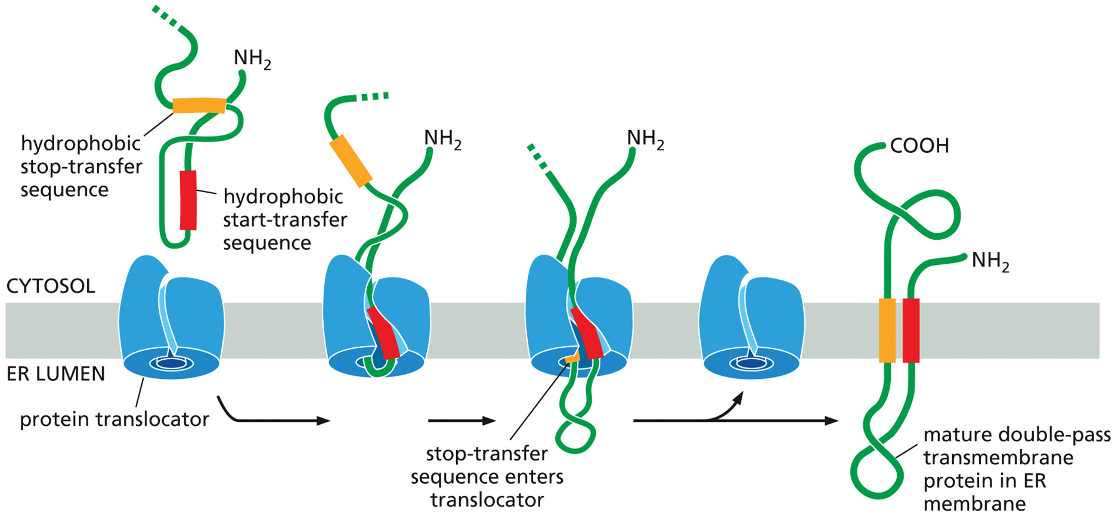
In the figure you’ll see
An N terminal ER signal sequence(Red) initiates translocation
transfer process is halted by a hydrophobic amino acids — stop transfer sequence (orange)
protein translocator releases the growing peptide chain—- the N terminal signal sequence is cleaved off while the stop transfer sequence stays in the bilayer and forms a an alpha helical membrane segment that anchors the protein in the membrane
Result: the protein is a single pass transmembrane protein.
How is it oriented?
the N terminus (NH2) is on the ER lumen side
C terminus (COOH) is on the cytosolic side
Once inserted into the membrane will a transmembrane protein change its orientation?
No, its cytosolic portion will always remain in the cytosol even if the protein is transported to another organelle via vesicle budding or fusion
For a protein that functions at the plasma membrane the portion of the protein that faces the ER lumen will ultimately be exposed to the _________ (outside or inside …choose one) of the cell
outside
multipass transmembrane protein
from ppt: Internal signals result in multi-pass proteins
in some transmembrane proteins an internal signal sequence ( rather than an N terminal) is used to start the protein transfer
internal signal sequence= start- transfer seqeunce (never removed from the polypeptide)
arrangement happens in multipass transmembrane proteins
the hydrophobic signal sequences work in pairs:
internal start transfer sequence (initiates translocation until it a stop transfer sequence is released into the bilayer— they then remain as membrane spanning alpha helices) — no the star transferred nor the stock transfer sequences are cleaned off and the entire polypeptide chain remains anchored in the membrane (in the figure you can see a double pass transmembrane protein)
Proteins that spend the membrane more than twice contain additional pairs of start and stop transfer sequences and the same process is repeated for each pair
Which organelle cannot receive proteins directly from the cytosol?
A. nucleus
B.mitochondria
C. chloroplast
D. golgi app
D. golgi app
First step on the pathway: ER
next stop: golgi apparatus
function of the golgi?
golgi app: proteins and lipids are modified and sorted for shipment to other destinations
Transport vesicles
membrane vesicle that carries proteins from one intracellular compartment to another
carry soluble proteins and membrane between compartments
Vesicular Transport
what is it?
what is the difference between exocytosis and endocytosis
allows materials to exit or enter the cell
movement of material between organelles in the eukaryotic cell via membrane enclosed vesicle
Figure explanation
exocytosis: vesicle fuses with the plasma membrane— releasing its content to the cells surroundings
endocytosis: extracellular materials are captured by vesicles that bud inward from the plasma membrane and are carried into the cell
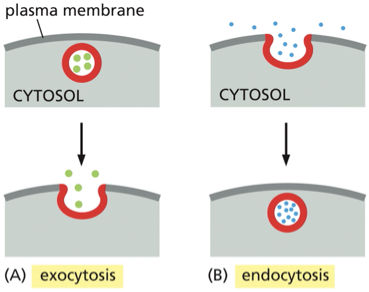
True or false transport vesicles carry soluble proteins and membrane between compartments
true
true or false vesicular transport between membrane enclosed compartments of the endomembrane system highly disorganized
false it is highly organized
Outward secretory pathway vs inward endocytic pathway
major outward secretary pathway starts w/ the synthesis of proteins on the ER membrane and their entrance into the ER — leads to through the golgi app to the cell surface— @ the golgi a side branch leads off through endosomes to lysosomes
major inward endocytic pathway- responsible for the ingestion and degradation of extracellular molecules, moves materials from the plasma membrane, through endosomes to lysosomes
some rules/ regulations…
transport vesicle that buds off from a compartment has to take ONLY proteins that are supposed to go to that location
transport vesicles have to fuse with only the appropriate target membrane
vesicle budding is driven by?
the assembly of a protein coat
coated vesicle
small membrane enclosed sac that wears a distinctive layer of proteins on its cytosolic surface. It is formed by pinching off a protein-coated region of a cell membrane
2 funcitons
helps shape the membrane into a bud
captures molecules for onward transport
after budding from its parent organelle what happens to the vesicles coat
the vesicle sheds its coat— allows its membrane that is underneath the coat to interact directly with the membrane that it wants to fuse with
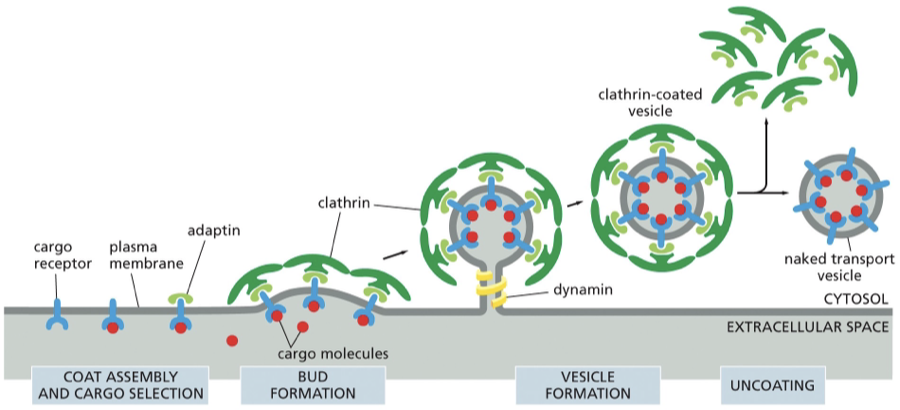
clathrin coated vesicles
what is clathrin
dynamin vs adaptins
describe this figure
clathrin: protein that makes up the coat of a transport vesicle that buds from either the golgi app (on the outward secretory pathway) or from the plasma membrane (on the inward endocytic pathway). Plays no part in choosing specific molecules for transport.
Dynamin: a GTP binding protein assembles as a ring around the neck of each deeply invaginated clathrin- coated pit — together with other proteins the dynamin causes the neck to restrict and pinches off
Adaptin: secure the clathrin coat to the vesicle membrane and help select cargo molecules for transport
from the powerpoint slides:
•Coat proteins bend the membrane to form vesicles
•Require adaptor molecules that recognize cargo receptor proteins
•Cargo is specific
vesicle docking depends on ?
tethers and SNARES
A transport vesicle buds from a membrane, it needs to find its way to its destination to deliver its contents what helps to actively transport the vesicles
motor proteins that move along skeletal fibers
once the transport vesicle reaches its target it needs to ?
recognize + dock with a specific organelle only then can the vesicle membrane fuse with its target membrane and unload the vesicles cargo
Rab proteins, tethering proteins, and SNAREs help direct transport vesicles to their target membranes.
specificity of vesicular transport once its gotten to its target protein suggests what about the transport vesicles
each type of transport vesicle in the cell displays molecular markers on its surface that identify the vertical according to its origin and cargo these markers must be recognized by complementary receptors on the appropriate target membrane including the plasma membrane
Rab proteins & tethering proteins
Rab proteins: one of a family of small monomeric GTPases proteins present on the surface of transport vesicles and organelles that serves as a molecular marker to help make sure that the transport vesicle fuse only with the correct membrane
the rab proteins are recognized by corresponding tethering proteins on the cytosolic surface of the target membrane
tethering proteins: are filamentous transmembrane proteins involved in the docking of the transport vesicles to membranes
rab and tethering help to make sure that the vesicles only fuse with the correct membrane— they serve as molecular markers
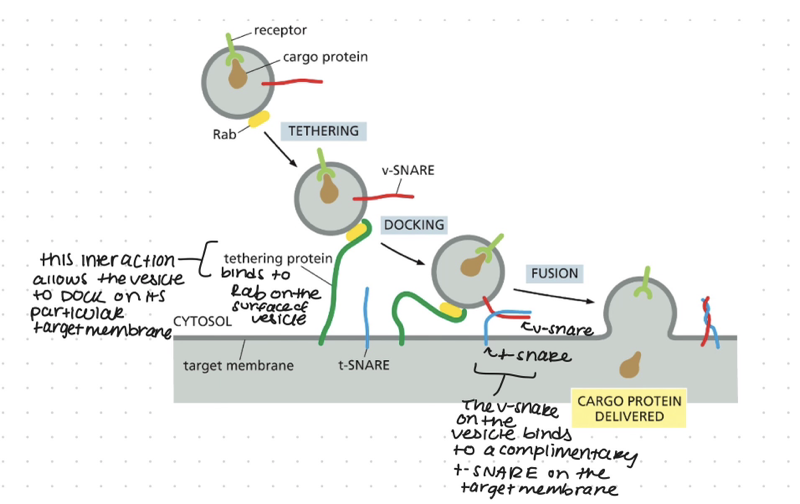
SNAREs
transmembrane protein
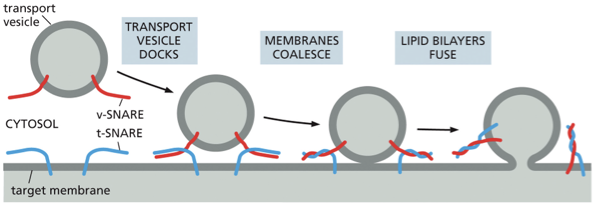
tethering protein grabs vesicle by the rab protein, the SNAREs on the vesicle called the v-SNAREs will interact with the SNAREs on the target membrane called the t-SNAREs this interaction allows the vesicle to dock firmly in place
catalyzes the membrane fusion thats needed to deliver the cargo that was inside the vesicle. this also adds the vesicles membrane to the membrane of the organelle
when fusion is triggered, the v-SNAREs and t-SNAREs wrap around each other tightly, thereby acting like a winch that pulls the two lipid bilayers into close proximity
figure description:
Once appropriately triggered, the tight pairing of v-SNAREs and t-SNAREs draws the two lipid bilayers into close apposition. The force of the SNAREs winding together squeezes out any water molecules that remain trapped between the two membranes, allowing their lipids to flow together to form a continuous bilayer. In a cell, other proteins recruited to the fusion site help to complete the fusion process. After fusion, the SNAREs are pried apart so that they can be used again.
Rab and tethering proteins provide the initial recognition between the vesicles and its target membrane the SNARE proteins?
ensure that transport vesicles dock at the correct target membranes they also catalyze the final fusion of the 2 membrane
Newly made proteins, lipids, and carbohydrates are delivered from the ER, via the Golgi apparatus, to the cell surface by transport vesicles that fuse with the plasma membrane in the process of __________
exocytosis
Proteins synthesized on the rough ER take the secretion pathway:
endoplasmic reticulum (ER): protein processing
ER- golgi intermediate complex (ERGIC): protein sorting
Golgi apparatus: more processing and protein sorting
possible destinations
- to the lysosomes (lysosomal proteins)
or
- to the plasma membrane ( transmembrane proteins)
or
-out of the cell through secretory vesicles (secreted pathways)
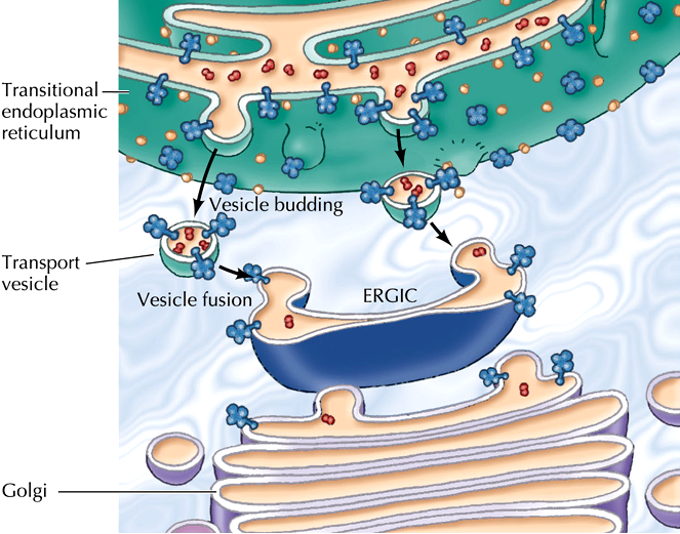
ER-Golgi intermediate complex (ERGIC)
is a system of membrane stacks located between the Endoplasmic reticulum and the Golgi.
Function:
a checkpoint that ensures that proteins that should stay in the ER (ER resident proteins) do not reach the Golgi complex
•Traffic from the ER is non-selective
golgi apparatus structure
located near the cell nucleus
animals cells is close to the centrosome( small cytoskeletal structure near the cell center)
consists of a collection of flattened membrane- enclosed sacs called cisternae (like piled stacks pita bread that have been cut open)
each stack contains 3-20 cisternae
# of golgi stacks varies depending on the cell type
2 distinct faces
Cis face (entry)— adjacent to the ER
Trans face (exit) — points toward the plasma membrane
both cis and trans are important for protein sorting— proteins entering the cis golgi network can either keep on moving through the golgi stack OR the proteins could have a ER retention signal which and they are returned to the ER
Proteins exiting from the trans golgi network are sorted on their destination either to the lysosomes via endosomes or for the cell surface
Soluble proteins and pieces of membrane enter the _____ Golgi network via transport vesicles derived from the ER.
cis
The proteins then travel through the cisternae in sequence in two ways: (1) by means of transport vesicles that bud from one cisterna and fuse with the next; and (2) by a maturation process in which the Golgi cisternae themselves migrate through the Golgi stack.
Proteins finally exit from the _____ Golgi network in transport vesicles destined for either the cell surface or another organelle of the endomembrane system
trans
Golgi membrane proteins recognize ER proteins and
send them backwards
constitutive secretion vs regulated secretion
constitutive secretion
operates in all cells continuously, unregulated secretion
In all eukaryotic cells, a steady stream of vesicles buds from the trans Golgi network and fuses with the plasma membrane in the process of exocytosis. This constitutive exocytosis pathway supplies the plasma membrane with newly made lipids and proteins
Regulated secretion
operates only in cells that are specialized for secretion = secretory cells—these cell types release their secreted products only when stimulated by an external signal
secretory vesicles: membrane enclosed organelle in which molecules destined for secretion are stored prior to release
part of the endomembrane system bud off from the trans golgi network and accumulate near the plasma membrane
selected proteins in the trans Golgi network are put into secretory cells where the proteins are concentrates and stored until and extracellular signal stimulates their secretion— ex would be an increase in blood glucose which signals insulin -producing endocrine cells to secrete the hormone