CELL 201: Lecture 4 - Genetic Code + Transcription
1/106
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
107 Terms
What is the Central Dogma?
Direction of info flow from DNA - RNA - Protein
Replication —> Transcription —> translation —> assembly (protein)
Is protein always the final product?
NO
What else can be the product of the central dogma? Give examples
RNA
eg. Enzymes (rRNA Ribosome)
tRNA
Is DNA always the genetic material for central dogma?
NO
If DNA is not always the genetic material. What else can be the template for protein synth? be specific as to what type.
Virus RNA /vRNA
If RNA can be template for proteins, can protein be the template for nucleic acids?
NO you cannot go from Protein to RNA
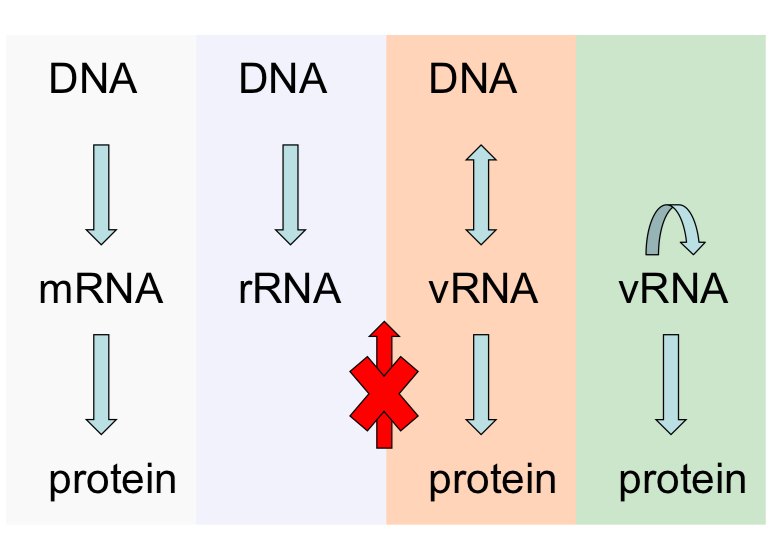
Can RNA be the template for DNA?
Yes
What is the only type of RNA can be the template for DNA?
vRNA
Define reverse transcriptase
enyzmes that catalyze the making of DNA from RNA
Where is reverse transcriptase most often require?
Virus RNA eg. HIV
A RETRO VIRUS
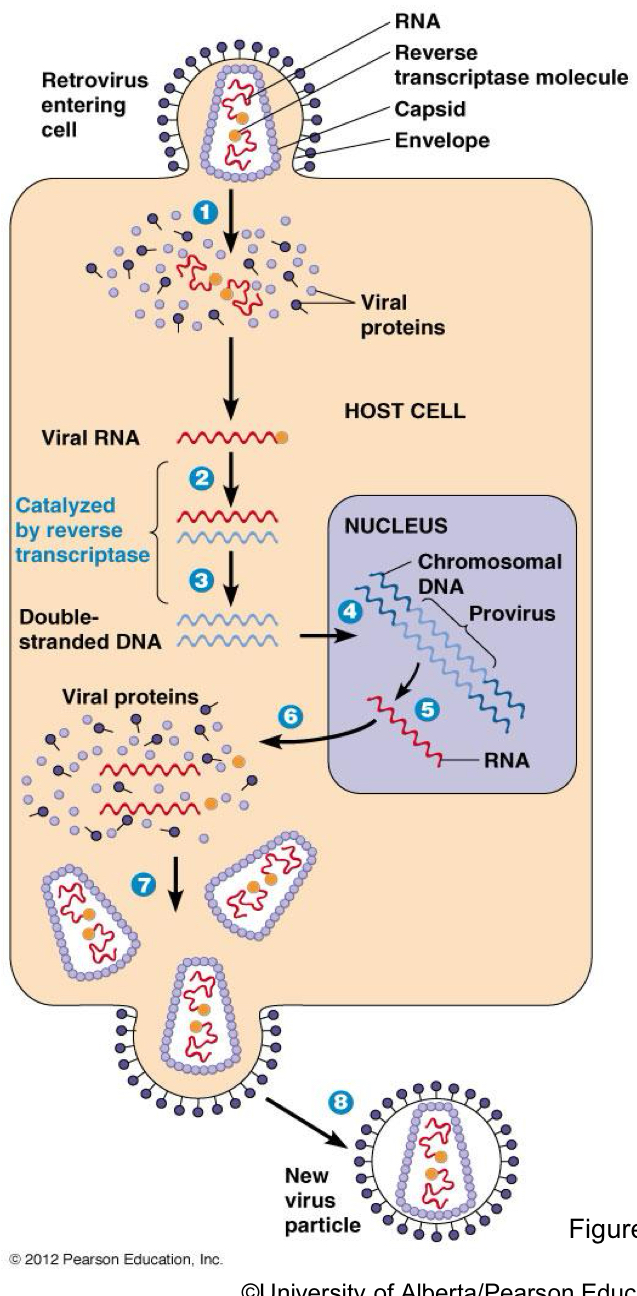
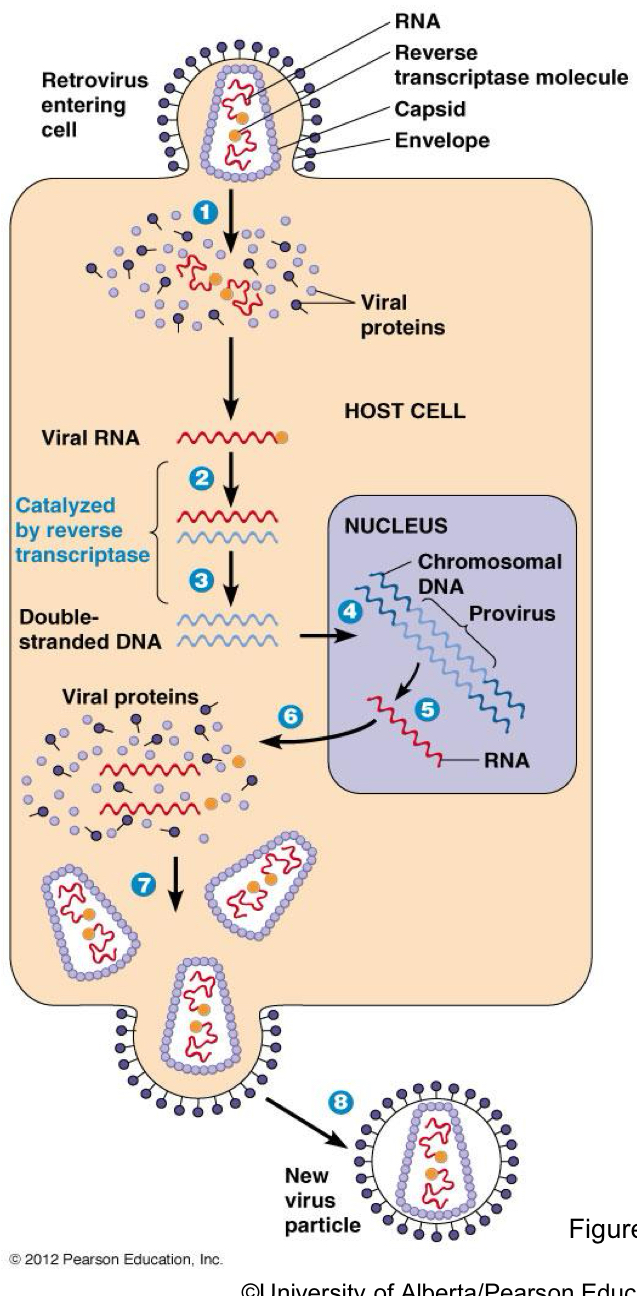
What is SELFISH DNA
Genetic Elements that copy themselves in our genomes.
They only exist to copy themselves and do not benefit the organism
has retrovirus like activity
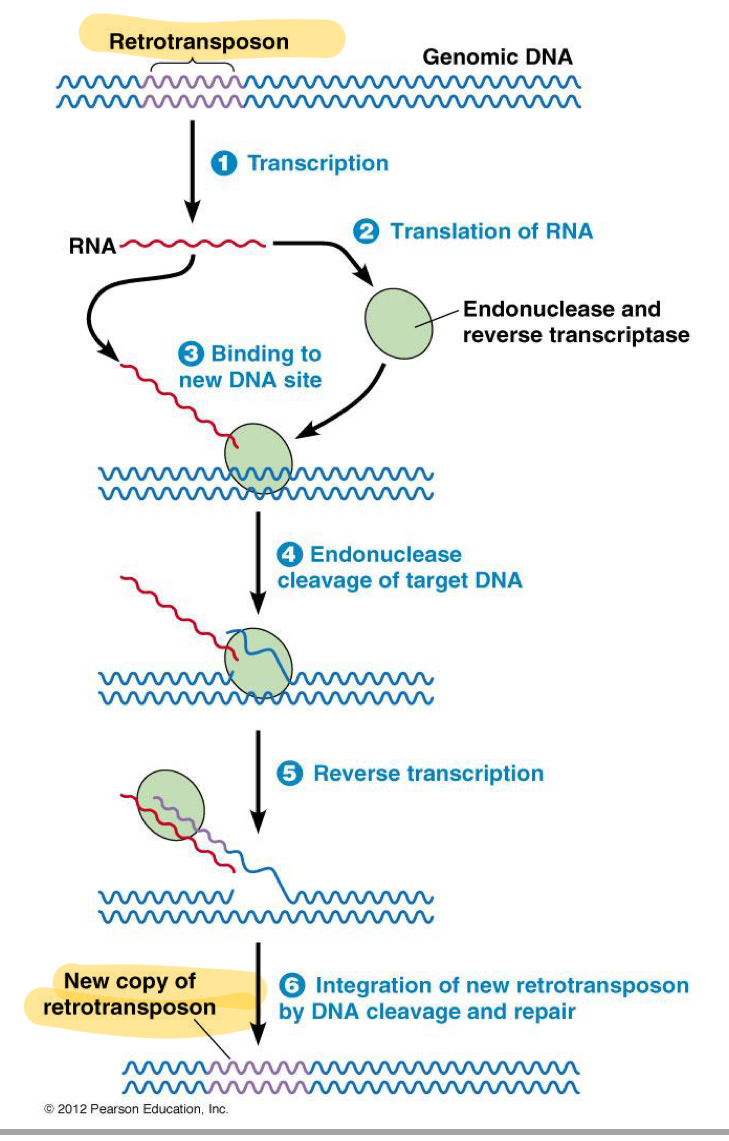
*****What 2 ACTIVITIES does the Selfish DNA Encode?
Reverse transcriptase (Uses RNA as template to synth DNA)
Endonuclease
***How does Selfish DNA work, List the Steps
Transcription —> Translation of the RETROTRANSPOSON (has the 2 activities)
Endonuclease activity cuts in the genome
Uses RNA from transcription of retrotransposon as template
Reverse transcriptase makes the copy the RNA to make DNA hanging on the end of the existing genome
This hanging DNA triggers the host repair machinery
Repair machinery “Fixes” the lesion by copying the extra DNA onto the other strand
Result = insert Retrotransposon DNA that will beable to also do this process
What is the Genetic code?
a triplet code in which combinations of the three bases specify amino acids
Which strand of DNA is the Template strand for mRNA and which is the coding strand?
Template = 3’—>5’ end
Coding = 5’—>3’ = the same as mRNA (Except U instead of T)
How many total combinations of Nucleotide triplets are there?
64
4 amino acids ³ (3 = # of amino acids per code)
Define degenerate
64 code but only 20 amino acids
one amino acid can be specified by more than 1 triplet
Is the Genetic code overlapping or non-overlapping?
non-overlapping
define non-overlapping
Reading frame is advance 3 at a time,
nucleotides from the previous triplet will not be incorporated into the next triplet
Which is the start codon?
AUG
How many Stop codons are there? What are they?
3
UAG
UAA
UGA
Do the stop codons correlate to an amino acid?
NO
they only stop the synth of polypep
***WHO found the evidence for a TRIPLET CODE?
Crick + Brenner
****What method did Crick + Brenner use to research evidence for the triplet code. AKA how did they do it?
Generated mutations in T4 Phages
Found that Phenotype was only maintained with additions or deletions that were in 3 base increments
Showed Reading frame = 3
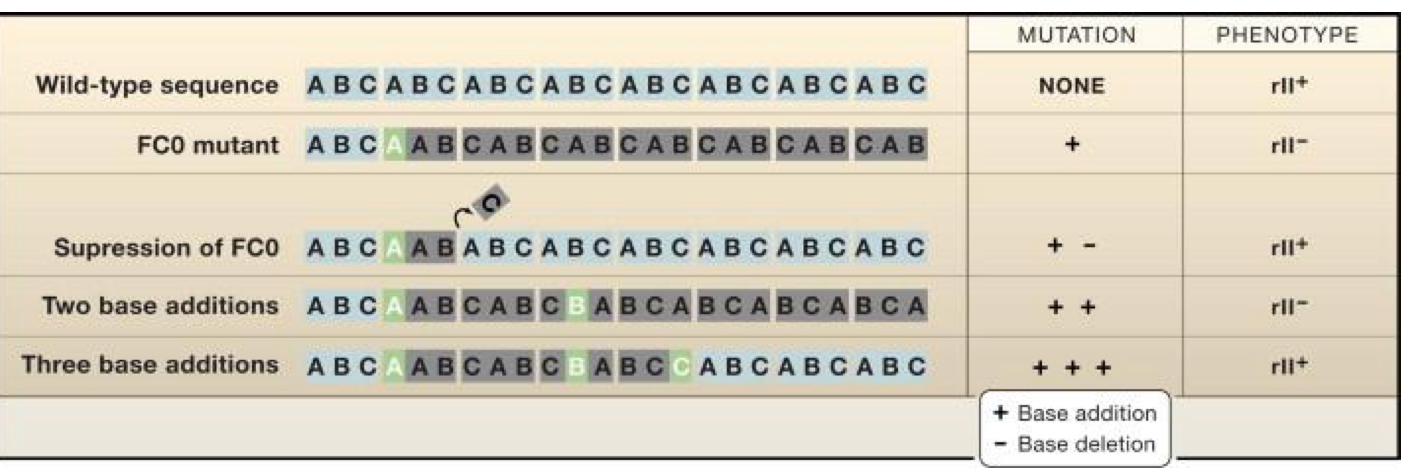
***What are the 3 key differences between DNA + RNA?
Sugar differences (2’OH)
OH give different shape and properties; physical + chemical
Base differences (U vs. T)
Single vs. Double strand
****How do Uracil and Thymine Differ?
Thymine is the METHYLATED version of Uracil
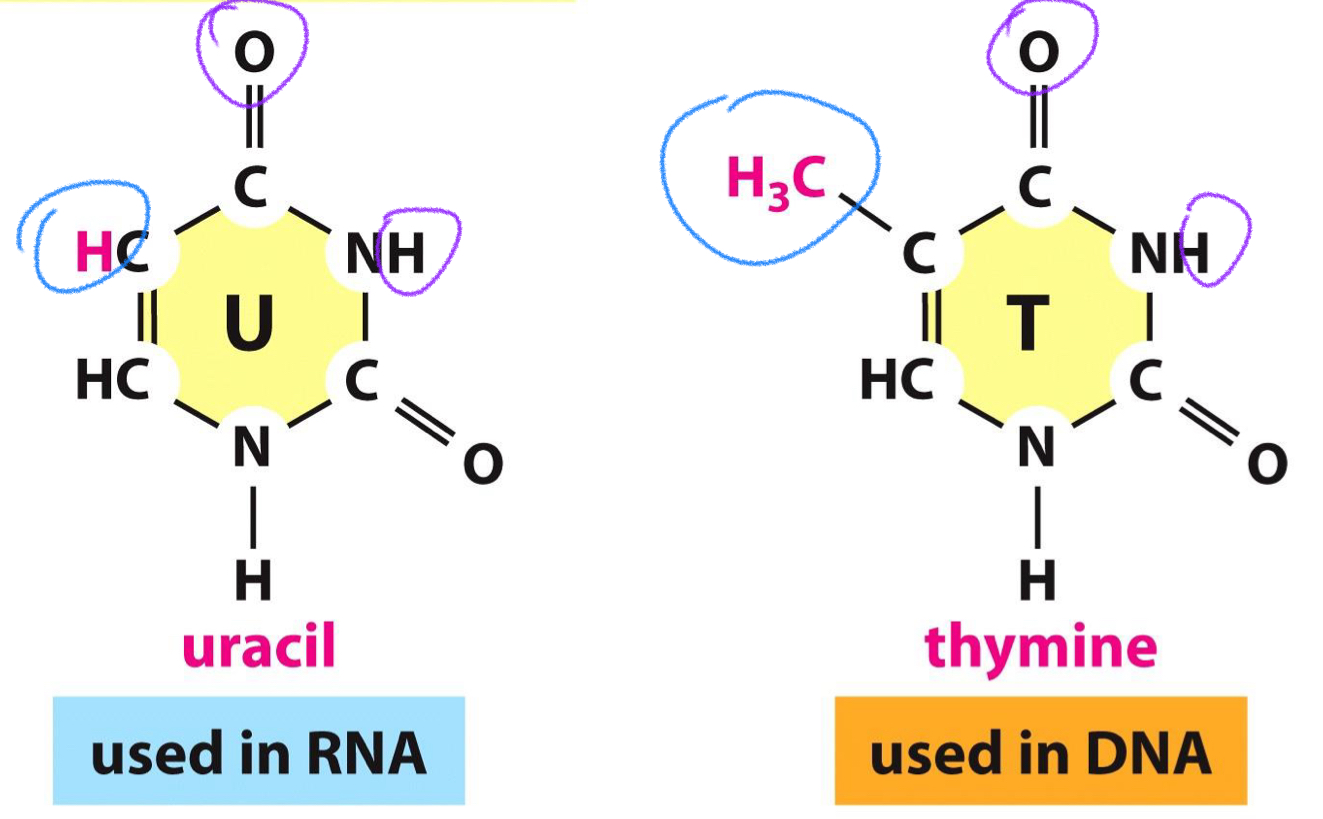
What is the main force in BASE PAIRING in DNA + RNA?
H-Bonds
****DNA is very uniform, why is that important?
Lets us detect if/ when something goes wrong with it.
Creates kinks
****What are the 3 parts of DNA that is needed for transcription of mRNA
Promoter
Coding sequence
Terminator
***What is the Role of the Promoter sequence?
Regulating gene expression: Assembles RNA Polymerase + Beginning of the the unwinding of DNA (aka. opening it up)
***What are the 4 General steps of Transcription?
Binding of RNA to promoter + opening of DNA/unwinding
Initiation of RNA synthesis
Elongation of mRNA
Termination of RNA Synthesis
***How is Transcription Controlled?
By regulating access
Base pairs must disrupted
DNA must be uncondensed
***What are transcription factors?
regulate gene expression by binding to specific DNA sequences
****True or false: Transcription is very similar between prokaryotes and eukaryotes
FALSE
translation is very similar
****True or false: Both prokaryotes and eukaryotes have Transcription factors (TF)
FALSE
Prokaryotes do not have TF
***What is the Prokaryotic equivalent of a TF?
Sigma factor
recognizes promoter sequence
****How many RNA poly are part of transcription for eukaryotes vs. Prokaryotes
Prokaryotes = 1 = RNA poly
Eukaryotes = 3 = RNA poly 1, 2 + 3
****What are all the TF’s involved in Initiation/Regulation of Eukaryotic transcription? List them in order of involvement
TF2D
TF2A + TF2B
TF2F
TF2E + TF2H
****Give an overview of the STEPS for INITIATION of transcription in eukaryotes
In the PROMOTER REGION of the gene
TF2D recognizes a TATA(easy to pull apart) box + binds = Conformational change in TF2D
conformational change = TF2A + TF2B binds to TF2D
RNA poly = in complex with TF2F in the nucleus. The TF2F binds to complex on DNA
Preinitiation complex = completed by recruiting of TF2H + TF2E
TF2E activates KINASE + HELICASE activity of TF2H
Phosphorylation of RNA poly 2 by kinase = begins transcription
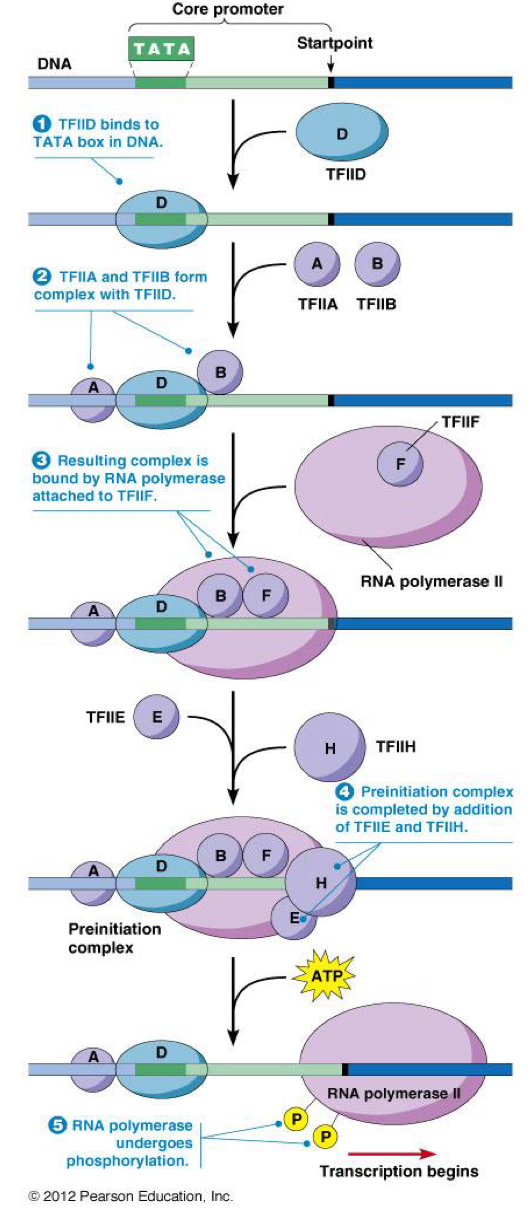
****What does the 2 in TF2D mean?
2 for RNA poly 2
*****Which TF is require to RECRUIT RNA poly?
TF2F is in complex with the RNA poly in the nucleus
***What is the point of TF2A + TF2B?
Provide stability
****Which TF’s complete the preinitiation complex?
TF2E TF2H
*****Which TF has Kinase + Helicase activity?
TF2H
****What does Kinase do?
Phosphorylate the RNA poly = begins transcription
****What is the source of the phosphates for phosphorylation by TF2H kinase?
ATP?
****Which AA are involved in Transcription regulation?
Serine
Tyrosine
Threonine
have OH = can be phosphorylated
****Which of the TF’s dissociate from the preinitiation complex after replication begins? What happens to them?
B, H, F + E
they get RECYCLED for future use
****What causes TF2E, H, B + D to Dissociate?
ATP = causes conformational change
****Why is TF2A left behind + does not dissociate with the other TF’s?
Makes it easier for RNA poly to transcribe the same gene again
Why is transcription and translation tightly coupled in prokaryotes but not eukaryotes?
Transcription + translation occur in the same space
segregated in eukaryotes
Where does Transcription occur and where does translation occur in eukaryotes?
Transcription = nucleus
Translation = cytoplasm + Rough ER
***What processes does the mRNA of Eukaryotes have to go through before translation?
mRNA processing
mRNA export from the nucleus
****How do the RNA poly’s in Eukaryotes (1, 2 + 3) differ? (2 differences)
Location in nucleus
TYPE OF RNA they synth.
How do we know the 3 RNA poly’s differ in location in the eukaryotic nucleus
Immunofluorescence microscopy
***What type of RNA do each RNA poly synthesize?
RNA poly 1 = rRNA
RNA poly 2 = mRNA + snRNA
RNA poly 3 = 5S rRNA + tRNA
****WHERE are each of the RNA poly’s located in the nucleus?
Poly 1 = Nucleolus
Poly 2 = Nucleoplasm (cytoplasm of the nucleus)
Poly 3 = Nucleoplasm
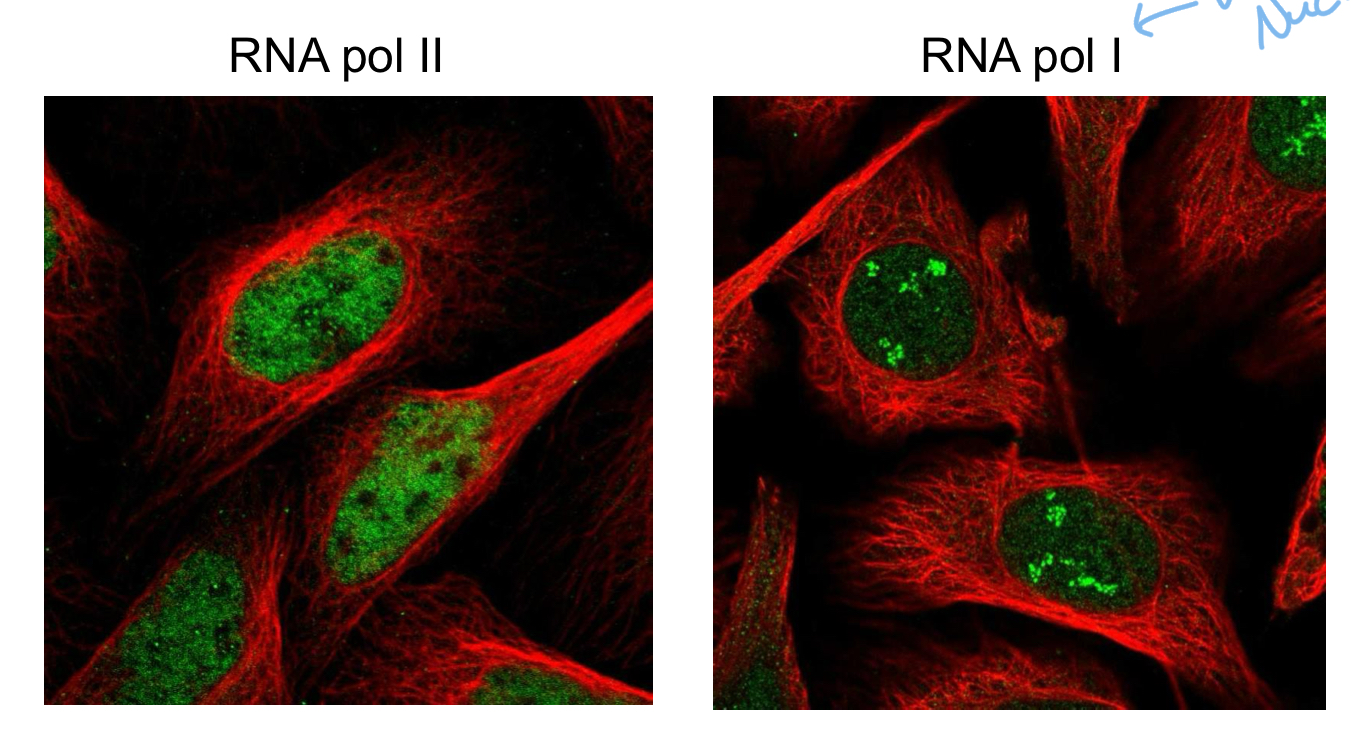
What is the nucleolus?
Part of the nucleus that is responsible for producing + Assembling RIBOSOMES
What is a newly produced RNA molecule called?
Primary transcript
****What are the 3 modifications that Nascent pre-mRNA has to undergo before becoming mRNA (Processing)
5’ Capping
Splicing
3’ Poly A tail
****What is the 5’ Cap? aka what is it composed of?
Methylated Guanosine (at position 7)
*****How is the 5’ cap bound to the RNA molecule and how does it differ from the usual bond?
It’s bound 5’ to 5’ instead of the usual 3’ - 5’
VERY DIFFERENT from the norm 3’-5’ end = very EASY TO RECOGNIZE
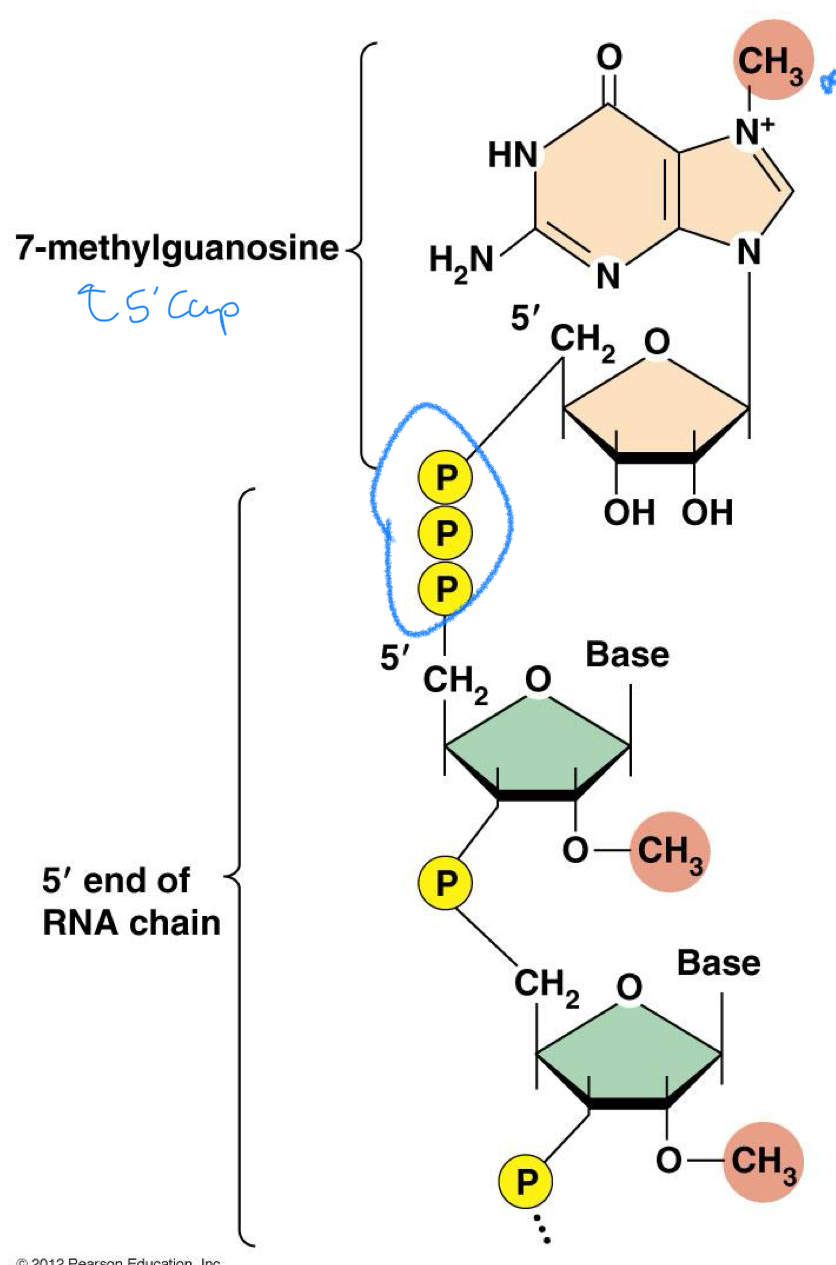
***When is the 5’ cap added?
Soon after Transcription is initiated
***What are the 2 ROLES of the 5’ cap?
PROTECT from nucleases = mRNA stability
Weird bonding + methylation = Distinguish from RNA of virus’
Positioning RNA on the Ribosome for initiation of translation
****WHERE is the poly A site located?
AAUAAA located upstream
U-rich site = down stream
****How is the POLY A - TAIL added?
a signal AAUAAA = upstream of the poly A site = recognized by CPSF + cleaved, then Poly-A transferase adds the bunch of A’s
****What are the 2 pieces of machinery associated with the addition of the Poly A tail?
CPSF
Polynucleotide adenylyl transferase
What does CPSF stand for + What does it do??
Cleavage and polyadenylation specificity factor
Recognizes AAUAAA signal upstream + CLEAVES it
Recruits Poly-A Transferase
*****What does Polynucleotide adenylyl transferase do?
Add Adenosine to the poly A tail + 3’ end using ATP
****What is the Function of the Poly-A tail? (3)
Protect mRNA from nuclease attack (length of tail influences stability)
required for EXPORT into cytoplasm (lets the machinery know mRNA is ready to go)
May help ribosomes recognize and bind mRNA
****Exons vs. Introns which is which
Exons = important coding stuff
Introns = Extra uncoding (removed from pre-mRNA)
What is typically the number of Introns related to the number of extrons?
Introns = # Extron’s -1
***What is the process of removing introns + joining exons known as?
RNA splicing
*****WHAT removes the introns from pre-mRNA?
the Spliceosomes
******FILL IN THE BLANK: Spliceosomes consist of __#_different _____ molecules and many ______
5 different RNA molecules + many PROTEINS
*****How are Spliceosomes assembled?
on transcripts from snRNPs each containing 1 or 2 snRNA molecules
*****What are snRNPs?
small nuclear ribonucleoprotein complexes that assemble into the spliceosome on the pre-mRNA and preform the splicing
******What are snRNAs?
small nuclear RNA: main job = recognition — they base-pair with pre-mRNA at splice sites (like the start and end of introns) to mark where cutting and joining should happen.
******What are ALL the snRNPs Involved in splicing?
U1
U2
U4
U5
U6
****What are the STEPS of Splicing?
U1 binds to pre-mRNA at 5’ Splice site
U2 binds to the branch point sequence (an adenine inside the intron). around 3’ splice site
U4/U6 complex + U5 join = form full spliceosome
Binding of U4/U6/U5 = brings GU(5’ splice U1) and A(3’ splice U2) close together forming a loop
RNA = Cleaved @ 5’ splice site
RNA = Cleaved @ 3’ splice site + the 2 exons join forming a LARIAT and EXONS are joined together + marked with and EJC
Intron lariat degrades
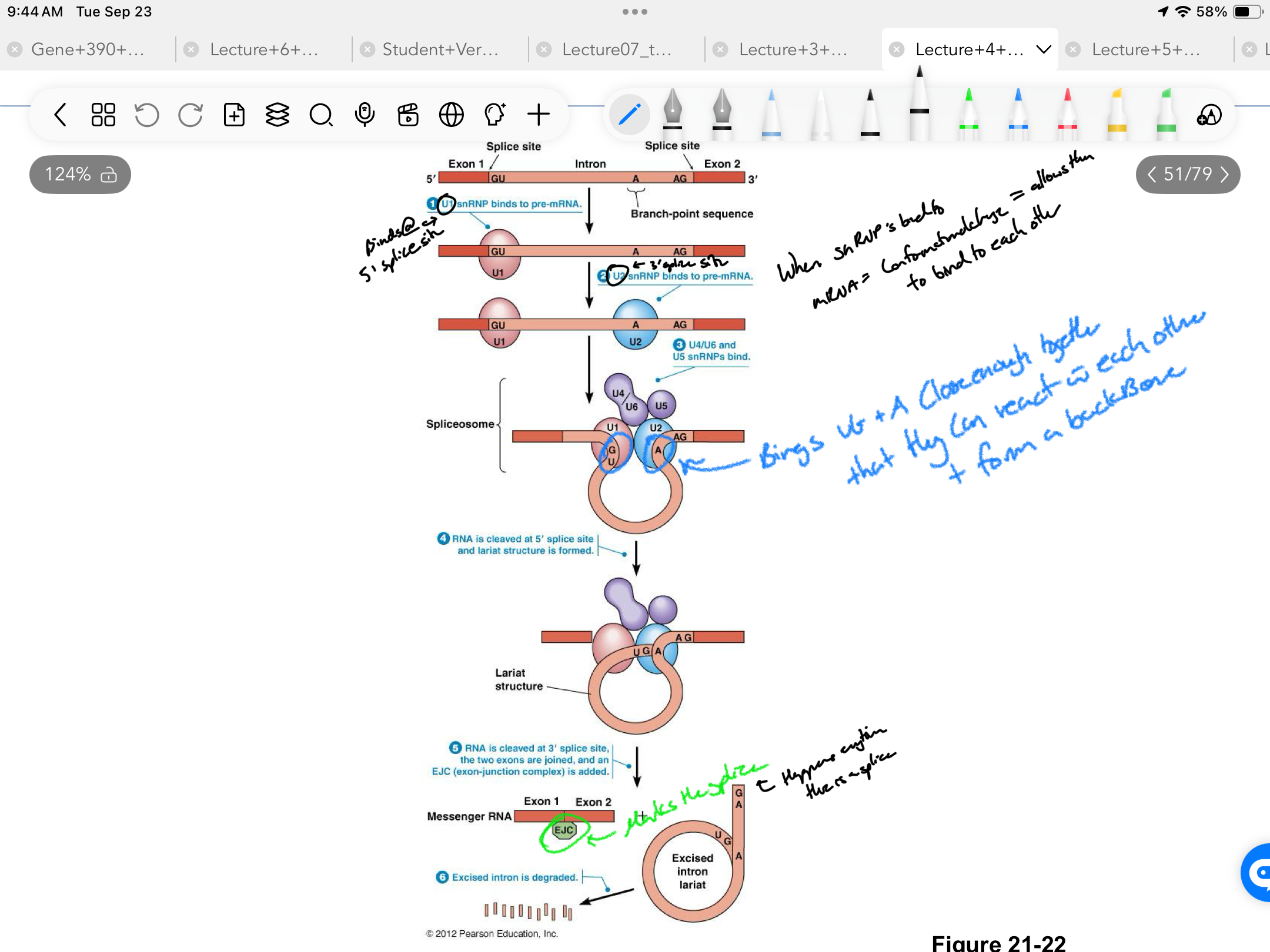
****What is the EJC what does it do?
Exon-Junction Complex
Marks every single splice
TRUE or FALSE: Some introns can splice without the Spliseosome?
TRUE
What types of genes have self-splicing RNA introns?
Group 1 + 2
*****How do these genes self splice without the help of any proteins?
the RNA INTRON itself catalyzes the process
****What are the RNA molecules that Function as Catalysts Called? AKA RNA that are ENZYMES
Ribozymes
****What does the existence of Introns Permit?
Alternative splicing + Exon shuffling
allow diff protein products from the same mRNA
****HOW is alternative splicing achieved?
mechanisms allowing certain splice sites to be ACTIVATED or SKIPPED
****Which TWO machines are involved int he mechanism of Alternative splicing?
Regulatory proteins + snoRNAs
bind to splicing enhancers or silencer sequences.
DO mRNAs have short or long half lives?
Short life span
What is the typical half life of mRNA for Eukaryotes vs. bacteria
Eukaryotes = hours - a few days
Bacteria = minutes
What is the amplification of Genetic information + how is it achieved?
1 mRNA = Multiple proteins
1 mRNA = synthesized many times
Each mRNA = translated multiple times
****What are Missense vs. Silent mutation vs. Nonsense mutations. Where are they found?
Found on mRNA = cause errors in polypep. chain
Missense = 1 aa = alter
Silent = mutation has no effect on aa
Nonsense = Premature stop codon
****How do eukaryotic cells deal with mRNAs containing PREMATURE STOP codon? What about mRNAs with Stop codon REMOVED?
Premature stop = Non-sense mediated decay
No stop = Nonstop Decay
*****What occurs during translation when the stop codon is removed from the mRNA?
Cause translation to STALL when ribosome reaches end of transcript
*****What is SUPPOSED TO HAPPEN on a non-mutated mRNA?
Ribosomes REMOVE EJC + ADD TC (termination complex) at the STOP CODON at the first round of translation
****Should an mRNA ever have BOTH TC + EJC?
NO
Only one
Brand new = EJC
Old viable mRNA = TC
*****What occurs during NON SENSE MEDIATED DECAY? How does it work?
FIRST ROUND OF TRANSLATION
Premature stop = Ribosome only removes some of the EJCs + not all + places the TC at the premature codon
Presence of BOTH EJC + TC = trigger cascade of events that drive degradation
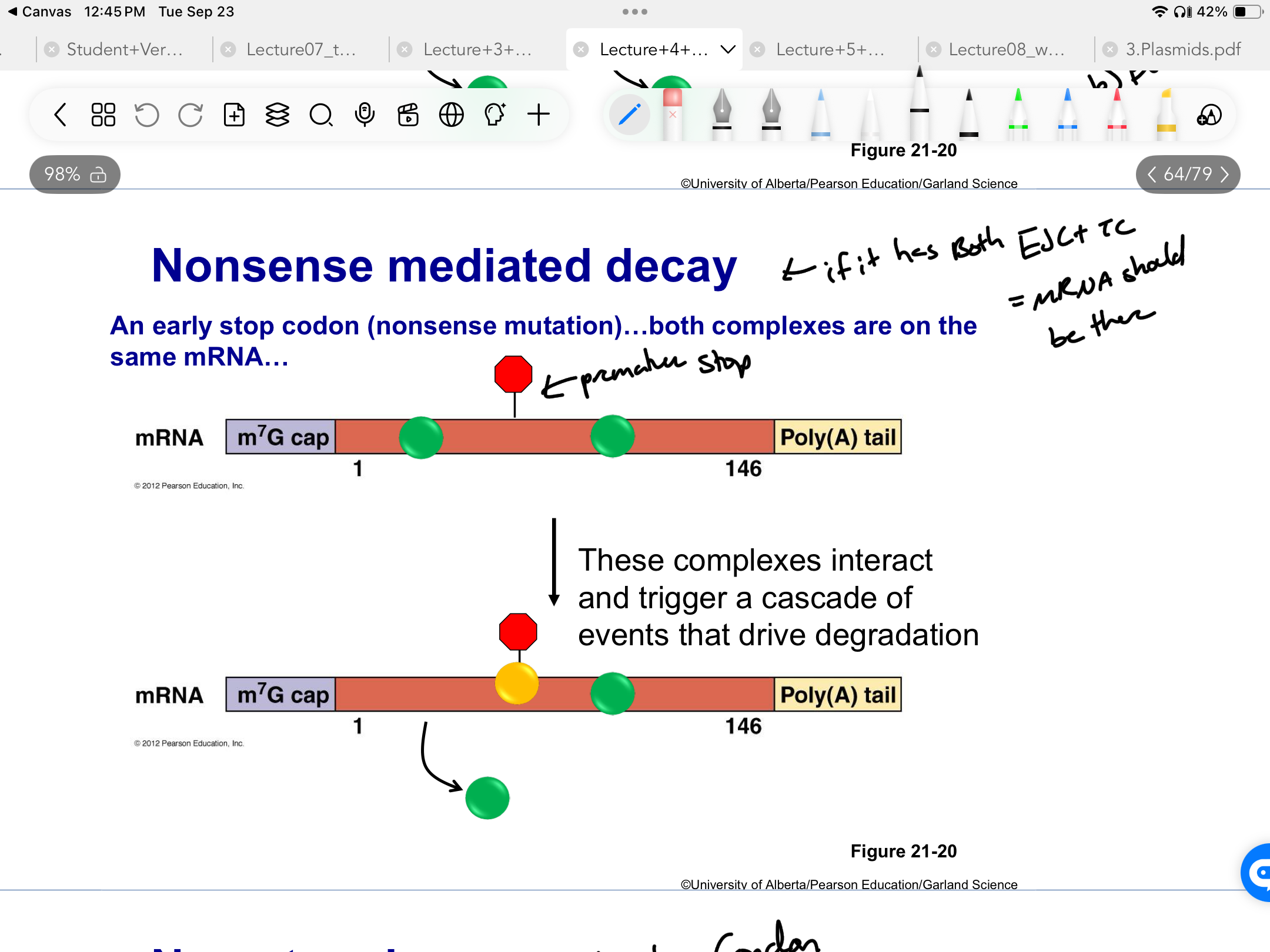
*****What is the only scenario in which Nonsense mediated decay does not work?
Premature stop = AFTER the last EJC but still before the actual end of the mRNA
In the LAST EXON
Really important stuff = not stored in last exon bc there is no quality control
*****What occurs during NON-STOP DECAY? How does it work?
No stop codon = ribosome move into POLY A TAIL
Possible mechanisms:
Bump into PABPs (poly a binding protein)
Start incorporating the same residue over + over
Run out of tRNA for that aa
= TRIGGERS DEGREDATION
*****What is the the MOST ADBUNDANT + STABLE form of RNA in the cells?
rRNA