Section 4: Nucleic Acids
1/72
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
73 Terms
What do nucleic acids consist of? What is it defined by?
They consist of bases linked to a sugar phosphate backbone
form of linear information
Each nucleic acid is defined by the sequence of bases
Each monomeric unit contains a sugar, base and phosphate
What are info,fed in the formation of the nucleic acid backbone on the sugar? What’s the difference between DNA and RNA?
3’ OH and 5’OH
DNA lacks 2’OH
How are sugars linked? What’s the directionality?
they’re linked by phosphodiester linkage, 5’-3’
The 2’OH in RNA can hydrolyze the phosphodiester backbone
What are purines vs pyrimidines?
Purine (double ring) - adenine, guanine
Pyrimidine (single ring) - cytosine, uracil, thymine
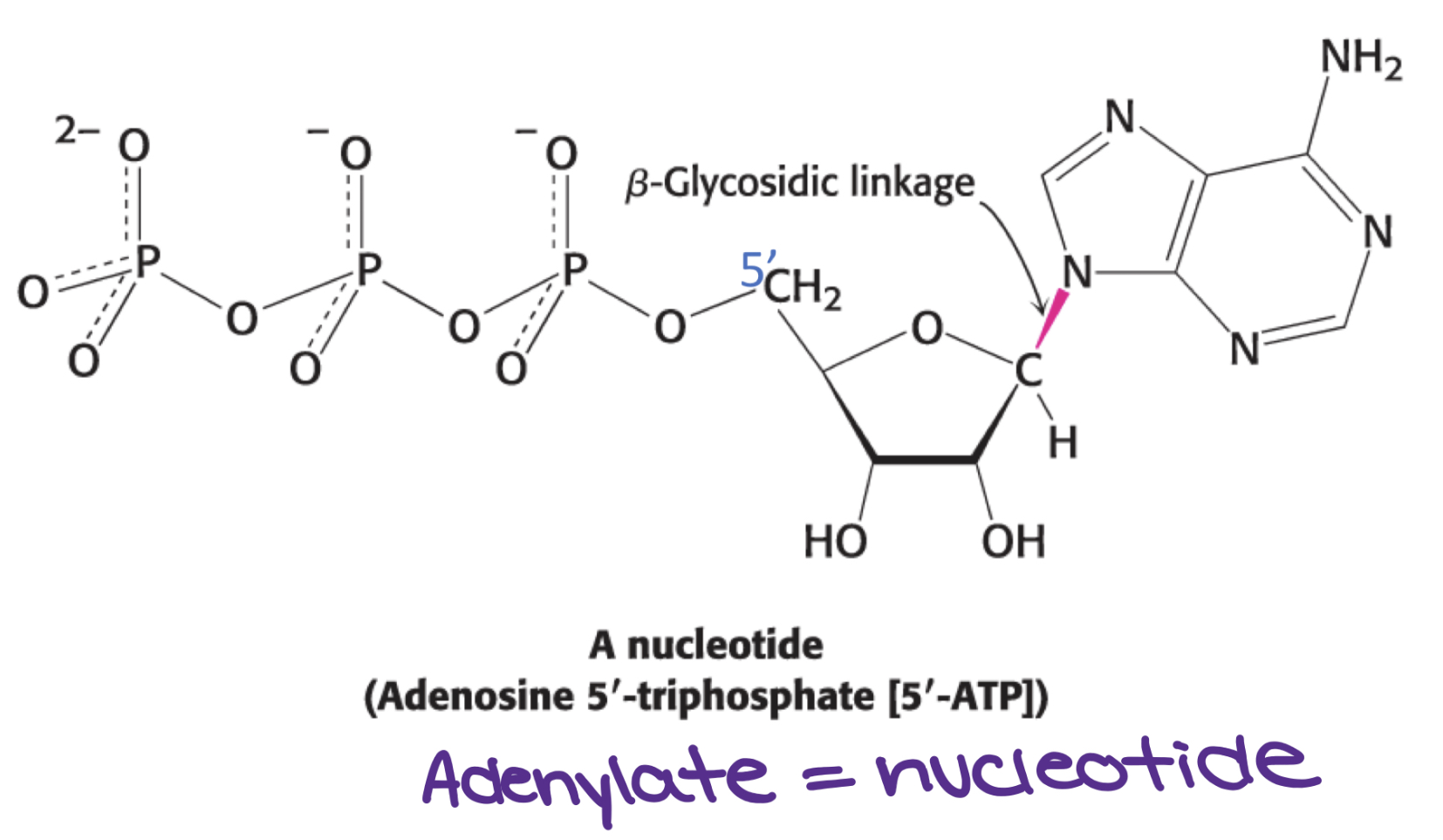
What’s a nucleotide?
Consists of a base, pentose sugar, one or more phosphates (nucleoside joined to phosphoryl group by ester linkage)
Ex. Adenylate = nucleotide
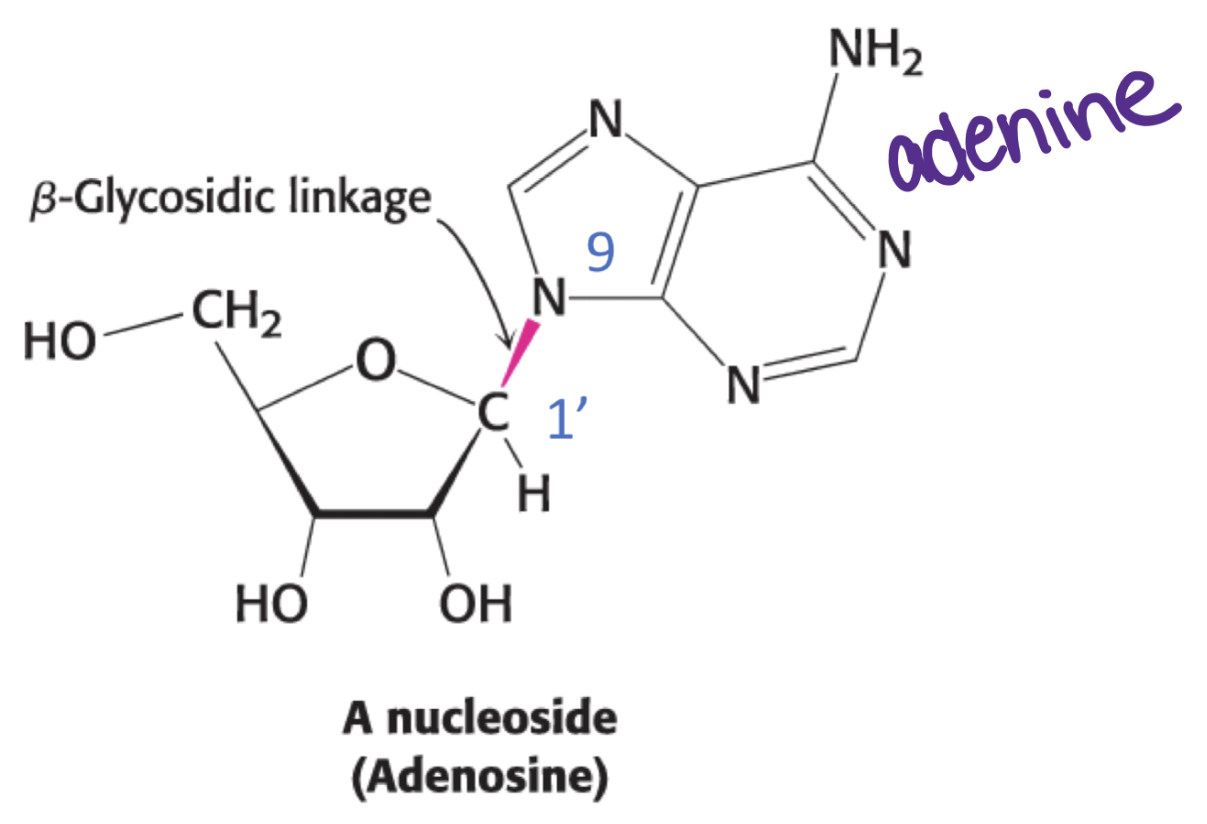
What’s a nucleoside?
Only the base and pentose sugar
Ex. Adenosine
How many bases per turn of double helix? How long? How much each base separated by?
34 Angstroms, 10.4 bases per turn, each base is 3.4 angstroms and 36 degrees
Four properties of double helix chain?
Two DNA chains of opposite directionality intertwine to form a right-handed double helix
Sugar-phosphate backbones are on outside, bases are inside
Bases perpendicular to axis of helix with bases separated by 3.4 angstroms and 10.4 bases/turn
Helix is 20 Angstroms wide
How many hydrogen bonds does AT and GC have?
GC-3
AT-2
What causes variations in base pairing?
rotation of a base in RNA or DNA ends (reverse Watson crick)
Tautomerization (wobble)
Rotation around C1-N glycosidic bond (Hoogsteen)
What contributes to the stability of the helix?
Hydrophobic effect: hydrophobic interactions drive bases to inside of the helix, more polar residues outside
Base stacking: stacked bases attract each other through van der waals forced
many weak non covalent interactions provide stability but still allow for strand separation
What is the major and minor grooves?
proteins recognize patterns of H-bond donors/acceptors in the major and minor grooves
Adenine-Thymine = (NHO/NO)
Guanine-Cytosine = (NOH/NHO)
Why does the major and minor grooves exist?
Glycosidic bonds of each base in a pair are not diametrically opposite each other
Lined by hydrogen bonds and acceptors
What are supercoiled structures?
Relaxed circle (260 bp, 10.4 bp/turn, 25 turns
Negative superhelix (23 regular turns and two negative turns
Unwound circle
Supercoiled species travel fastest through agarose gel
What does 3D RNA structure allow for?
Some RNA act as catalysts (ribosomes)
How does DNA replicate?
DNA polymerase: catalyses addition of deoxyribonucleotides to a DNA strand
E Coli polymerases=
polymerase I: primer removal, DNA repair
3-5 exonuclease
5-3 exonuclease
Polymerase III: replicative polymerase
3-5 exonuclease domain
What induces a conformational change in DNA pol?
binding of the correct incoming dNTP
Mg2+ stabilizes negative charges on phosphates, dNTP is added and helix clamps down on active site
What does DNA pol III catalyze during replication?
-strand elongation reaction
Mg2+ stabilize negative charges and deprotonates hydroxyl so it can attack alpha phosphate
Pyrophosphate is released, two bases are now connected by phosphate group
What would happen if ddNTP was added instead of dNTP?
It would still base pair and the O would bind phosphate of previous dNTP, however synthesis would stop following the ddNTP since there would be no 3’ hydroxyl to attach alpha phosphate
5 requirements/abilities of strand-elongation reaction by DNA pol
Requires dNTPs and Mg2+
New DNA strand is assembled on a pre-existing strand
Requires a primer to begin synthesis
Elongation is in 5-3’ direction
Can correct mistakes using 3-5’ exonuclease activity (proofreading)
How are mistakes corrected by Polymerase?
3-5 exonuclease domain removes incorrect nucleotides from 3’ end of the growing strand by hydrolysis
Mismatch results in a stall
Pause gives additional time for the incorrect region to flop into the exonuclease active site where it’s removed
What are the four components of DNA replication in E Coli?
Origin of Replication (OriC): region of DNA that is the start site of replication
DnaA: binds to OriC to initiate pre-priming complex
SSB: single strand binding protein that keeps 2 single stranded regions separate
DnaB: helicase that seperates duplex DNA
How do helicases unwind ddDNA?
Hydrolysis of ATP causes ratcheting of the subunits of the hexamer, pulling ssDNA through the center
What does primase do?
rna pol that creates a short primer (10 nt) (directs dna pol where to go)
Describe the DNA replication fork
Parental DNA is getting unzipped by helicase, and held separate by SSB proteins
DNA pol works in 5’-3’ direction, so one strand can be synthesized continually (leading strand)
Whereas the other is synthesized in fragments (lagging strand), as helicase moves forward primase has to lay new primers for dna pol III
What does B2 do?
Sliding clamp improves processivity
Describe process of leading and lagging strand synthesis?
Leading strand:
made continuously in 5-3’ direction by pol III
Lagging strand:
looped out
Starting from an RNA primer, pol III adds ~1000 nt in 5-3’ direction
Released B2 sliding clamp
New loop formed, sliding clamp added
Primase creates new RNA primer
Pol III synthesizes new Okazaki fragment
Pol I fills gaps between fragments and removes RNA primer with 5-3’ exonuclease activity
Ligase seals fragments
How does ligase join fragments
Joins the 3’ OH and the 5’ phosphate group of two fragments (with ATP)
Describe the process of ligase sealing fragments
Initial charging:
catalytic lysine attacks the alpha phosphate of ATP, generating Lysyl-AMP adduct and releasing pyrophosphate
Transfer:
adenylated ligase transfers AMP from lysine to the 5’ phosphate at the nicked backbone
Phosphodiester bond formation:
3’OH at the nick site attacks adenylated 5’ phosphate, making a phosphodiester bond and releasing AMP
What are mutagens? How can DNA be damaged?
mutagens = chemical agents that alter DNA bases
DNA bases can be a,termed through oxidation, sea inaction, alkylation, uv radiation, x-ray exposure
What is guanine oxidation?
hydroxyl radical reacts with guanine to form 8-Oxoguanine
8-oxoguanine base pairs with adenine; mismatch since one strand will have AT base pair
What’s cytosine deamination?
occurs around 500x per cell/day
Removal of an amino group through the reaction with water
Cytosine + water → uracil + ammonia
What’s adenine deamination?
removal of amino group with water → hypoxanthine
Hypoxanthine base pairs to cytosine; AT→ CG
What’s 5-methylcytosine Deamination
cytosine is methylated at C5 to regulate gene transcription
Deamination of 5-methylcytosine results in thymine
Can thymine undergo deamination?
No, thymine lacks an exocyclic amine group
What’s guanine alkylation?
alkylation = addition of hydrocarbon to the base
Alfotoxin B1 (crop mold) epoxide alkylates N-7 of guanine
Epoxide = very reactive 3 atom ring structure
GC→AT transversion
What’s a thymine dimer?
UV lights covalentky links adjacent pyrimidines along the dna strand
pyrimidine dimer (cyclobutane ring) that creates a bulge in the DNA double helix
What are 3 principles of DNA repair?
Damage and repair is occurring constantly
Often repair will restore genetic info
Sometimes, it is not possible to restore original info so the cell uses approximate repairs and or undergoes apoptosis
What are the three steps for DNA repair
Recognize the damage
Remove the damage
Repair the damage
What is mismatch repair?
Mismatch is recognized by MutS
MutL binds and recruits MutH (endonuclease)
An exonuclease excises incorrect region
DNA pol III fills the gap
DNA ligase seals the backbone
Eukaryotes and prokaryotes
What’s direct repair?
repairs without removing fragments of DNA
Photochemical cleavage of pyrimidine dimmers by DNA photolyase
uses energy of visible light to break cyclobutane ring! Doesn’t require nucleotides
Not in mammals
What’s nucleotide excision repair?
“dark repair”, doesn’t require blue light
E. coli, eukaryotes, mammals
Recognizes distortions in helix like bulge created by pyrimidine dimer
UvrABC excinuclease cuts DNA at two sites
DNA pol I fills the gap
DNA ligase repairs the phosphodiester backbone
What’s base excision repair?
non helix distorting damage
Defective base flipped into DNA glycosylase active site (goycosidic bond cleavage)
AP endonuclease nicks phosphodiester backbone
Deoxyribose phosphodiesterase removes deoxyribose phosphate unit
DNA pol I inserts correct nucleotide
DNA ligase seals strand (creation of phosphodiester backbone)
What is needed for transcription to happen?
Template DNA
NTPs (ATP, GTP, UTP, CTP)
RNA polymerase
Mg2+
What are the subunits and function of E. Coli RNA polymerase
alpha 1 and 2: Assembly of core enzyme; interacts with regulatory factors
Beta and beta’: Catalysis, interactions with DNA, RNA
w: required for structure/folding
sigma 70: promoter recognition
How is transcription in E. Coli initiated?
Sigma 70 decreases affinity of RNA pol for DNA
Sigma 70 recognizes and binds the promoter sequence (-35 consensus sequence and -10 pribnow box TATAAT)
After several nucleotides of RNA are synthesized, sigma falls off the core enzyme
NO primer needed
When does elongation begin?
after formation of the first phosphodiester linkage
What’s the elongation reaction for RNA transcriptase?
3’ ON of terminal ribose of growing chain attacks inner most phosphoryl group of the incoming ribonucleotide triphosphate
Synthesis begins de novo; no primer needed
How is transcription intrinsically terminated?
Hairpin in RNA product causes RNA pol to pause
rU-dA base pairs are weak, so that RNA dissociated from DNA template and enzyme
Protein dependent termination = Rho Protein (helicase)
What are differences in transcription between eukaryotes and bacterium?
translation and transcription occur simultaneously in the cytoplasm
Simple control elements
Termination signal is GC rich hairpin poly U structure (not polyA tail)
mRNA is not transported across a membrane (nuclear envelope in eukaryotes)
What are promoter elements for eukaryotic RNA pol II
cis acting element = DNA sequences that regulate expression of a gene located on the same molecule of DNA
TATA -25
Inr = initiator +1
DPE = downstream core promoter element +30
Enhancer (can be more than 1 kb from start site)
CAAT box and GC box (-40 to -150)
What’s the Pre-Initiation Complex?
Transcripotion factors bind cis-acting elements to recruit RNA polymerase II
TATA box is recognized by TFIID by the TATA binding protein
TFIID is a dynamic protein complex
What are the 5 steps of forming the pre initiation complex
TFIID binds DNA and the TBP domain moves between multiple conformation until it binds the TATA box
TFIIA binds and stabilizes the complex and promoter region
TBP fully engages promoter, bending DNA
TFIIB recognized the TBP/DNA complex and recruits Pol II and TFIIF and TFIIE
TFIIH (helicase) unwinds DNA and phsophorylates the C-terminal domain of pol II, which triggers the elongation and recruitment of RNA processing enzymes
Does Pre-Initiation Complex initiate transcription at high levels? How much influence does one TF have?
PIC initiates transcription at low levels
TFs that bind other sites stimulate high levels of transcription
One TF has little influence, many are needed to form a complex that influences transcription (combinatorial control)
How is the mRNA transcript modified?
a 5’ 7-methyl guanosine cap
A 3’ polyadenylate tail
What occurs during splicing?
Introns (non coding regions) are removed and exons (coding regions) are joined together
When does the 5’ cap occur?
Very early on in transcription, after 20-30 nucleotides of RNA are synthesized
what is the cap synthesizing complex composed of?
Four enzymes that associate with phosphorylated RNA pol II
What purpose does the 5’ cap serve?
protect from degradation by nucleases
Interacts with ribosome to enhance translation
What’s the steps in adding the 5’ cap?
Removal of the terminal gamma phosphate at the 5’ end by phosphohydrolase
Diphosphate 5’ end attacks alpha phosphate of a GTP to form a 5’ to 5’ triphosphate linkage
NT of guanine is methylated
Methylation of 2’OH of adjacent riboses
What is the poly A tail?
~ 250 adenylates at the 3’ end of the RNA
What are the enzymes involved in building the poly A tail associated with?
C terminal domain of RNA pol II
What role does PolyA tail have?
Enhance stability and translation efficiency (and nuclear transport)
How is the polyA tail added?
CPSF (cleavage and polyadenylation specificity factor) recognized the cleavage signal and an endonuclease cleaves the mRNA transcript
Poly(A) polymerase adds ~ 259 adenylate to the 3’ end
How is the splice site marked?
Introns begin with GU and ends with AG, conserved from yeast to humans
What’s the spliceosome?
complex of protein and RNA that excises introns and joins exons
What type of reaction does spliceosome catalyze?
2 transesterification reactions (reaction of an alcohol and ester to make a different alcohol and ester)
Describe the splicing process
branch site adenylate 2’OH attacks phosphoryl at 5’ splice site
exon 1 is released
Lariat intermediate is formed
3’OH of exon 1 attacks phosphoryl group at the 3’ splice site
splices product exon1-exon2
Lariat intron
What is the spliceosome made up of?
~300 proteins and five critical small nuclear RNAs (snRNA)
U1,U2,U4,U5,U6
SnRNA + protein = snRNPs (small ribonucleoprotein)
Where does U1 and U2 bind
U1 binds the 5’ splice site
U2 binds the branch site
What does U4,5,6 do?
the U4-U5-U6 complex joins and displaces U1 and U4
Extensive interaction between U2 and U6 brings the 5’ splice site and branch site close together
What facilitates the first and second tranesterification?
1st = facilitated by the “catalytic core” of the spliceosome
2nd = facilitated by U5; requires Mg2+ for catalysis
What products are formed from this?
spliced RNA product and lariat intron