chromosomes and cytogenetics
1/13
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
14 Terms
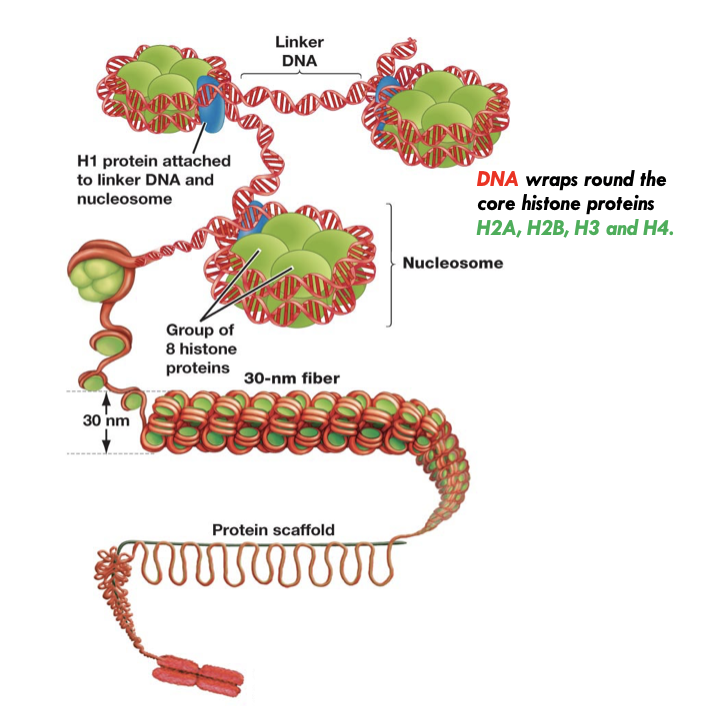
chromatin
In chromosomes, DNA is compacted by forming complexes with histone proteins
Most condense form
Can exist at euchromatin or heterochromatin
Occurs during metaphase
Regulated by interaction with histone proteins
Wraps around twice
Forms a nucleosome
Can form a 30 nm fibre
30 nm fibre loops around a protein scaffold
euchromatin
Most of the genome is in the form of 'euchromatin'
The degree of compaction varies and can be very dynamic
Can regulate association with H1, which in turn regulates rate of gene transcription
Contains all of the genes
Forms 90% of chromosomes
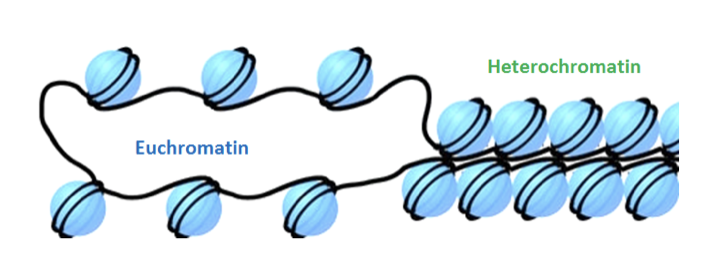
heterochromatin
A smaller proportion of human chromosomes is in the form of heterochromatin, where the chromatin is more permanently compacted
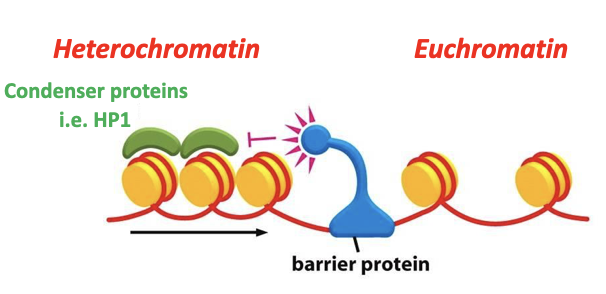
Forms via condenser proteins
Not dynamic (cannot reverse compactness)
Barrier elements recruit barrier proteins
Prevent heterochromatin spilling into euchromatin
Can be lost following a chromosomal translocation
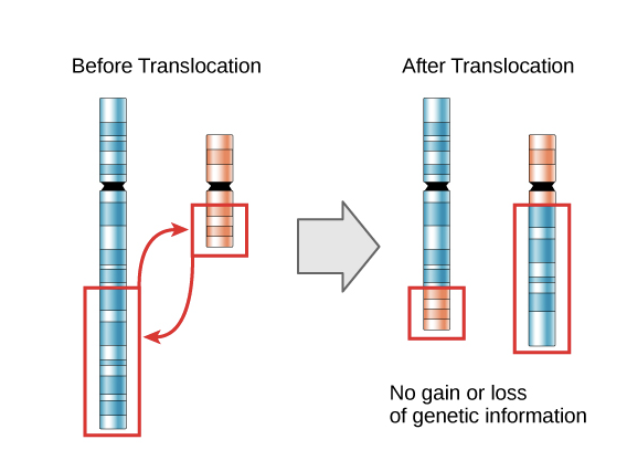
spilling of heterochromatin
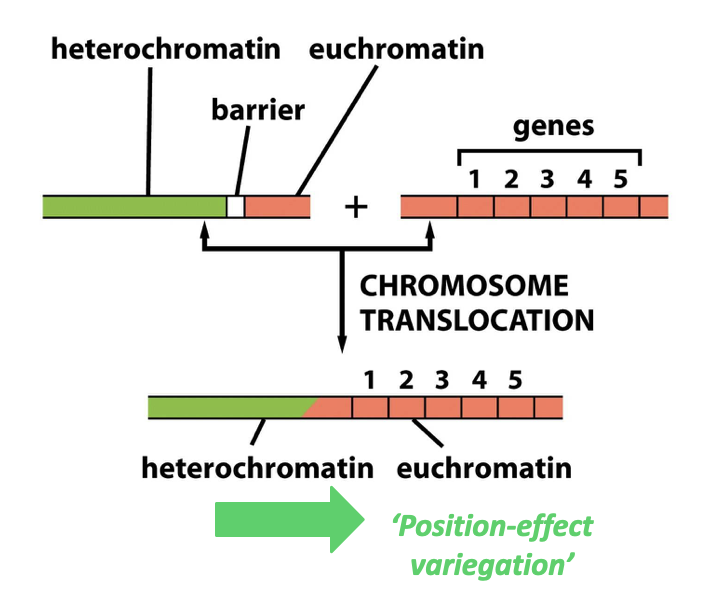
If heterochromatin spills = position effect variegation
Shuts off all the genes in the euchromatin
Heterochromatin has a tendency to spread (e.g. into neighbouring euchromatic regions)
Molecular mechanisms exist to prevent this
structural heterochromatin
Structural heterochromatin usually contains repetitive DNA sequences known as satellite DNA
It is found in structural elements such as centromeres and importantly in telomeres
Prevent fusion of chromosomes
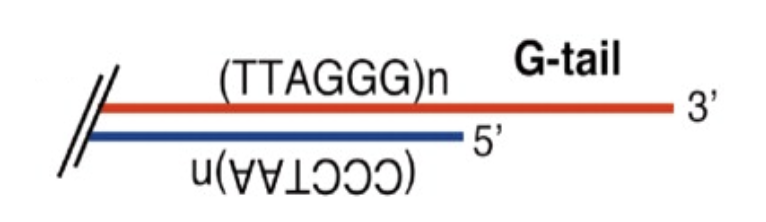
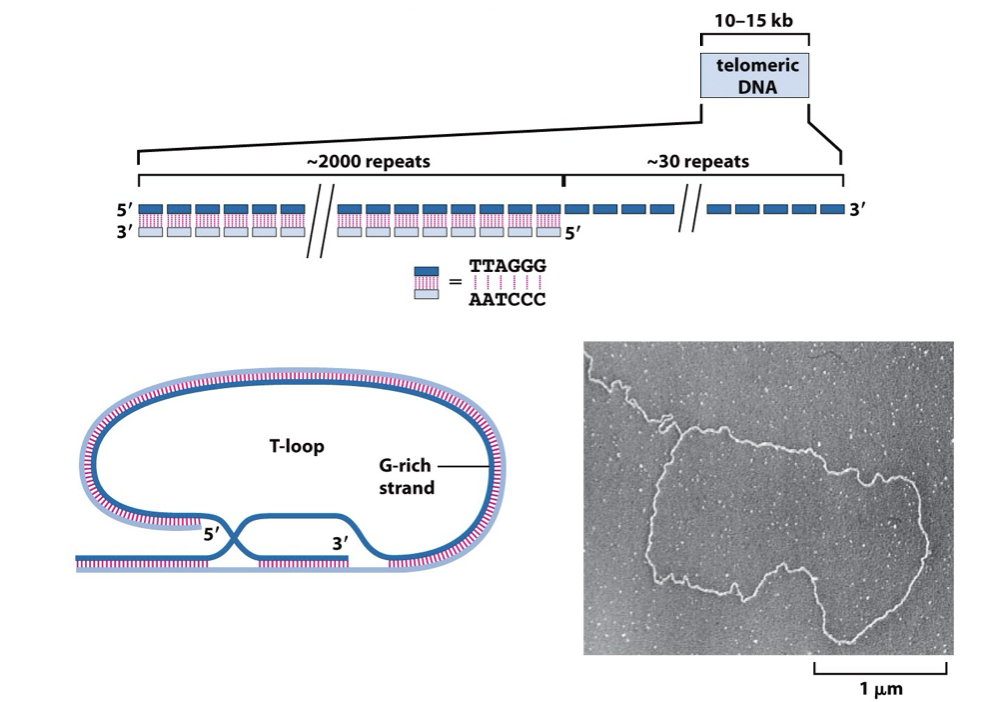
Telomeric DNA sequences are composed of long arrays (10-15 kilobases) of short tandem repeats
G rich strand = longer, resulting in an overhang called the G-tail (around 30 repeats)
C rich strand = shorter
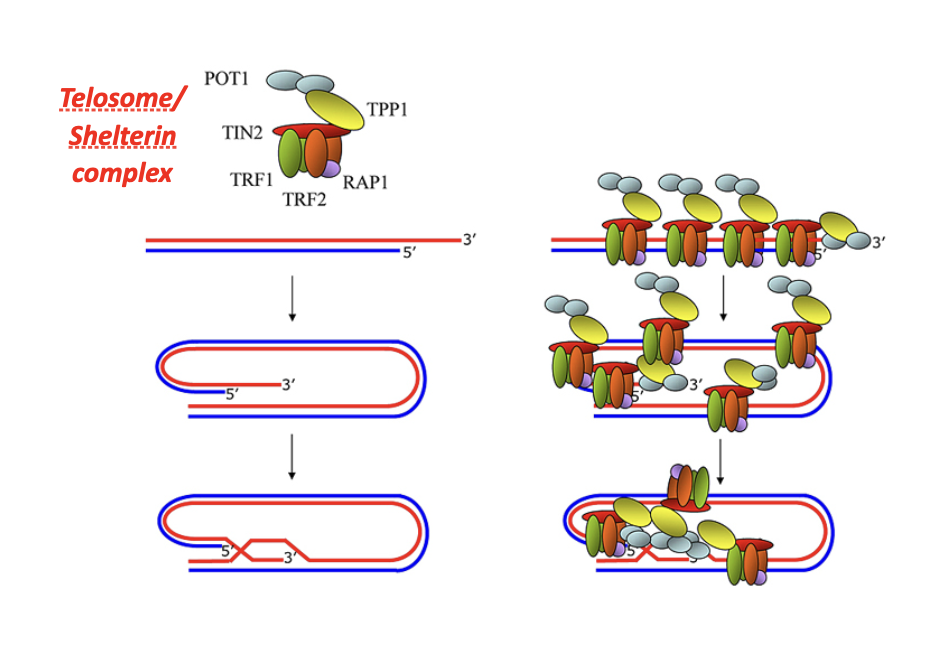
G-tail
Protects chromosome ends
Prevents chromosome ends from being recognized as DNA double-strand breaks
Avoids activation of DNA damage response pathways
Enables telomere capping
G-tail folds back to form a T-loop
The T-loop physically hides the chromosome end
Recruits shelterin complex
Binds telomere-specific proteins (e.g. TRF1, TRF2, POT1)
Shelterin stabilizes telomere structure and maintains heterochromatin
Prevents end-to-end fusions
Blocks non-homologous end joining (NHEJ) at chromosome ends
Maintains genomic stability
Ensures proper chromosome maintenance across cell divisions
Facilitates telomere replication
Provides a substrate for telomerase to extend telomeres
function of structural heterochromatin
Maintains chromosome structural integrity
Provides mechanical stability to chromosomes
Located at repetitive DNA regions
Found at centromeres, telomeres, and other repeat-rich regions
Ensures proper chromosome segregation
Centromeric heterochromatin is essential for kinetochore formation
Prevents mis-segregation during mitosis and meiosis
Suppresses recombination
Prevents homologous recombination between repetitive sequences
Reduces chromosomal rearrangements
Silences repetitive DNA
Transcriptionally inactive
Prevents expression of transposons and satellite DNA
Protects genome stability
Limits insertional mutagenesis by mobile elements
Maintains nuclear organization
Anchors chromatin to the nuclear periphery
Contributes to higher-order chromatin architecture
Defines chromosomal domains
Acts as a boundary separating active euchromatin regions
Epigenetically maintained
Characterized by DNA methylation
Enriched in H3K9me3 and HP1 proteins
Evolutionarily conserved
Present in most eukaryotes with similar structural roles
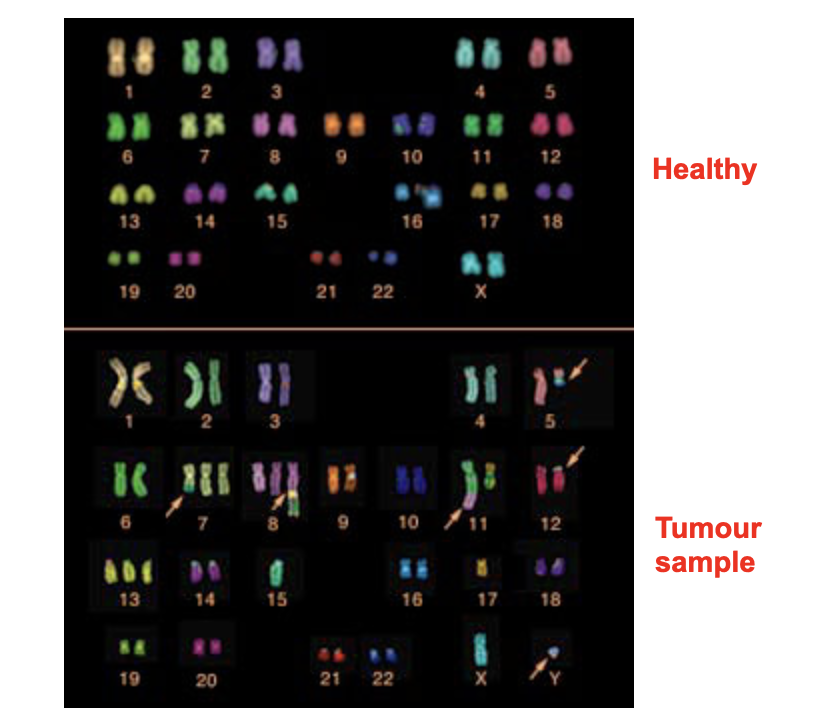
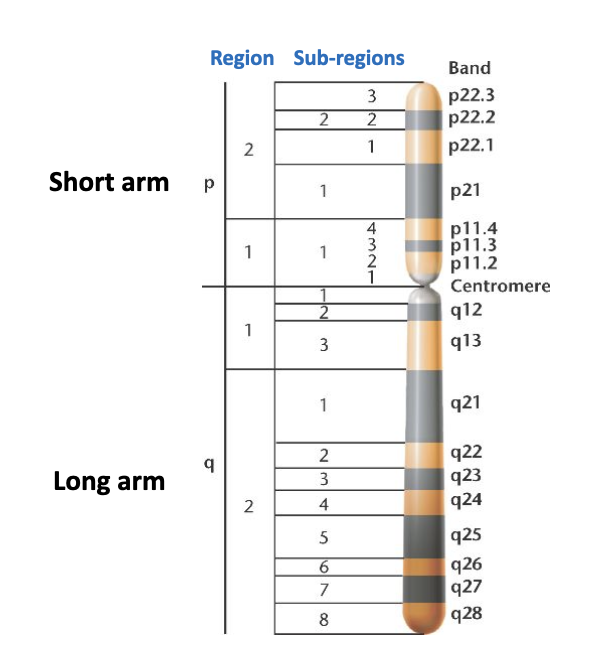
cytogenetics
Chromosomes from cell preparations can be 'spread' on a microscopic slide for analysis
Want to collect them in metaphase
As they are more condensed
Can be distinguished from each other
Chromosomes in prometaphase are less compacted and thus reveal more detail
Can find structural features
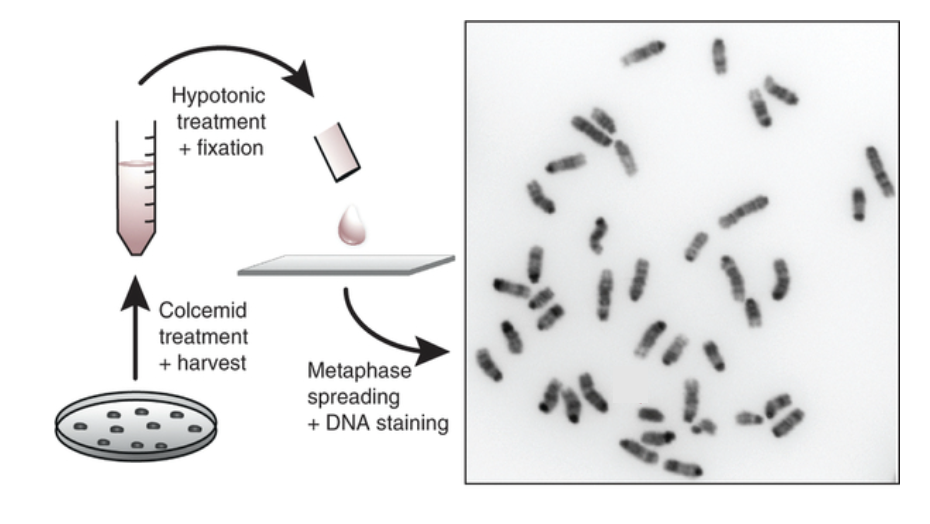
hypotonic treatment
Cells are placed in a hypotonic solution (e.g. 0.075 M KCl)
Water enters cells by osmosis
Cells swell due to increased internal pressure
Nuclear membrane weakens and ruptures
Chromosomes spread apart within the swollen cell
Cell is then fixed (e.g. methanol:acetic acid) to preserve chromosome structure
Chromosomes are dropped onto a slide for staining and analysis
Stains create a banding pattern
Can distinguish one chromosome from another
Using karyotyping
molecular cytogenetics
Higher resolution analysis (e.g. detection of specific DNA sequences within chromosomes)
Achieved by designing and chemically synthesising an 'oligonucleotide probe' directed against a target DNA sequence
Labelled with a fluorophore
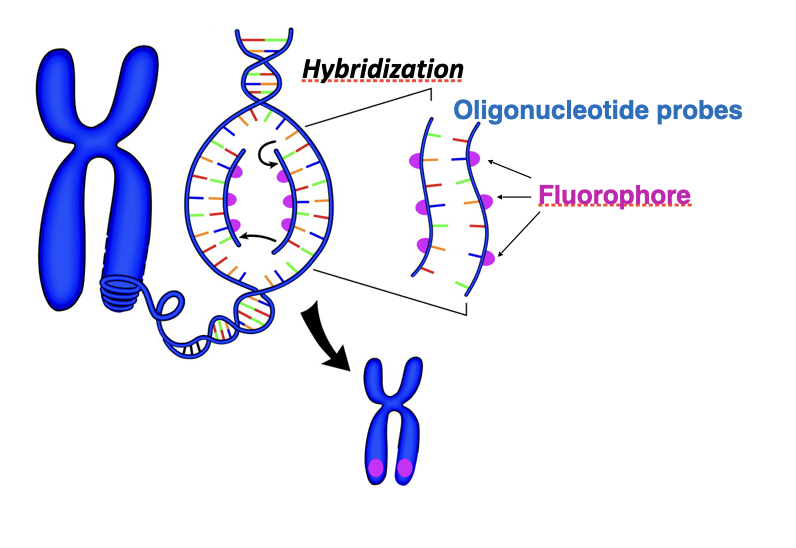
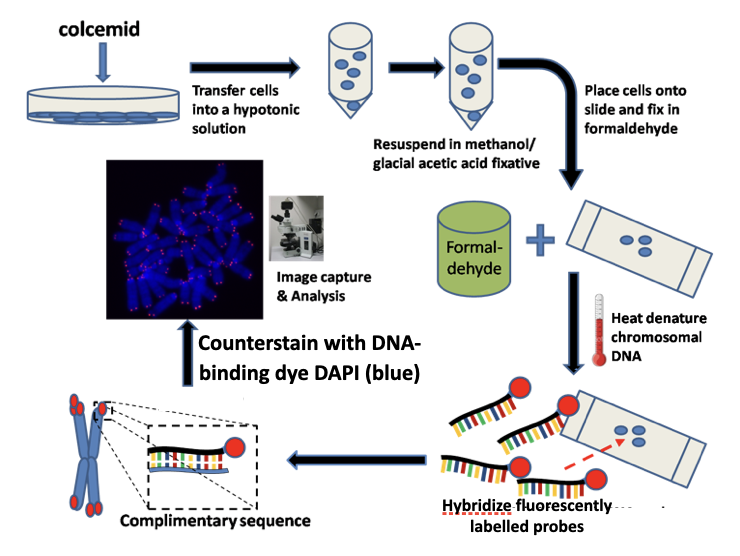
how does FISH (fluorescence in situ hybridisation) work
Purpose:
Detects and localizes specific DNA (or RNA) sequences on chromosomes or in cells
Starting material:
Metaphase chromosomes or interphase nuclei fixed on a slide
Denaturation:
Chromosomal DNA is heated or chemically treated
Double-stranded DNA separates into single strands
Probe preparation:
Short, single-stranded DNA oligonucleotide probes
Labelled with a fluorescent dye
Hybridization:
Probe is applied to the slide
Incubation allows probe to find and bind its complementary sequence
Washing:
Removes unbound or weakly bound probes
Ensures specific binding
Detection:
Slide viewed under a fluorescence microscope
Bound probes appear as bright fluorescent signals
how does the oligonucleotide probe attach
Base pairing
Probe binds by complementary base pairing
A–T and G–C hydrogen bonds form between probe and target DNA
Hybridization conditions
Temperature, salt concentration, and time control stringency
High stringency → only perfect matches bind
Low stringency → partial mismatches may bind
No covalent bonds
Attachment is non-covalent (hydrogen bonds only)
Binding is reversible under denaturing conditions
Probe length matters
Short oligos → high specificity
Longer probes → stronger binding, lower specificity
chromosome painting
Uses a large cocktail of probes that will bind to many different locations along the length of a single type of chromosome
Spectral karyotyping (SKY) can allow for the entire set of chromosomes to be analysed