MIC 105 Module 2
0.0(0)
0.0(0)
Card Sorting
1/106
Earn XP
Description and Tags
Study Analytics
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
107 Terms
1
New cards
Mobile Genetic Elements (MGEs)
A type of DNA or RNA that can move around in the genome, or be transferred to a recipient organism
2
New cards
Mobile Genetic Element Examples
plasmids, viruses, transposable elements
3
New cards
Mechanism of MGE transfer
1. Conjugation
2. Transformation
3. Vesicles
4. Nanotubes
5. Capsids
2. Transformation
3. Vesicles
4. Nanotubes
5. Capsids
4
New cards
Plasmid + types
term for any extrachromosomal heredity determinant
- can have broad or limited host ranges; range of low to high copy numbers
1. episome \= non-essential genetic element replicating autonomously or integrated into the chromosome
2. conjugative \= can transfer horizontally, carry transfer genes
3. mobilizable \= can transfer horizontally, carry mobilization genes
- can have broad or limited host ranges; range of low to high copy numbers
1. episome \= non-essential genetic element replicating autonomously or integrated into the chromosome
2. conjugative \= can transfer horizontally, carry transfer genes
3. mobilizable \= can transfer horizontally, carry mobilization genes
5
New cards
Benefits of plasmids
may confer genes w/ beneficial traits to hosts
eg. bacterial produced toxins, antibiotic resistance, virulence, degradative enzymes
eg. bacterial produced toxins, antibiotic resistance, virulence, degradative enzymes
6
New cards
Plasmid replication
-rolling circle replication
-depends on a protein that guides DNA and assists replication in recipient
-ori determines the number of copies
-depends on a protein that guides DNA and assists replication in recipient
-ori determines the number of copies
7
New cards
Cloning Vector Features
1. origin of replication
2. selectable marker
3. multiple cloning site
4. insert
2. selectable marker
3. multiple cloning site
4. insert
8
New cards
Phage Characteristics
structure: capsid, tail, proteases
genome replication: helicase, primase, dna pol
packaging: terminase, protal
regulation: repressor, anti-repressor
integration, excision: integrase, excisionase
tRNAs
*lots of "accessory" genes (bidirectional genome)
genome replication: helicase, primase, dna pol
packaging: terminase, protal
regulation: repressor, anti-repressor
integration, excision: integrase, excisionase
tRNAs
*lots of "accessory" genes (bidirectional genome)
9
New cards
Phage lifecycle(s)
lytic cycle: productive phage infection --\> lysis of host
horizontal transmission: transmission of phage genome b/w contemporary hosts
lysogenic cycle: replication of the phage genome occurs with the replication (includes integration)
integration: physical insertion of the phage genome into the host
prophage \= integrated phage
horizontal transmission: transmission of phage genome b/w contemporary hosts
lysogenic cycle: replication of the phage genome occurs with the replication (includes integration)
integration: physical insertion of the phage genome into the host
prophage \= integrated phage
10
New cards
Integration
- uses short pieces of homology recognized by integrase to cut and ligate
- maintained over generations
- maintained over generations
11
New cards
Phage-Host Arms Race
hosts: under constant and strong selection to inhibit phage infection
- phage adsorption + surface resistance
- restriction modification systems
- CRISPR/Cas9
phage: under constant and strong selection to infect hosts
- phage adsorption + surface resistance
- restriction modification systems
- CRISPR/Cas9
phage: under constant and strong selection to infect hosts
12
New cards
Giant Viruses
- complex, mobile, monophyletic, broad host spectrum
- can reprogram host
- include virophages
- can reprogram host
- include virophages
13
New cards
Transposons
flanked by direct repeats and include terminal inverted repeats
14
New cards
Class I Transposons: Retrotransposons
- transpose by a copy-paste mechanism by copying as RNA transcripts and then using a reverse transcriptase --\> integrate (considered to be replicative)
15
New cards
Class II Transposons: DNA Transposons
- non-replicative cut and paste by excising themselves and integrating somewhere else
- have long terminal repeats on both ends
- have long terminal repeats on both ends
16
New cards
Conjugative Transposons
are able to transfer from one cell to another by conjugation (enable both mobilizable plasmids and nonconjugative transposons to be transferred)
17
New cards
Autotrophy vs Heterotrophy
auto: makes complex carbs from CO2 (need external source of energy)
hetero: makes energy from complex/organic carbon molecules --\> cellular carbon
hetero: makes energy from complex/organic carbon molecules --\> cellular carbon
18
New cards
Photoautotroph CO2 Fixation Pathways
1. Calvin Cycle
2. Reductive TCA Cycle
3. 3HP Bi-cycle
(L8 Slide 7)
2. Reductive TCA Cycle
3. 3HP Bi-cycle
(L8 Slide 7)
19
New cards
Calvin Cycle
a biochemical pathway of photosynthesis in which carbon dioxide is converted into glucose using ATP
("dark reactions" in cyanobacteria + plants)
essential enzymes: rubisco and phosphoribulokinase
("dark reactions" in cyanobacteria + plants)
essential enzymes: rubisco and phosphoribulokinase
20
New cards
Calvin Cycle Steps
1. CO2 fixation by rubisco
2. reduction to the oxidation state of carbohydrate + cell material
3. regeneration of CO2 acceptor by phosphoribulokinase
2. reduction to the oxidation state of carbohydrate + cell material
3. regeneration of CO2 acceptor by phosphoribulokinase
21
New cards
Bacterial Autotrophs
1. purple nonsulfur bacteria*
2. ammonia oxidizing bacteria*
3. purple sulfur bacteria*
4. sulfate reducing bacteria
5. green sulfur bacteria^
6. green nonsulfur bacteria"
7. some clostridia
2. ammonia oxidizing bacteria*
3. purple sulfur bacteria*
4. sulfate reducing bacteria
5. green sulfur bacteria^
6. green nonsulfur bacteria"
7. some clostridia
22
New cards
Reverse/Reductive TCA Cycle
uses 4-5 ATPs to fix four molecules of CO2 and generate one oxaloacetate
- requires less energy
- same enzymes as in forward except there are three key enzymes that will send reaction into reverse (fumarate reductase, a-ketoglutarate synthase, citrate lyase)
- reverse glycolysis supports biomass building
- only pathway used by both bacteria and archaea
- requires less energy
- same enzymes as in forward except there are three key enzymes that will send reaction into reverse (fumarate reductase, a-ketoglutarate synthase, citrate lyase)
- reverse glycolysis supports biomass building
- only pathway used by both bacteria and archaea
23
New cards
3-hydroxypropionate bi-cycle
A carbon fixation process that condenses hydrated CO2 and acetyl-CoA to form 3-hydroxy propionate (5ATP)
Key enzymes: acetyl-CoA + proprionyl-CoA carboxylases
(anaerobic as one step is O2 sensitive)
Key enzymes: acetyl-CoA + proprionyl-CoA carboxylases
(anaerobic as one step is O2 sensitive)
24
New cards
The 3-Hydroxypropionate/4-Hydroxybutyrate Pathway
(thermoacidophilic crenarchaeota)
same intermediates different enzymes
same intermediates different enzymes
25
New cards
Dicarboxylate/4-hydroxybutyrate cycle
an autotrophic pathway found in thermophilic crenarchaeota
-patchwork pathway from rTCA and 3HP/4HB cycle
-patchwork pathway from rTCA and 3HP/4HB cycle
26
New cards
Reductive Acetyl-CoA Pathway
- Used by anaerobic soil bacteria, autotrophic sulfate reducers, and methanogens
- Two CO2 molecules are condensed through converging pathways to form the acetyl group of acetyl-CoA.
- Carbon monoxide is an intermediate.
- Reducing agent is H2 instead of NADPH.
- Two CO2 molecules are condensed through converging pathways to form the acetyl group of acetyl-CoA.
- Carbon monoxide is an intermediate.
- Reducing agent is H2 instead of NADPH.
27
New cards
Why so many CO2 fixation pathways
- energy requirements
- oxygen sensitivity of enzymes
- temp
- requirements of metals
- products formed
- oxygen sensitivity of enzymes
- temp
- requirements of metals
- products formed
28
New cards
Phototrophy
involves light capture by chlorophyll, usually coupled to splitting of H2S or H2O or organic molecules
chlorophyll-based (auto + heterotrophs) vs retinal-based (heterotrophs)
chlorophyll-based (auto + heterotrophs) vs retinal-based (heterotrophs)
29
New cards
Chlorophyll-based phototrophy
1. photopigments \= absorb different wavelengths of light
2. reaction centers \= PS I (Fe-S protein) or II (mobile quinone)
3. membranes \= house pigments, rc, + photosystems --\> connect to ETC
2. reaction centers \= PS I (Fe-S protein) or II (mobile quinone)
3. membranes \= house pigments, rc, + photosystems --\> connect to ETC
30
New cards
Light-Harvesting Pigments
1. Chlorophylls \= tetrapyrrole + Mg2+ (cyanobacteria)
2. Bacteriochlorophylls \= side groups differ (purple,green phototrophs)
2. Bacteriochlorophylls \= side groups differ (purple,green phototrophs)
31
New cards
Accessory Light Harvesting Pigments
carotenoids \= long chain hydrocarbons w/ extensive conjugated double bonds
- determine wavelengths absorbed
- determine wavelengths absorbed
32
New cards
Cyanobacteria
monophyletic w/ similar physiologies
metabolism: oxygenic photoautotrophs (some organoheterotrophs in the dark)
habitats: anywhere there's light
carboxysome: contains rubisco --\> sequesters CO2 for efficiency and to protect rubisco from O2
spp. prochlorococcus \= most abundant photosynthetic organism; fix 50 billion tons of CO2; large pangenome
metabolism: oxygenic photoautotrophs (some organoheterotrophs in the dark)
habitats: anywhere there's light
carboxysome: contains rubisco --\> sequesters CO2 for efficiency and to protect rubisco from O2
spp. prochlorococcus \= most abundant photosynthetic organism; fix 50 billion tons of CO2; large pangenome
33
New cards
Cyanobacteria pigments
chlorophyll a, carotenoids, phycobilins
phycobilin \= phycocyanin (dark blue-green)
phycobilin \= phycocyanin (dark blue-green)
34
New cards
Electron Transport (oxygenic phototrophy)
PS I and PSII involved
(L9 slide 50)
(L9 slide 50)
35
New cards
Purple Phototrophs
proteobacteria
metabolism: anoxygenic photoauto/heterotrophs
metabolism: anoxygenic photoauto/heterotrophs
36
New cards
Electron Flow (anoxygenic phototrophy)
PSII; reverse electron flow needed to generate NADH
(L9 slide 61)
(L9 slide 61)
37
New cards
Purple Nonsulfur Bacteria vs Purple Sulfur Bacteria
PNS: only phototrophic under anaerobic conditions
PS: mostly obligate photoautotrophs using H2S --\> S ; also use S as e- source; variety of metabolisms
habitats: anaerobic environments w H2S and light
both can use a variety of electron donors
PS: mostly obligate photoautotrophs using H2S --\> S ; also use S as e- source; variety of metabolisms
habitats: anaerobic environments w H2S and light
both can use a variety of electron donors
38
New cards
Green Sulfur Bacteria (Chlorobi)
physiology: anoxygenic photoautotrophs (some photoheterotrophs)
- use H2S or H2
Habitat: freshwater + marine sedimens
Enzyme: PSI (efficient light gatherers)
CO2 Fixation: reductive TCA cycle
- use H2S or H2
Habitat: freshwater + marine sedimens
Enzyme: PSI (efficient light gatherers)
CO2 Fixation: reductive TCA cycle
39
New cards
Green Nonsulfur Bacteria (Chloroflexi)
physio: photoheterotrophs (sugars, organic acids or aa); photoautotrophs w/ H2 or H2S; can grow as aerobic organoheterotrophs
habitat: freshwater and marine environments; alkaline hot springs
Enzyme: PSII
CO2 Fixation: 3 HP Bi-Cycle
habitat: freshwater and marine environments; alkaline hot springs
Enzyme: PSII
CO2 Fixation: 3 HP Bi-Cycle
40
New cards
Heliobacteria
anoygenic photoheterotrophic endosporeformers (can fix nitrogen); chemoheterotrophs in dark
Enzyme: PSI w/ unique bCHLg
Substrates: organic acids
Habitats: anaerobic alkaline soils, rice paddies
Enzyme: PSI w/ unique bCHLg
Substrates: organic acids
Habitats: anaerobic alkaline soils, rice paddies
41
New cards
Aerobic anoxygenic phototrophs
marine a-proteobacteria
pigments: bchl a, carotenoids
enzyme: PSII w/ cyclic photophosphorylation
habitats: variety but mostly in marine environments
- use light as a supplement until conditions improve
*candidatus chloracidobacterium containing PSI and chlorosomes; very small genome
pigments: bchl a, carotenoids
enzyme: PSII w/ cyclic photophosphorylation
habitats: variety but mostly in marine environments
- use light as a supplement until conditions improve
*candidatus chloracidobacterium containing PSI and chlorosomes; very small genome
42
New cards
Retinal-based phototrophy
Light driven proton pump
Bacteriorhodospin - light sensitive pigment retinal
Halorhodopsin - pumps Cl- in
Use lights, and protons pumped out to create ATP
Bacteriorhodospin - light sensitive pigment retinal
Halorhodopsin - pumps Cl- in
Use lights, and protons pumped out to create ATP
43
New cards
Haloarchaea
most obligate aerobic organoheterotrophs (proteins + aa)
- require high NaCl concentrations for membrane stability
- use light to suppement energy when nutrients/O2 are low
-bacteriorhodopsin synthesized under low O2 conditions --\> absorbs green and reflects blue and red; makes cells buoyant so they can get to the surface for maximal light exposure
- require high NaCl concentrations for membrane stability
- use light to suppement energy when nutrients/O2 are low
-bacteriorhodopsin synthesized under low O2 conditions --\> absorbs green and reflects blue and red; makes cells buoyant so they can get to the surface for maximal light exposure
44
New cards
Proteorhodopsin
A bacterial membrane-embedded protein that contains retinal and acts as a light-driven proton pump; it is homologous to the archaeal protein bacteriorhodopsin
- widespread in many marine bacteria and archaea
- apparently horizontal gene transfer from haloarchaea
- widespread in many marine bacteria and archaea
- apparently horizontal gene transfer from haloarchaea
45
New cards
Acetogens
-Anaerobic bacteria, most are firmicutes
-Use carbon dioxide as electron acceptor. reduces to acetate
-Multiple donors are possible but H2 is common for reducing CO2
-Pathway is called Reductive Acetyl-CoA pathway
habitats: anoxic (soils, termite hindgut, cow rumen)
-Use carbon dioxide as electron acceptor. reduces to acetate
-Multiple donors are possible but H2 is common for reducing CO2
-Pathway is called Reductive Acetyl-CoA pathway
habitats: anoxic (soils, termite hindgut, cow rumen)
46
New cards
Reductive Acetyl-CoA Pathway (Acetogenesis)
C1 Carrier \= Tetrahydrofolate (THF)
Key enzymes: CO dehydrogenase/acetyl-CoA synthase; electron bifurcating hydrogenase
Key enzymes: CO dehydrogenase/acetyl-CoA synthase; electron bifurcating hydrogenase
47
New cards
Methanogenesis
- Methane from CO2, acetate, or methanol
- Produces Coenzyme M, Coenzyme F420, Coenzyme F430, MP, MF, CoM, CoB (C1 careers)
- Creates a sodium motive pump for ATP production
- **H is e- donor and CO2 is e- acceptor + carbon source
- Strictly anaerobic process
- Obligate anaerobes
- thermophiles, mesophiles, etc
Habitats: anoxic enviro, wetlands, animal digestive tracts, landfills, wastewater, hydrothermal vents
- Produces Coenzyme M, Coenzyme F420, Coenzyme F430, MP, MF, CoM, CoB (C1 careers)
- Creates a sodium motive pump for ATP production
- **H is e- donor and CO2 is e- acceptor + carbon source
- Strictly anaerobic process
- Obligate anaerobes
- thermophiles, mesophiles, etc
Habitats: anoxic enviro, wetlands, animal digestive tracts, landfills, wastewater, hydrothermal vents
48
New cards
Methanogenesis Pathway
- like reductive acetyl-coa pathway
- energy conserved by converting CO2 --\> CH4 coupled to H+ or Na+ transport
- energy conserved by converting CO2 --\> CH4 coupled to H+ or Na+ transport
49
New cards
Reductive Acetyl-CoA Pathway (Methanogens)
formation of acetyl-coa from two CO2 molecules that are independently reduced by two different mechanism
key enzymes: acetyl-coa synthase (bifunctional)
key enzymes: acetyl-coa synthase (bifunctional)
50
New cards
Methylotrophic Pathway
(L10 slide 23)
4 CH3OH --\> 3 CH4 + CO2 + 2H2O
4 CH3OH --\> 3 CH4 + CO2 + 2H2O
51
New cards
Acetoclastic Pathway
Acetate + H + --\> CO2 + CH4
52
New cards
Methanogens + Syntrophy
methanogens + secondary fermenters
- common fermentation end products: CO2, H2, acetate, ethanol
- allows for interspecies hydrogen transfer as it 'pulls' the fermentation reactions
(L10 slide 28)
- common fermentation end products: CO2, H2, acetate, ethanol
- allows for interspecies hydrogen transfer as it 'pulls' the fermentation reactions
(L10 slide 28)
53
New cards
Filamentous Methanogens (wastewater treatment)
- bacterial waste products from aerobic respiration + fermentation --\> methane
- bacteria are packed together by filamentous methanogens (settle out of liquid)
- bacteria are packed together by filamentous methanogens (settle out of liquid)
54
New cards
Methane Hydrates
Small bubbles or individual molecules of methane (natural gas) produced by psychrophilic methanogens trapped in a crystalline matrix of frozen water
55
New cards
Methylotrophy
The metabolic oxidation of single-carbon compounds such as methanol, methylamine, or methane to yield energy
(all C-C bonds synthesized de novo)
- facultative and obligate
(all C-C bonds synthesized de novo)
- facultative and obligate
56
New cards
Methanotrophy
located at oxic/anoxic interface in aquatic sediments and soil environments
obligate anaerobes
obligate anaerobes
57
New cards
Methane monooxygenase (MMO)
- introduces oxygen into methane to produce methanol
two forms: solume and membrane bound (most common)
CH4 + O2 + NAD(P)H + H --\> CH3OH + NAD(P) + H2O
two forms: solume and membrane bound (most common)
CH4 + O2 + NAD(P)H + H --\> CH3OH + NAD(P) + H2O
58
New cards
Type I Methanotrophs
- gamma proteobacteria
- obligate methylotrophs, no complete TCA cycle
- assimilate C1 via ribulose monophosphate pathway (key enzyme \= hexulose-P synthase)
- obligate methylotrophs, no complete TCA cycle
- assimilate C1 via ribulose monophosphate pathway (key enzyme \= hexulose-P synthase)
59
New cards
Type II Methanotrophs
- alpha proteobacteria
- assimilate C1 via serine pathway (THF) (key enzyme \= serine transhydroxymethylase)
- complete TCA cycle
- many are facultative and can utilize simple organic compound
- assimilate C1 via serine pathway (THF) (key enzyme \= serine transhydroxymethylase)
- complete TCA cycle
- many are facultative and can utilize simple organic compound
60
New cards
Nonproteobacterial methanotroph (verrumicrobia)
- live at oxic/anoxic interface of geothermally released methane
- hyperthermophilic, acidophilic
- obligate methylotroph
- use serine pathway
- hyperthermophilic, acidophilic
- obligate methylotroph
- use serine pathway
61
New cards
Methane to formaldehyde
- occurs in methano or methylotrophs
- CH3OH oxidized by methanol dehydrogenase
- CH3OH oxidized by methanol dehydrogenase
62
New cards
Anaerobic methane oxidation
- members of euryarchaeota in syntrophic association w/ sulfate reducing bacteria
- reverse methanogenesis with oxidization of methane in anoxic ocean sediments
- not energetically favorable --\> syntrophy required
- ANME oxidizes CH4 as electron donor for SRB
- slow growth but important for capturing greenhouse gas
- MMO for conversion + reduces nitrite to produce O2 (candidatus methylomirabilis oxyfera)
habitat: anoxic freshwater and marine sediments, soils, peat bogs, wastewater treatment plants
- reverse methanogenesis with oxidization of methane in anoxic ocean sediments
- not energetically favorable --\> syntrophy required
- ANME oxidizes CH4 as electron donor for SRB
- slow growth but important for capturing greenhouse gas
- MMO for conversion + reduces nitrite to produce O2 (candidatus methylomirabilis oxyfera)
habitat: anoxic freshwater and marine sediments, soils, peat bogs, wastewater treatment plants
63
New cards
Respiration
oxidation of organic compounds coupled to transfer of electrons to an external electron acceptor
(38 ATP/glucose)
reoxidizes NADH, coupled to energy conservation
(L11 slide 5)
(38 ATP/glucose)
reoxidizes NADH, coupled to energy conservation
(L11 slide 5)
64
New cards
Fermentation
internally balanced oxidation-reduction reactions
excretion of products (some C wasted)
energy conserved via substrate level phosphorylation
*requires reversed function of ATPase to have an energized membrane/generate PMF
(2 ATP/ glucose)
(L11 slide 8 + 9 + 13)
excretion of products (some C wasted)
energy conserved via substrate level phosphorylation
*requires reversed function of ATPase to have an energized membrane/generate PMF
(2 ATP/ glucose)
(L11 slide 8 + 9 + 13)
65
New cards
Anaerobic Respiration
* variety of alternative anaerobic respiration
* carried out by specific microorganisms
* higher energy yielding respirations are used preferentially through gene regulation --> depends on O2 presence, presence of alternative electron acceptor \*e coli (O2 > nitrate > fumarate > fermentation)
* carried out by specific microorganisms
* higher energy yielding respirations are used preferentially through gene regulation --> depends on O2 presence, presence of alternative electron acceptor \*e coli (O2 > nitrate > fumarate > fermentation)
66
New cards
Nitrate Reduction
The process by which nitrate (NO3-) is reduced to ammonia (NH3) using membrane bound or periplasmic enzymes (also generates PMF)
67
New cards
Degradation of Polymers
degradation of polymers into utilizable compounds by exoenzymes that are secreted or attached to the surface of bacteria (production carefully regulated)
68
New cards
Degradation of plant biomass
degradation of cellulose by cellulases (relatively rare among bacteria) that are secreted or bound --\> starch, xylans, pectin which have their own exoenzymes
*cellulosomes in clostridium thermocellum
*cellulosomes in clostridium thermocellum
69
New cards
Degradation of proteins
degradation of proteins by proteases --\> deaminated to form organic acids for TCA cycle or fermentation
70
New cards
Degradation of lipids
cleavage of lipids by lipases (secreted) to release fatty acids --\> degrated by B-oxidation
71
New cards
Degradation of lignin
polymer of aromatic compounds
- extremely hydrophobic
- low aqueous solubility
- tend to partition into membranes
- very stable
--\> funnel into common intermediates *catechol is key intermediate
- extremely hydrophobic
- low aqueous solubility
- tend to partition into membranes
- very stable
--\> funnel into common intermediates *catechol is key intermediate
72
New cards
Aerobic Aromatic Degradation Pathways
addition of oxygen to the ring for destabilization and ring cleavage by mono or dioxygenases and ring cleavage dioxygenases --\> convert to compounds that enter central metabolis
73
New cards
Aromatic Ring Cleavage
Cleavage of Catechol
- intradiol (ortho) \= between hydroxylated carbons
- extradiol (meta) \= adjacent to one hydroxylated carbon
eg P Putida benzoate degradation uses oxygen as a substrate
- intradiol (ortho) \= between hydroxylated carbons
- extradiol (meta) \= adjacent to one hydroxylated carbon
eg P Putida benzoate degradation uses oxygen as a substrate
74
New cards
Xenobiotic Chemicals
unnatural or synthetic chemicals such as herbicides, pesticides, refrigerants, solvents, organic compounds
75
New cards
Bacterial Degradation of Xenobiotic Aromatic Compounds
- reactions are catalyzed by monooxygenases or dioxygenases that use oxygen as a substrate w/ catechol as a key intermediate
- enzymes for different aromatic compounds are often homologous to other oxygenases
- selection allows for utilization of new carbon, nitrogen
- compounds may be cooxidized but not provide anything
- compounds are funneled to common intermediates and pathways are modular
- pathways are STRAIN specific not species specific
- enzymes for different aromatic compounds are often homologous to other oxygenases
- selection allows for utilization of new carbon, nitrogen
- compounds may be cooxidized but not provide anything
- compounds are funneled to common intermediates and pathways are modular
- pathways are STRAIN specific not species specific
76
New cards
BTEX
Benzene, Toluene, Ethylbenzene, Xylene
most soluble aromatic hydrocarbons in gasoline
most soluble aromatic hydrocarbons in gasoline
77
New cards
Nitroaromatic compounds
used in tnt, dyes, pesticides ,polymers
- synthetic
- high stability
- toxic, mutagenic, carcinogenic
- synthetic
- high stability
- toxic, mutagenic, carcinogenic
78
New cards
Anaerobic Aromatic Funneling Pathways
central intermediate \= benzoyl-CoA
- occurs under denitrifying, sulfate-reducing, photoheterotrophic conditions
- involves ring reduction and B-oxidation like reactions (addition of CoA to destabilize)
- much slower than aerobic and very oxygen sensitive
eg fumarate addition to toluene --\> benzylsuccinate
- occurs under denitrifying, sulfate-reducing, photoheterotrophic conditions
- involves ring reduction and B-oxidation like reactions (addition of CoA to destabilize)
- much slower than aerobic and very oxygen sensitive
eg fumarate addition to toluene --\> benzylsuccinate
79
New cards
Sulfate/Sulfur Reducing Bacteria
- inorganic sulfur compounds serve as electron acceptors in anaerobic respiration
- end product of respiratory reduction is H2S (excreted)
- end product of respiratory reduction is H2S (excreted)
80
New cards
Sulfate Reducing Bacteria + Archaea
many are gamma-proteobacteria, thermophiles
(endosporefomers)
all can also respire with sulfur
archaeoglobus \= hyperthermophilic euryarchaeote, oxidize organic carbon sources w/ SO4 as terminal electron acceptor (anoxic conditions)
bacteria \= obligate anaerobes; H2 (lithoautotrophs) lactate or pyruvate, other fermentation products as electron donors; most use reductive acetyl-CoA pathway; compete for H2 with methanogens but conserve more energy and use a wider range of substrates
(endosporefomers)
all can also respire with sulfur
archaeoglobus \= hyperthermophilic euryarchaeote, oxidize organic carbon sources w/ SO4 as terminal electron acceptor (anoxic conditions)
bacteria \= obligate anaerobes; H2 (lithoautotrophs) lactate or pyruvate, other fermentation products as electron donors; most use reductive acetyl-CoA pathway; compete for H2 with methanogens but conserve more energy and use a wider range of substrates
81
New cards
Sulfate-Reduction
habitat: anoxic aquatic and terrestrial enviro w/ sufficient sulfur sources
- can be enriched in anoxic medium containing lactate and ferrous iron
- less energetically favorable than more common respiration or fermentation
- ATP is invested to activate sulfate and drive reaction
- reduction requires 8 electrons; form of anaerobic respiration
(1 ATP/sulfate)
key enzymes \= ATP sulfurylase, APS reductase, sulfite reductase, hydrogenase
(L12 S17)
- can be enriched in anoxic medium containing lactate and ferrous iron
- less energetically favorable than more common respiration or fermentation
- ATP is invested to activate sulfate and drive reaction
- reduction requires 8 electrons; form of anaerobic respiration
(1 ATP/sulfate)
key enzymes \= ATP sulfurylase, APS reductase, sulfite reductase, hydrogenase
(L12 S17)
82
New cards
Sulfur/sulfide oxidizing bacteria
anaerobes: PS and GS use H2S/S as e donor
83
New cards
"colorless" sulfur oxidizing bacteria
aerobic sulfur oxidizing bacteria that oxidize reduced sulfur compounds H2S and S as energy sources
chemolithotrophic w reduced sulfur compounds while most are obligate chemolithoautotrophs using inorganic compounds + TCA cycle
habitat: soil, sediments, freshwater, and marine enviro (typically in narrow zones where sulfide + oxygen coexist); symbionts w/ riftia at hydrothermal vents
chemolithotrophic w reduced sulfur compounds while most are obligate chemolithoautotrophs using inorganic compounds + TCA cycle
habitat: soil, sediments, freshwater, and marine enviro (typically in narrow zones where sulfide + oxygen coexist); symbionts w/ riftia at hydrothermal vents
84
New cards
'Large' colorless sulfur oxidizing bacteria
gamma-proteobacteria
large cell size + internal sulfur deposits (sparkly inclusions)
habitat: both freshwater and marine (difficult to culture)
some are lithoautotrophs
eg thiomargita nambiensis: large vacuole containing nitrate
large cell size + internal sulfur deposits (sparkly inclusions)
habitat: both freshwater and marine (difficult to culture)
some are lithoautotrophs
eg thiomargita nambiensis: large vacuole containing nitrate
85
New cards
colorless sulfur bacteria examples
beggiatoa: filamentous gliding, typically in microoxic water enviros @ surface of sediments; chemotactic + repellent to O2 and H2S; can form thick mats
thioploca: large filamentous sulfur bacteria, filaments in polysaccharide sheaths; motile by gliding w/ positive chemotaxis to nitrate + negative taxis to oxygen; -response to high sulfide; oxidize H2S --\> S --\> SO4
sulfolobus: hyperthermophilic, acidophilic sulfur-oxidizing chrenarchaeote; aerobic lithoautotroph that oxidizes H2S or S to H2SO4 + fixes CO2 w/ 3HP/4HB cycle
thioploca: large filamentous sulfur bacteria, filaments in polysaccharide sheaths; motile by gliding w/ positive chemotaxis to nitrate + negative taxis to oxygen; -response to high sulfide; oxidize H2S --\> S --\> SO4
sulfolobus: hyperthermophilic, acidophilic sulfur-oxidizing chrenarchaeote; aerobic lithoautotroph that oxidizes H2S or S to H2SO4 + fixes CO2 w/ 3HP/4HB cycle
86
New cards
Oxidation States of Nitrogen
NH3, NH4 = incorporated into cells
N2 = most abundant form
HNO3, NO3- = oxidized to
N2 = most abundant form
HNO3, NO3- = oxidized to
87
New cards
Nitrogen Cycle Reactions
Assimilatory Nitrate Reduction = NO3- → NH4+
Denitrification = NO3- → N2
Nitrogen Fixation = N2 → NH4+
\
Nitrification/Comammox = NH4+ → NO3-
Ammamox = NH4+ + NO2- → N2
\
Denitrification = NO3- → N2
Nitrogen Fixation = N2 → NH4+
\
Nitrification/Comammox = NH4+ → NO3-
Ammamox = NH4+ + NO2- → N2
\
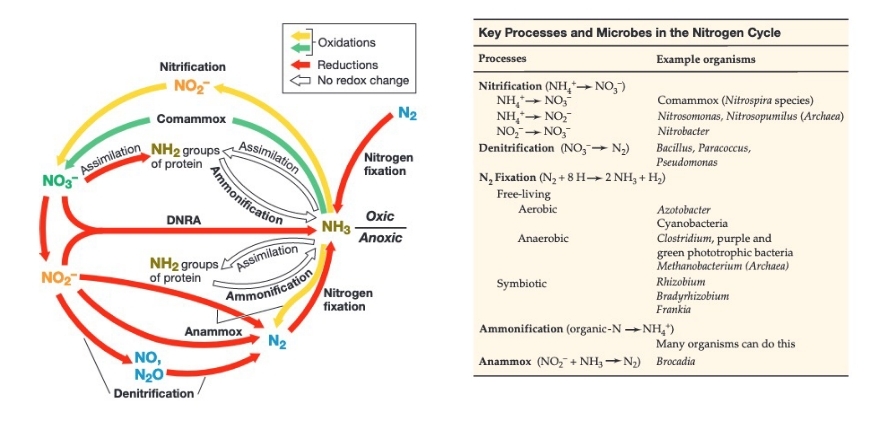
88
New cards
N2 Fixation
widespread among bacteria and archaea; NN bond very stable
\
fixers: proteobacteria, firmicutes, cyanobacteria, archaae, chlorobi, actinobacteria, spirochetes
N2 + 8e- + 8H+ + \`16ATP → 2NH3 + H2 + 16 ADP + 16 Pi
Key enzyme = nitrogenase which reduces N2 → NH3 (very oxygen sensitive; irreversibily inactivated)
\*H2 always a byproduct
\
fixers: proteobacteria, firmicutes, cyanobacteria, archaae, chlorobi, actinobacteria, spirochetes
N2 + 8e- + 8H+ + \`16ATP → 2NH3 + H2 + 16 ADP + 16 Pi
Key enzyme = nitrogenase which reduces N2 → NH3 (very oxygen sensitive; irreversibily inactivated)
\*H2 always a byproduct
89
New cards
Nitrogenase
complex enzyme with \~20 genes required for its production (only 3 make up the components)
homologous in all orgs as it evolved once
homologous in all orgs as it evolved once
90
New cards
Dealing with O2 while fixing N2
Control expression of nitrogenase genes: when NH4+ is limiting and O2 absent
\
microaerophiles + obligate aerobes: high respiration rate that is often faster or as fast as O2 diffusion to protect nitrogenase (azotobacter have polysaccharide capsule slows O2 diffusion + harbors aerobes)
\
microaerophiles + obligate aerobes: high respiration rate that is often faster or as fast as O2 diffusion to protect nitrogenase (azotobacter have polysaccharide capsule slows O2 diffusion + harbors aerobes)
91
New cards
Separation of O2 and N2
temporal separation of photosynthesis from N2 fixation based on cycles with N2 fixation in dark
\
spatial separation of photosynth + N2 fixation: filamentous cyanobacteria have development of heterocysts when N2 is limiting (5-10%); heterocysts limit diffusion of O2 w/ thick cell wall
\
spatial separation of photosynth + N2 fixation: filamentous cyanobacteria have development of heterocysts when N2 is limiting (5-10%); heterocysts limit diffusion of O2 w/ thick cell wall
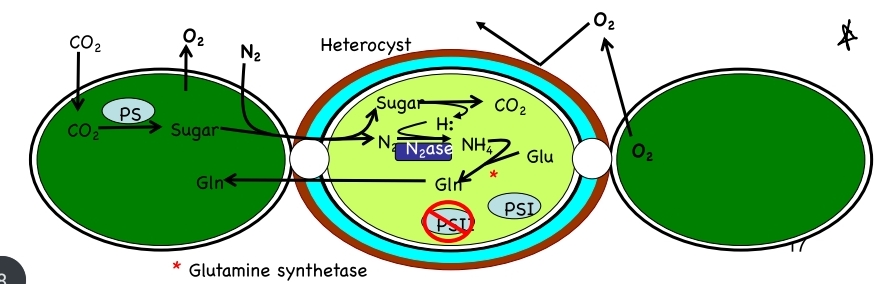
92
New cards
Symbiotic Nitrogen Fixers
Rhizobia = gram- proteobacteria that are symbiotic with eukaryotes (aerobic heterotrophs)
\
form root nodules and are significant N2 fixers
* nitrogen is provided to the plant + rhizobium gets C-source and protected environment
different species form specific associations with certain plants
\
form root nodules and are significant N2 fixers
* nitrogen is provided to the plant + rhizobium gets C-source and protected environment
different species form specific associations with certain plants
93
New cards
Root Nodule Formation
1. Recognition of correct partner and attachment of the bacterium to the root hair
1. secrete flavonoids (chemoattractants) that induce Nod factor production (host-microbe specificity)
2. Invasion of the rhizobia into the plant root
1. nod factors stimulate production of infection thread → rhizobia colonize root cells through infection thread
3. Differentiation of plant cells into nodule tissues and bacterial differentiation into bacteroids
1. nod factors stimulate nodule formation (O2 binding protein leghemoglobin controls O2 levels)
2. sym plasmid expressed for nif/fix gene
94
New cards
Diversity in symbiotic nitrogen fixation
Frankia = actinomycete alder symbiote
* can fix nitrogen at full oxygen tension + when in symbiosis
* sequesters nitrogenase in vesicles (protect from O2)
* can fix nitrogen at full oxygen tension + when in symbiosis
* sequesters nitrogenase in vesicles (protect from O2)
95
New cards
Nitrification
oxidation of ammonia to nitrate: carried out by ammonia oxidizers and nitrite oxidizers
\
aerobic chemolithoautotrophs that use calvin cycle
\
aerobic chemolithoautotrophs that use calvin cycle
96
New cards
Ammonia oxidizing bacteria (AOB)
beta and gamma proteobacteria + nitrospira
NH3 + 1/2 O2 → NO2- + H2O (energetically favorable)
\
Key enzymes:
1. ammonia monooxygenase (converts ammonia to hydroxylamine; related to MMO)
2. hydroxylamine oxidoreductase (catalyzes hydroxylamine to nitrite + coupled to ETC)
NH3 + 1/2 O2 → NO2- + H2O (energetically favorable)
\
Key enzymes:
1. ammonia monooxygenase (converts ammonia to hydroxylamine; related to MMO)
2. hydroxylamine oxidoreductase (catalyzes hydroxylamine to nitrite + coupled to ETC)
97
New cards
Nitrite oxidizing bacteria (NOB)
alpha, beta, gamma, and delta proteobacteria + nitrospira
NO2- + H2O → NO3- + 2H+ + 2e- (energetically favorable)
\
Key enzyme: nitrite oxidoreductase (catalyzed nitrite to nitrate; coupled to ETC w/ PMF)
NO2- + H2O → NO3- + 2H+ + 2e- (energetically favorable)
\
Key enzyme: nitrite oxidoreductase (catalyzed nitrite to nitrate; coupled to ETC w/ PMF)
98
New cards
AOB + NOB
obligate aerobes in a tight mutualistic symbiosis
\
habitats: soil + water where ammonia is high, sewage treatment plants
\
habitats: soil + water where ammonia is high, sewage treatment plants
99
New cards
Ammonia oxidizing archaea
chemolithoautotrophic ammonia oxidizing marine archaea that can grow on low NH3 concentrations and also solely on ammonia as energy source
\
fix CO2 using 3HP/4HB cycle
\
fix CO2 using 3HP/4HB cycle
100
New cards
Comammox
nitrospira has genes for all the key enzymes and can completely oxidize ammonia to nitrate by itself as the reactions are energetically favorable