Week 6 - Gene Mutation, DNA Repair, and Homologous Recombination
1/106
Earn XP
Description and Tags
Chapter 11 & Chapter 13
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
107 Terms
What is a point mutation?
Substitution, insertion, or deletion of a single base pair in a gene.
When do point mutations often occur?
Due to mistakes during DNA replication. Rare per cell cycle, but common across large populations.
How frequent are mutations at the phenotype level?
The mutation rate is approximately 10⁻⁶ to 10⁻⁸ per individual.
How frequent are mutations at the DNA sequence level?
The mutation rate is approximately 10⁻⁹ per base per replication (referred to as error rate of DNA polymerase).
What experiment provided evidence that mutations are random?
The Luria and Delbrück experiment (1943).
What were the two hypotheses tested by Luria and Delbrück?
Mutations are random.
Mutations arise from environmental triggers.
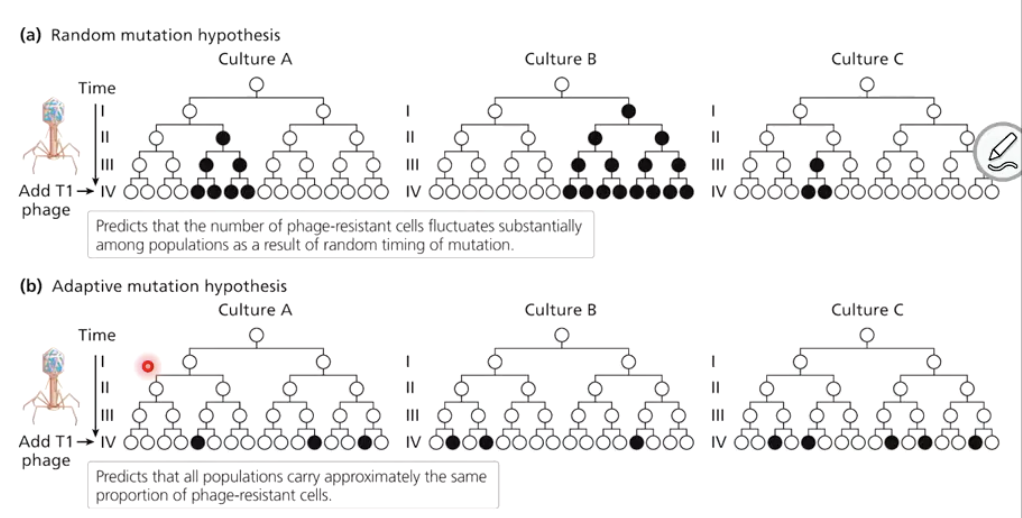
How did Luria and Delbrück test their hypotheses?
They cultivated bacteria, exposed them to T1 phage, and observed that resistance patterns followed the random mutation hypothesis.
Black circle is culture resistant to virus
Added virus in gen 4
If data looked like a) indicates that mutations occurred in previous generations and were passed on
If data looked like b) indicates that a change in environment led to mutations
Result: Resistance bacteria developed in a way consistent with random mutation hypothesis (top graph).
What are germ-line mutations?
Mutations that occur in gametes and can be passed on to the next generation.
Ex: Mutations during meiosis
What are somatic mutations?
Mutations that occur in body cells (somatic cells) and are not passed on to offspring but can affect the individual.
Ex: Mutations during mitosis, cancerous tumours
What are common point mutations?
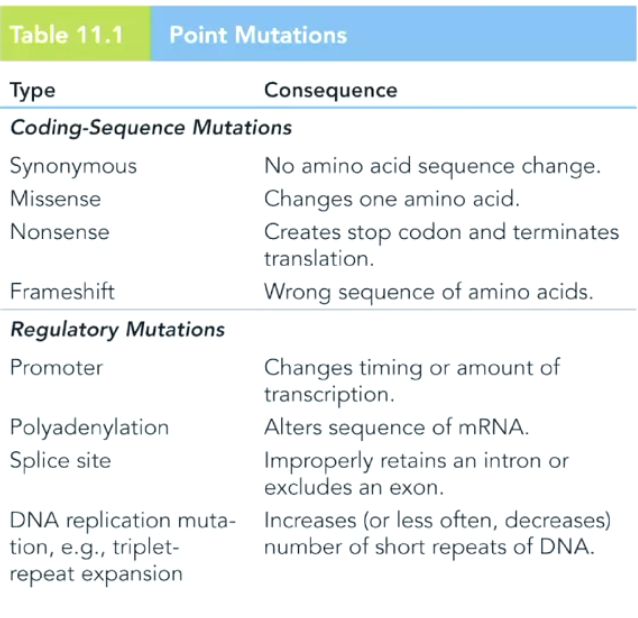
**Don’t need to know regulatory mutations
What is the third type of classification for point mutations?
Transition mutations (A↔G, T↔C)
1 purine turns into another purine; 1 pyrimidine turns into another pyrimidine
Transversion mutations (A↔T, A↔C, G↔T, G↔C)
A purine turns into a pyrimidine
Which type of point mutation is more common: transition or transversion?
Transition mutations.
Review genetic code table, most third bases in codon are redundant. Either purine/pyrimidine leads to the same amino acid.
What is a wild-type sequence?
The original, non-mutated DNA sequence.
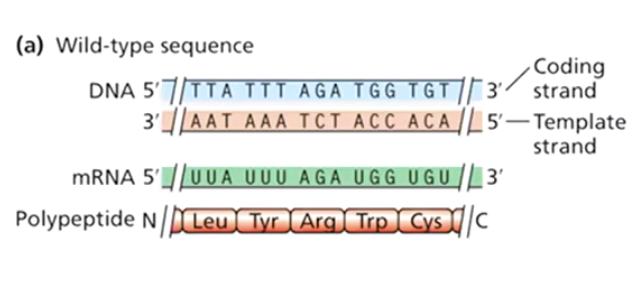
What is a synonymous mutation?
A mutation that changes a codon but still encodes the same amino acid (also called a silent mutation).
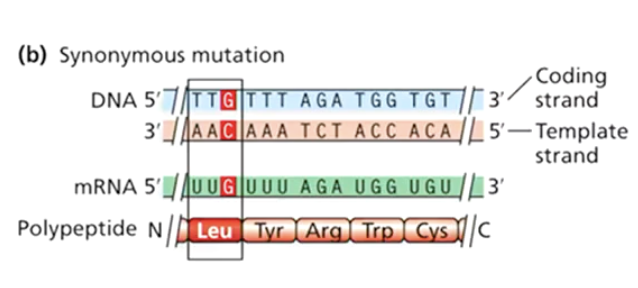
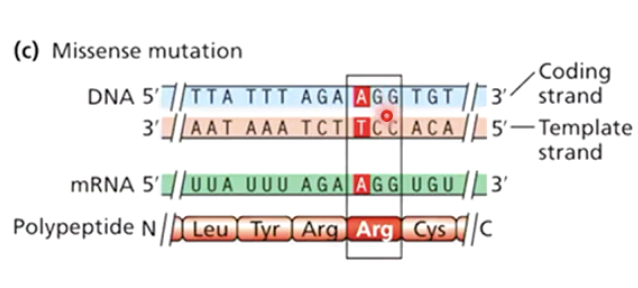
What is a missense mutation?
A mutation that changes a codon, resulting in a different amino acid being incorporated into the protein.
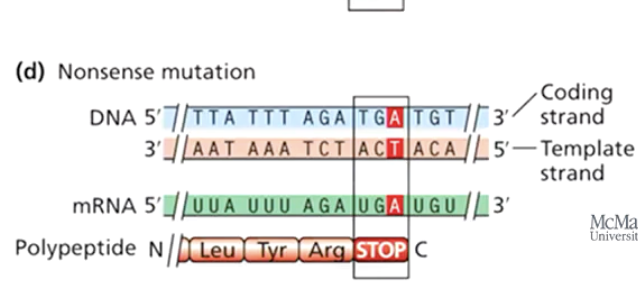
What is a nonsense mutation?
A mutation that changes a codon into a stop codon, leading to premature termination of translation.
Do all insertions and deletions cause frameshift mutations?
No. If the insertion or deletion occurs in multiples of three, the reading frame remains unaffected.
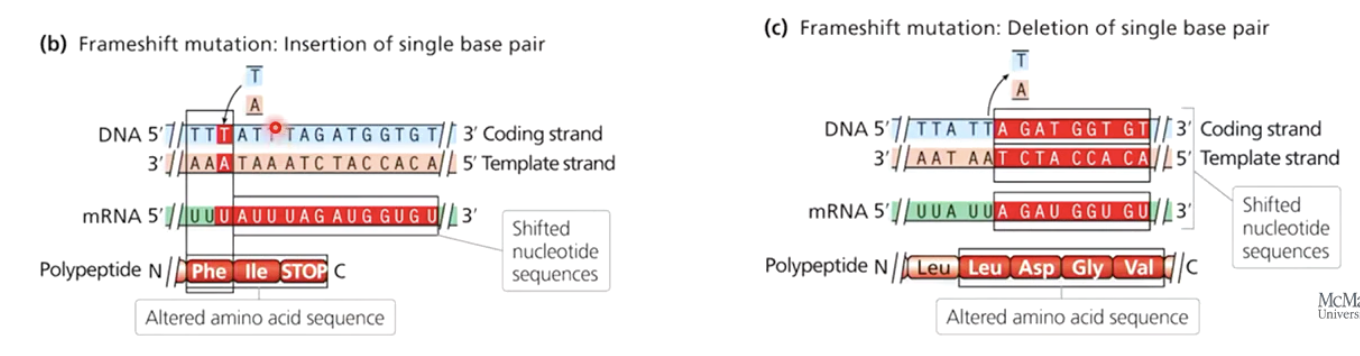
Give an example of an insertion that does not cause a frameshift mutation.
A 6 base pair insertion adds two extra amino acids without altering the reading frame.
In 1997 it was found that short stem length in Mendel’s pea plants was the result of a recessive point mutation in the Le gene. Specifically, the mutation caused alanine to change to threonine in one region of the translated polypeptide. What kind of point mutation is this?
a. Synonymous mutation
b. Nonsense mutation
c. Missense mutation
d. Frameshift mutation
c. Missense mutation
What is a forward mutation?
A mutation that changes a wild-type allele into a mutant allele.
What is a reverse mutation (reversion)? Types (3)?
A mutation that restores a mutant allele back to the wild-type sequence or function.
3 Types
True reversion
Intragenic reversion
Second-site reversion
What is a true reversion?
A mutation that restores the exact wild-type DNA sequence. Can also be a mutation in same codon, restoring wild-type amino acid.
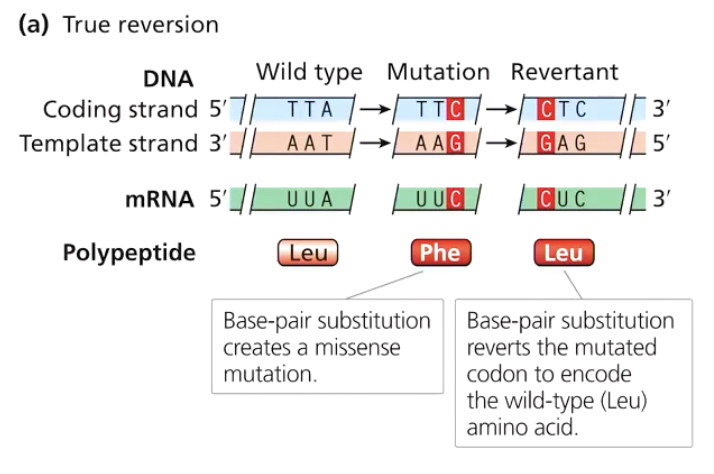
What is an intragenic reversion?
A mutation in the same gene that compensates for the original mutation, restoring function.
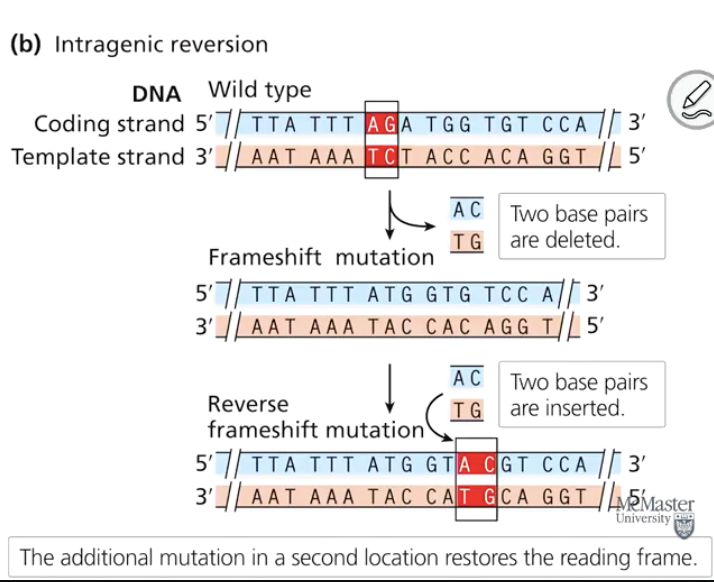
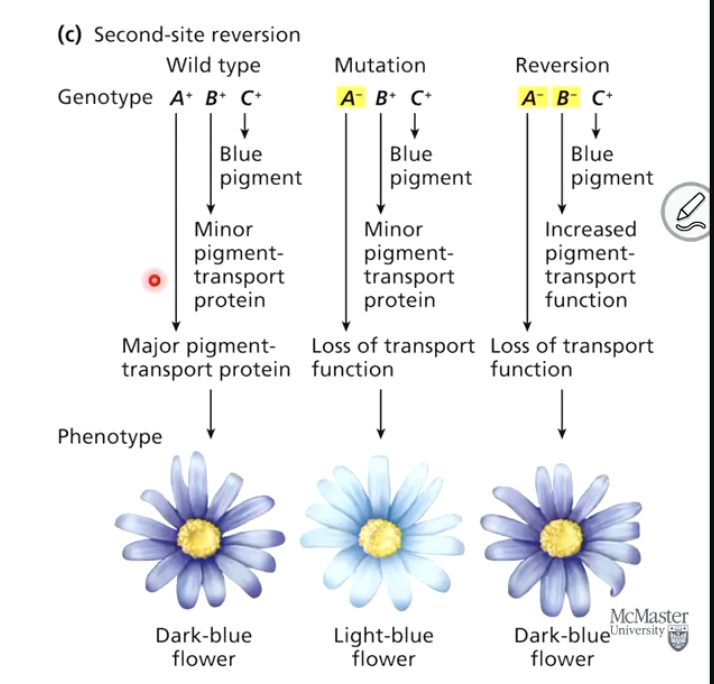
What is a second-site reversion (suppressor mutation)?
A mutation in a different gene that compensates for the original mutation, restoring the wild-type phenotype. Also called suppressor mutation.
Give an example of a second-site reversion.
If a mutation in one gene reduces pigment transport, a mutation in a different gene may compensate by upregulating another transport protein, restoring normal pigmentation.
What are three mechanisms that cause point mutations?
Mispaired nucleotides during replication
Spontaneous nucleotide base changes
Mutagens (chemicals or radiation)
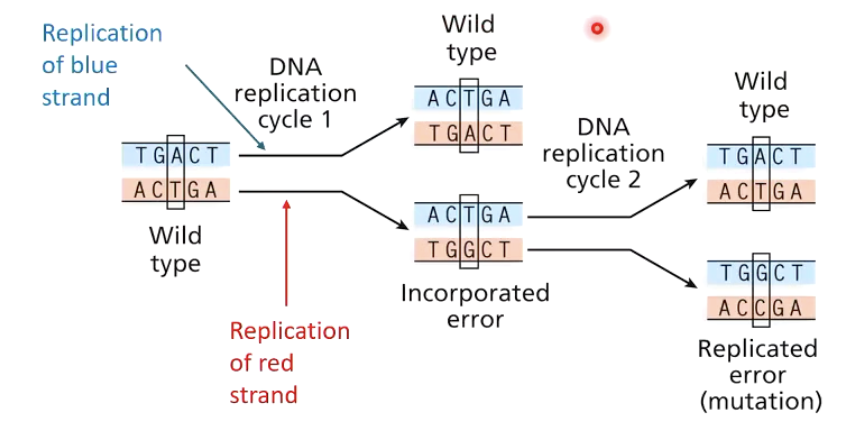
What happens when mispaired nucleotides occur during replication?
Non-complementary base pairing (incorporating error) can occur. If not repaired, this leads to mutations.
e.g., G:T or C:A pairing
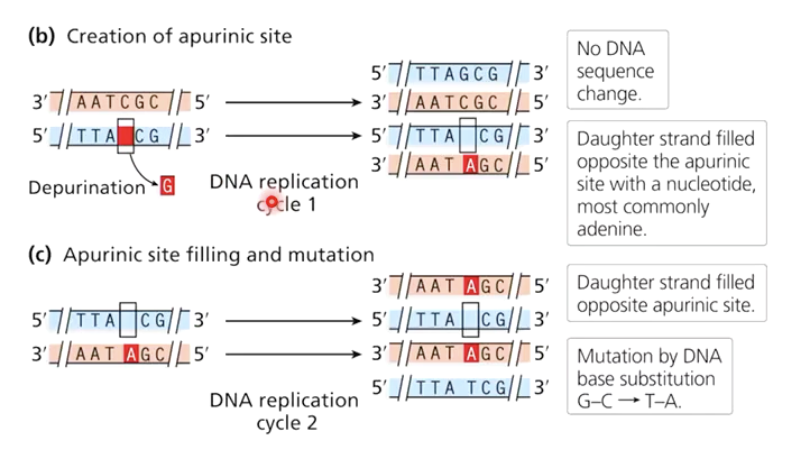
What is depurination?
The loss of a purine base (adenine or guanine) from DNA, creating an apurinic site.
What happens if an apurinic site is not repaired?
DNA polymerase often inserts an adenine, leading to G → A substitutions.
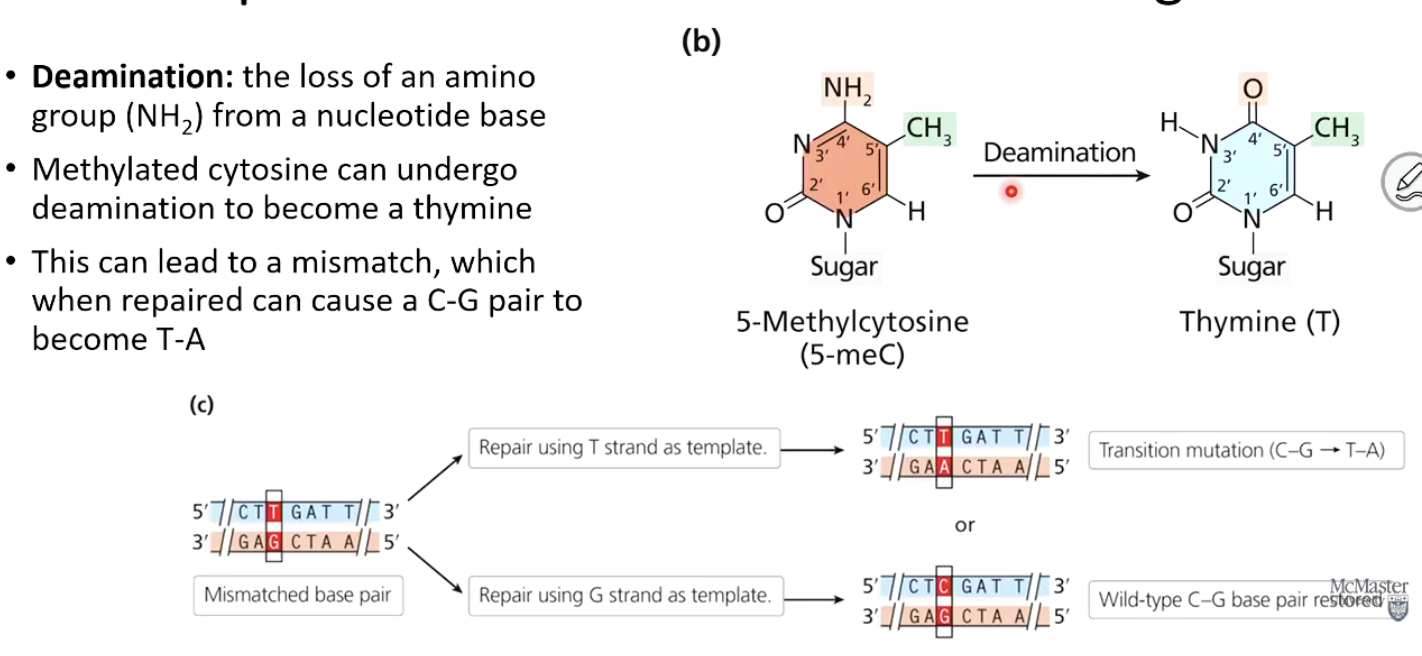
What is deamination?
The removal of an amino group (NH₂) from a nucleotide base.
How can deamination lead to a mutation?
Methylated cytosine can deaminate into thymine.
If not repaired, this can cause a C-G to T-A mutation.
What are the six main types of chemical mutagens?
Nucleotide base analogs – mimic DNA bases, causing mutations.
Deaminating agents – remove amino groups, leading to C-G → T-A changes.
Alkylating agents – add methyl/ethyl groups, distorting the DNA helix.
Oxidizing agents – oxidize nucleotide bases, causing transversion mutations.
Hydroxylating agents – add hydroxyl groups, resulting in modified cytosine pairing with A.
Intercalating agents – molecules that fit between bases, distorting DNA and leading to lesion that may cause frameshift mutations.
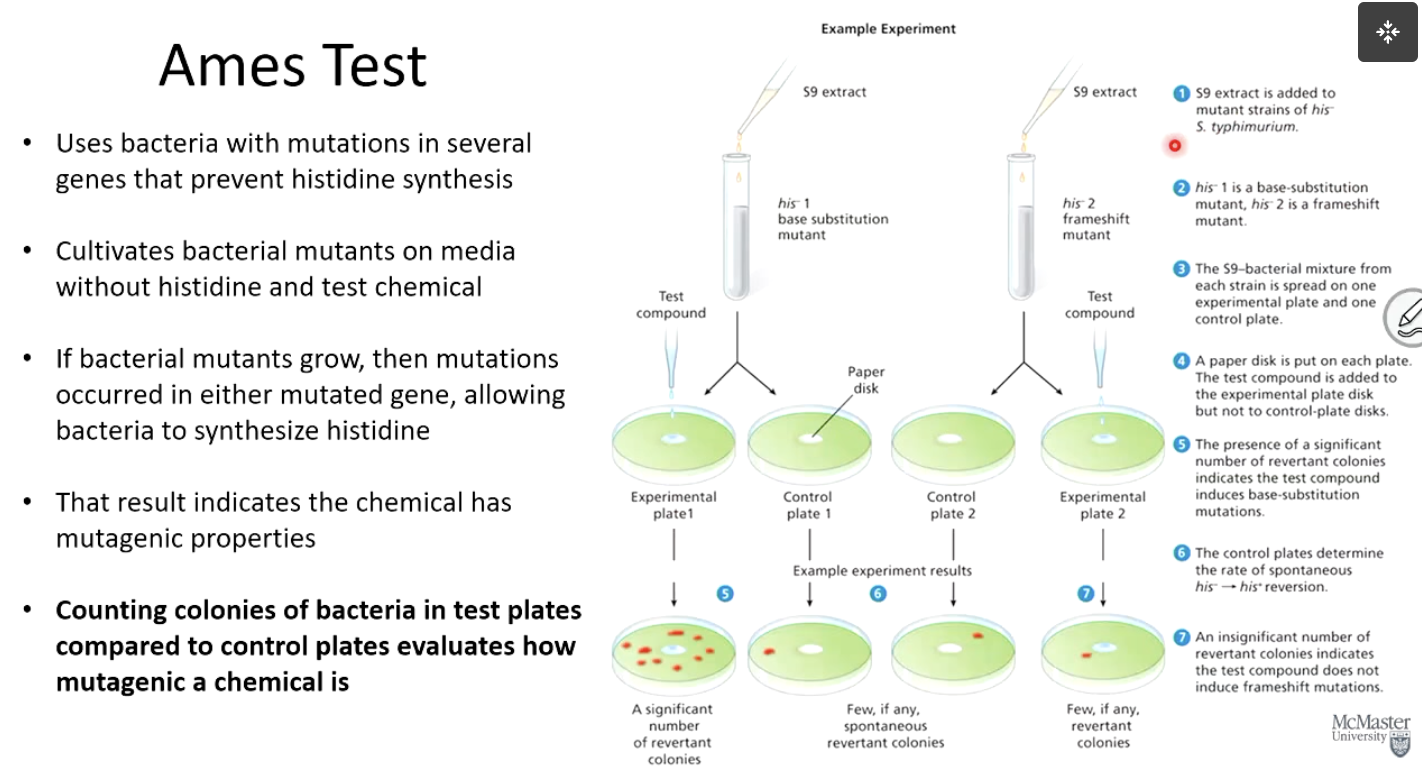
What is the Ames Test used for?
It determines whether a chemical is a mutagen by exposing bacteria to the chemical in the presence of mammalian liver enzymes.
Liver enzymes break down toxins into various byproducts for detoxification.
Ames test mimics this process.
Identifies whether chemical or various detoxifying byproducts are mutagenic.
How does the Ames Test work?
Uses bacteria with mutations that prevent histidine synthesis.
If the bacteria grow without histidine, mutations must have occurred, suggesting the test chemical is mutagenic.
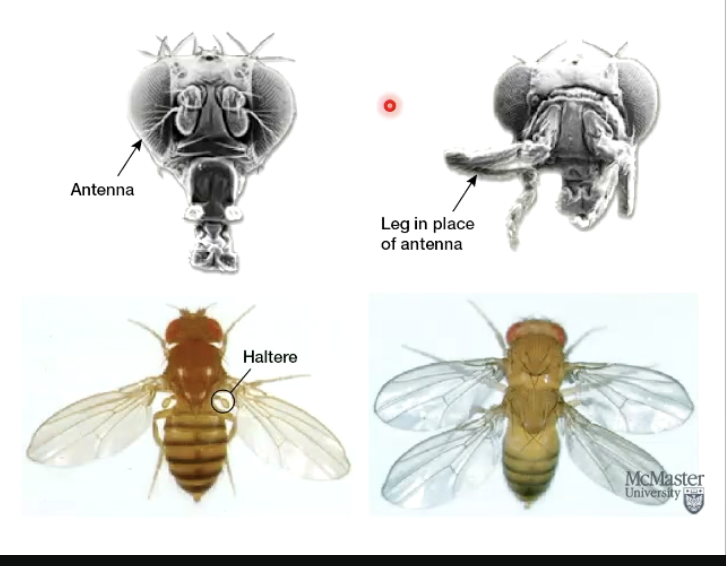
What did Hermann Muller discover about radiation?
He showed that X-rays can cause mutations in fruit flies.
Radiation exposure induces mutations in germ-line, which may get passed on to offspring.
What types of radiation are mutagenic?
All high-energy radiation is mutagenic:
Ultraviolet (UV) rays
X-rays
Gamma rays
Cosmic rays
How does UV radiation cause mutations?
UV exposure causes thymine dimers, where adjacent thymines form covalent bonds, disrupting DNA replication.
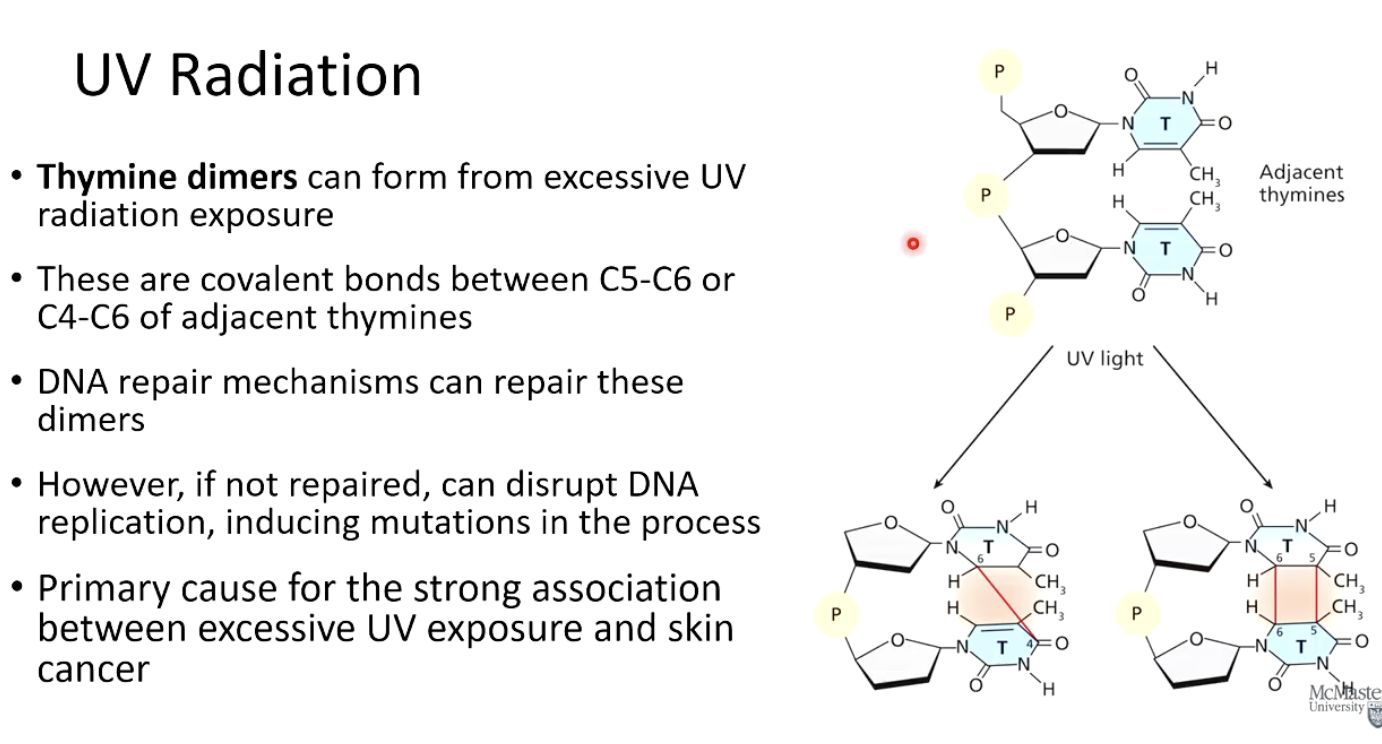

Why are thymine dimers harmful?
They distort the DNA structure and, if not repaired, can cause mutations, increasing the risk of skin cancer.
Can radiation-induced mutations be passed on to offspring?
Yes, if radiation damages germ-line cells, mutations can be inherited.
Common Systems for Direct DNA Repair
Base excision repair (BER)
Nucleotide excision repair (NER)
Mismatch repair
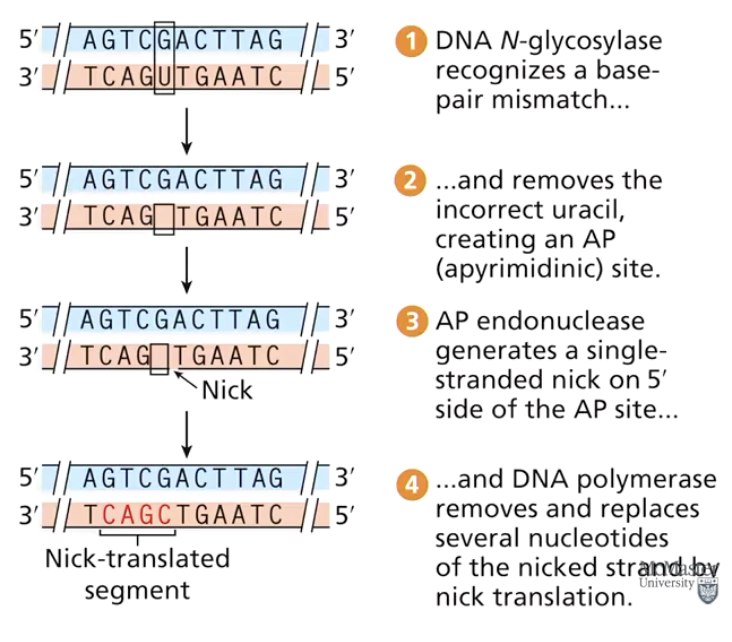
What is Base Excision Repair (BER)?
A repair mechanism that removes an incorrect or damaged DNA base and repairs by synthesis of a new strand segment (nick translation).
How does Base Excision Repair (BER) work?
Incorrect/damaged base is removed.
Nick translation: DNA polymerase removes and replaces nucleotides.
DNA ligase seals the sugar-phosphate backbone.
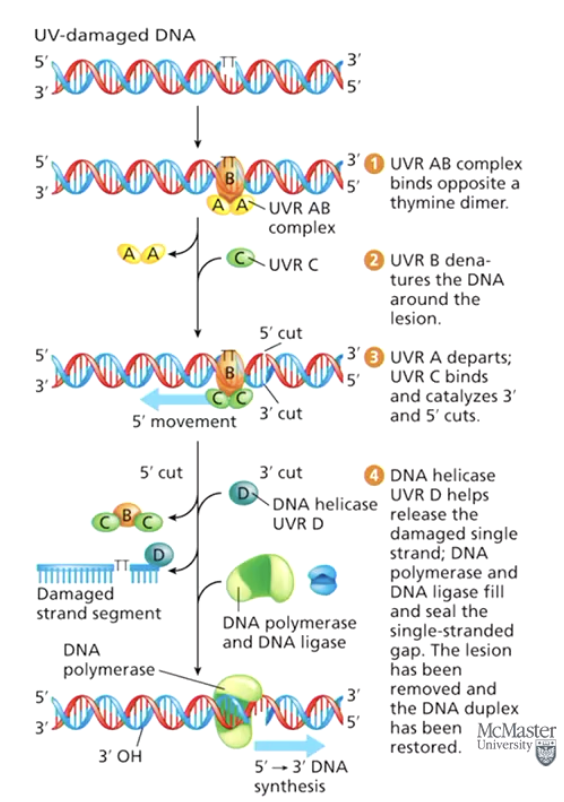
What is Nucleotide Excision Repair (NER)?
A repair system that removes a strand segment containing damaged DNA and replaces it with newly synthesized DNA.
What type of DNA damage does Nucleotide Excision Repair (NER) fix?
UV-induced DNA damage, such as thymine dimers.
What are the steps of Nucleotide Excision Repair (NER)?
Enzymes recognize and bind to the damaged DNA region.
Segment of nucleotides is removed.
DNA polymerase fills in the gap.
DNA ligase seals the sugar-phosphate backbone.
What is Mismatch Repair (MMR)?
A system that removes base-pair mismatches by excising a segment of the newly synthesized strand, then resynthesizing it.
How does Mismatch Repair (MMR) work in E. coli?
MutH binds to the unmethylated daughter strand.
MutS binds to the base-pair mismatch.
MutL connects MutH to MutS.
MutH cleaves the daughter strand.
DNA polymerase resynthesizes the excised segment.
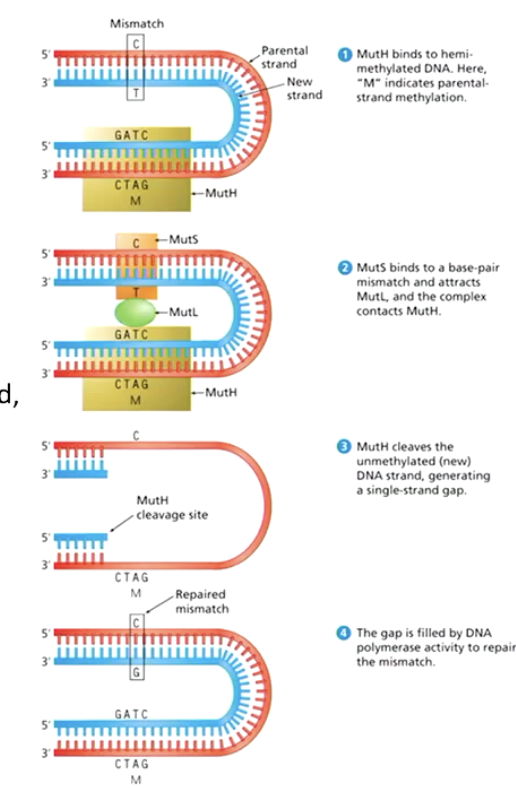
How does the cell differentiate between parental and newly synthesized DNA strands in Mismatch Repair?
The parental strand is usually methylated, while the newly synthesized strand is unmethylated.
What is Translesion DNA Synthesis?
Unrepaired DNA damage can block DNA polymerase III. Thus, this is an error-prone repair mechanism that allows DNA polymerase V to bypass lesions and synthesize short DNA segments.
What is SOS repair in E. coli?
A repair system activated in response to massive DNA damage, using translesion DNA polymerase (pol V) to bypass lesions.
Why is translesion DNA polymerase (pol V) error-prone?
It lacks proofreading abilities, leading to a high mutation rate.
What is Double-Strand Break (DSB) Repair?
A repair mechanism that fixes double-strand DNA breaks, preventing chromosome instability, cell death, and cancer.
What are the two main mechanisms for Double-Strand Break (DSB) Repair?
Nonhomologous End Joining (NHEJ) – error-prone
Synthesis-Dependent Strand Annealing (SDSA) – error-free
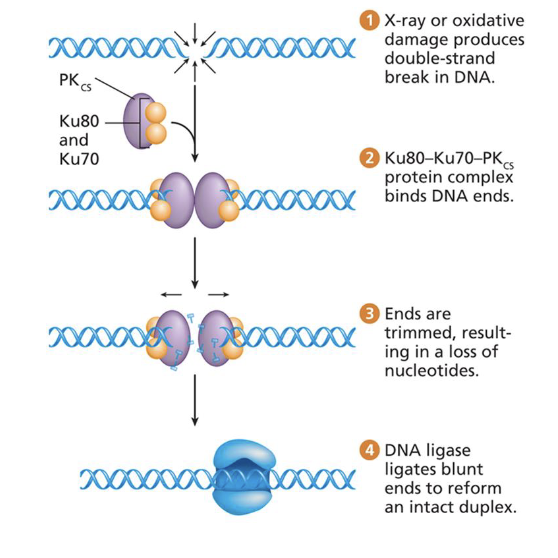
What is Nonhomologous End Joining (NHEJ)?
A mutagenic repair mechanism where DNA ends are trimmed into even “blunt ends” and rejoined with DNA ligase. Trimming causes nucleotide loss and possible frameshift mutations.
Blunt ends are overhangs that are not double-stranded
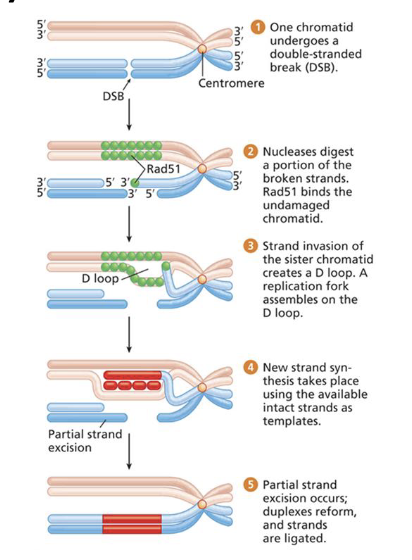
What is Synthesis-Dependent Strand Annealing (SDSA)?
An error-free repair mechanism where the intact sister chromatid provides a template for DNA repair after strand invasion. Similar process to homologous recombination but repairs DNA.
Why has CRISPR revolutionized gene editing protocols?
Simplicity: Relies on cell’s natural DNA repair mechanisms to facilitate genetic engineering
For CRISPR, inject an embryo with a plasmid or mRNA to express:
Cas9 nuclease enzyme
Guide RNA to guide Cas9 to genomic target
Donor template (synthetic DNA) if a “knock-in” is required
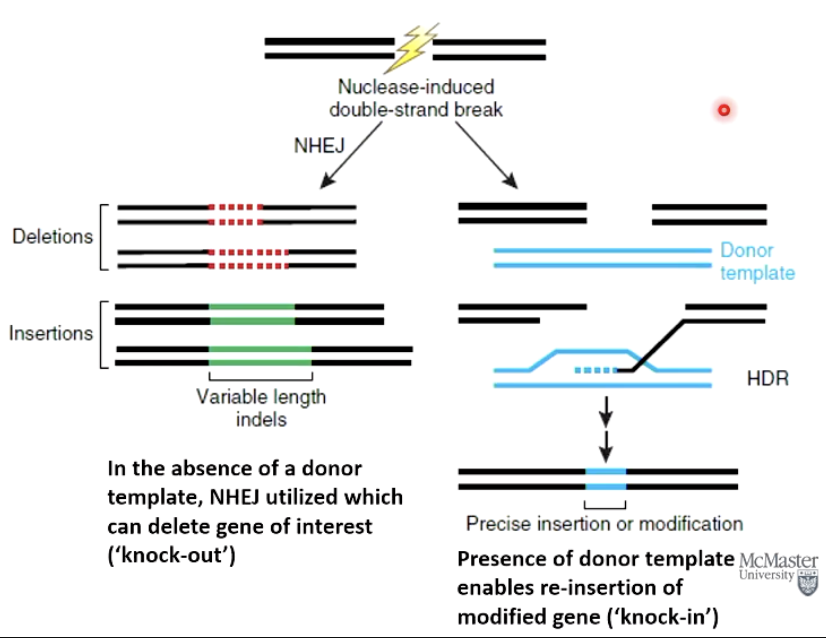
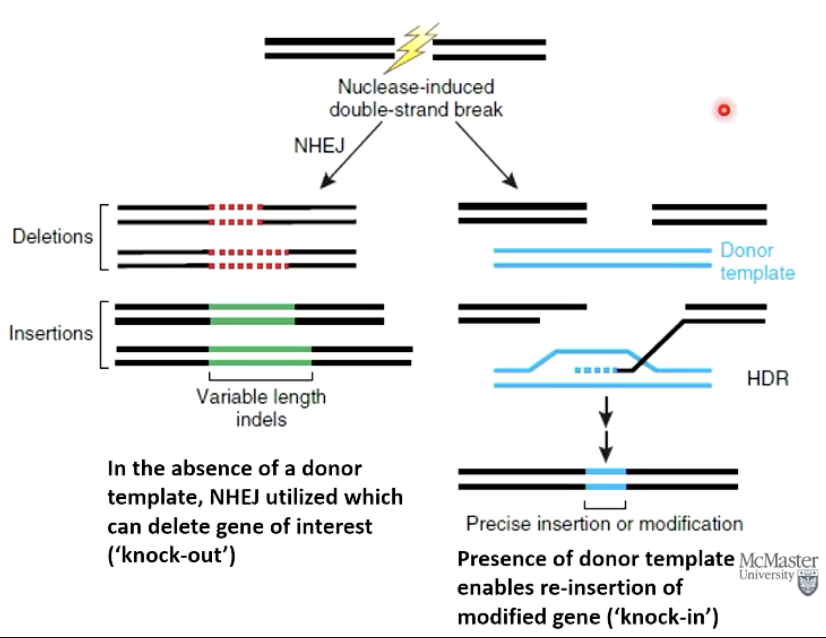
How does CRISPR gene editing utilize DNA repair mechanisms?
NHEJ - non-homologous end joining - can delete a gene (‘knock-out’).
SDSA - homologous directed repair - with a donor template can insert a modified gene (‘knock-in’).
What are Transposable Genetic Elements (TGE)?
“Selfish DNA” sequences that move within the genome via transposition, facilitated by transposase enzyme.
Different TGEs vary in length, sequence composition, and copy number.
What are the two types of transposition?
Non-replicative transposition – “cut & paste” (TGE moves to a new location).
Replicative transposition – “copy & paste” (TGE duplicates and inserts a copy elsewhere).
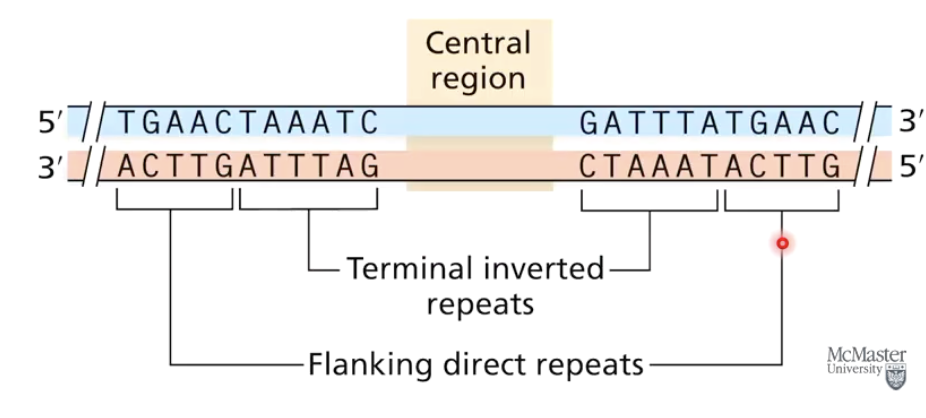
What are the shared structural features of Transposable Genetic Elements (TGEs)?
Terminal inverted repeats at both ends (part of TGE).
Flanking direct repeats surrounding the inserted TGE (not part of TGE).
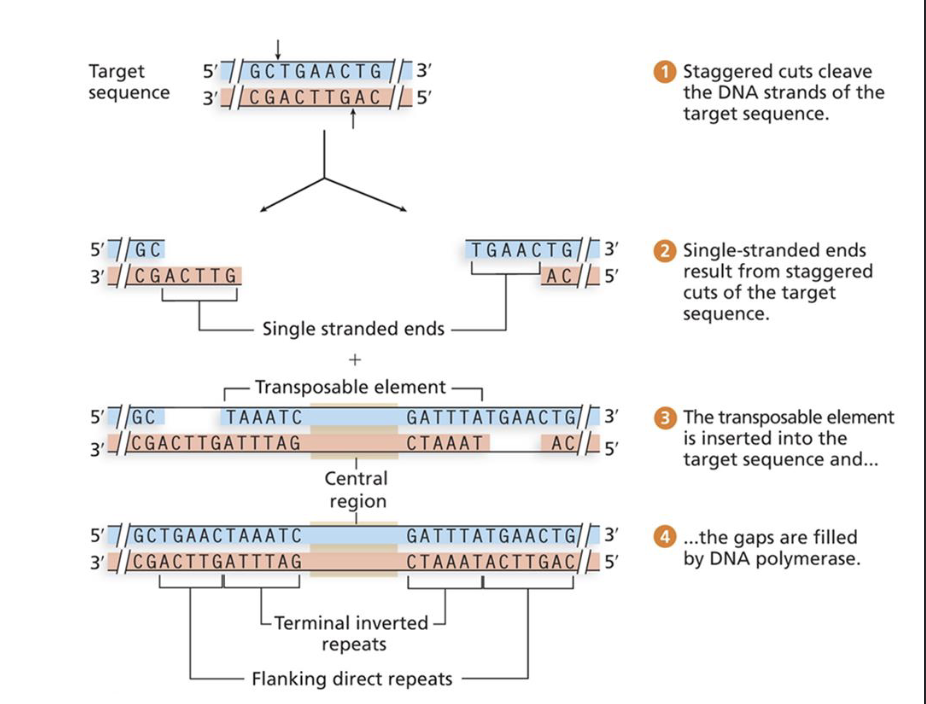
How does transposition work?
Common in areas that are not essential coding areas.
Transposase enzyme staggers cuts to leaves single stranded overhangs.
Transposable element inserts itself in between these overhangs but there’s still gaps.
Gaps repaired by DNA polymerase.
What are the two main categories of transposable elements?
DNA transposons – move as DNA sequences.
Replicative: “Copy & Paste”
Non-replicative: “Cut & Paste”
Retrotransposons – composed of DNA, but transpose via an RNA intermediate.
DNA → RNA → reverse transcribed into DNA
Reverse transcribed DNA inserts into new location
Uses reverse transcriptase (same enzyme as retroviruses).
How do retrotransposons transpose?
DNA → RNA
RNA is reverse transcribed into DNA
New DNA copy inserts into a new location
Why are TGEs mutagenic?
They can insert into crucial genetic regions (coding sequences, promoters, etc.), disrupting gene function.
Can cause diseases and mutations in various organisms.
Give an example of TGE-induced mutations in humans, plants, and flies.
Humans: Hemophilia A, Coffin-Lowry syndrome
Plants: Round vs wrinkled pea phenotype (Mendel’s study)
Drosophila melanogaster: P-elements introduced around 1960, rapidly spread
What are P-elements in Drosophila?
A type of transposable element found in fruit flies, used for genetic engineering before CRISPR.
Utilized in a technique to generate transgenic flies (before CRISPR).
How were P-elements used in biotechnology to generate transgenic flies?
Clone gene of interest into a plasmid with inverted repeats characteristic of TGF.
Inject embryo with plasmid + transposase enzyme.
Gene of interest randomly inserts into the fly genome.
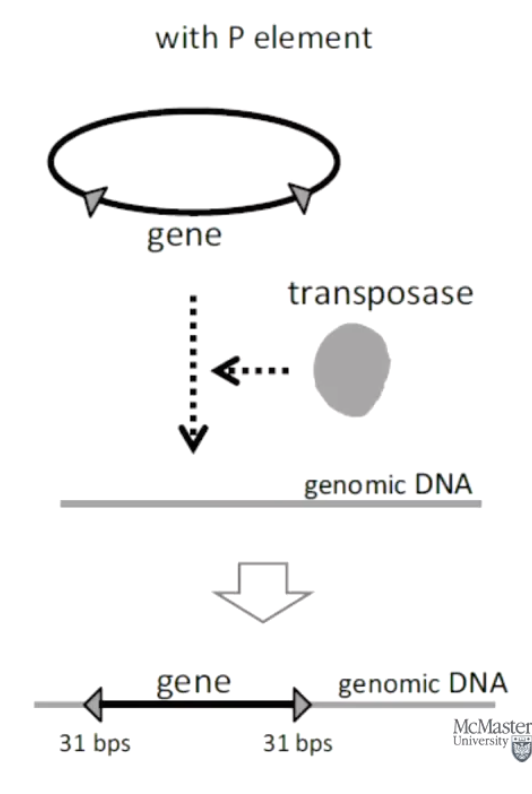
What is epigenetics?
The study of traits above inheritance; it does not change DNA sequences but affects gene expression.
What evidence suggests epigenetic modifications can be inherited? And why?
In mice, a modified agouti gene causes yellow coat color and obesity, but feeding pregnant females a specific diet led to wild-type offspring despite inheriting the agouti gene.
The modified diet was rich in methyl factors. This lead to increased methylation and silencing of the modified agouti gene.
What did studies on Dutch famine survivors reveal about epigenetics? And why?
Those born during the famine had a higher risk of heart disease, diabetes, and obesity compared to siblings born after the famine.
IGF2 gene less methylated in citizens born during the famine.
Siblings in the same families born after the famine have higher methylation of IGF2
What are the five features of epigenetic modifications?
Alter chromatin structure
Transmissible during cell division
Reversible
Directly impact gene transcription
Do not alter DNA sequence
Difference in chromatin structure (2)?
Euchromatin: Loosely packed, transcriptionally active.
Heterochromatin: Densely packed, transcriptionally inactive.
Types of heterochromatin (2)?
Constitutive: Always condensed and inactive.
Facultative: Can switch between active and inactive states.
What are position effects in gene expression?
Occur when a gene’s expression changes depending on its location in the genome. Some regions are “hot” (high expression) or “cold” (low expression). This is important when inserting transgenes to ensure they land in an active region for proper expression.
What is a nucleosome?
A structure of DNA wrapped around 8 histone proteins (H2A-H2B dimers + H3-H4 tetramers), condensing DNA into chromatin. Explains how chromatin can be compact (heterochromatin) vs less compact (euchromatin).
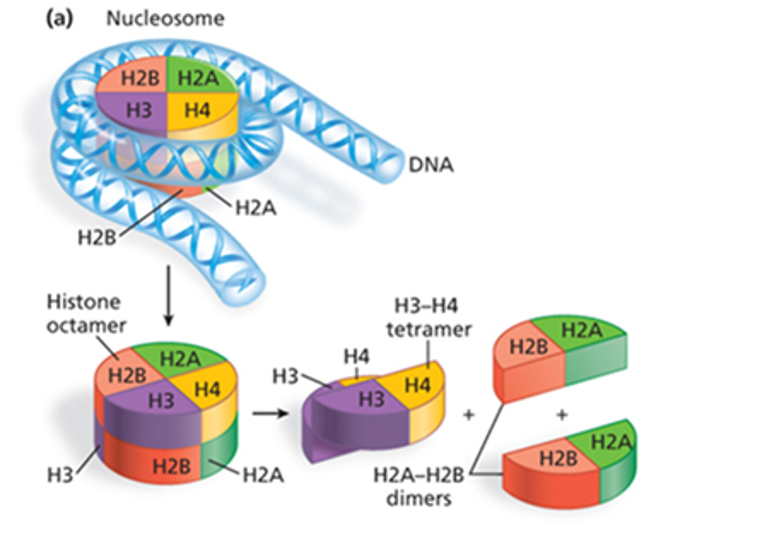
How can chromatin structure be modified? (2)
Acetylation
Methylation
How does acetylation affect chromatin?
Relaxes DNA-histone interaction by neutralizing positively charged histones, making chromatin more accessible for transcription.
HATs (Histone Acetyltransferases): Add acetyl groups → leads to euchromatin.
HDACs (Histone Deacetylases): Remove acetyl groups → leads to heterochromatin.
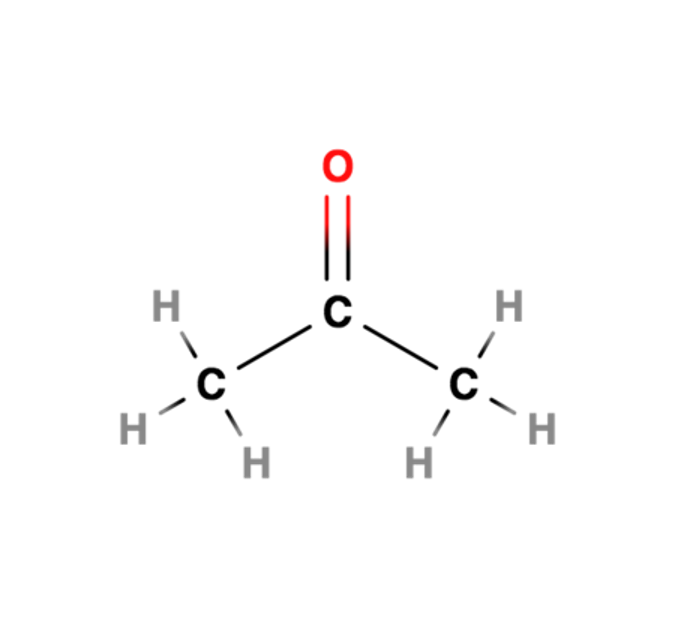
How does methylation affect chromatin?
Usually leads to heterochromatin but can also activate euchromatin.
HMTs (Histone Methyltransferases): Add methyl groups.
HDMTs (Histone Demethylases): Remove methyl groups.
What is Position Effect Variegation (PEV)?
The silencing of genes when heterochromatic regions spread into euchromatin.
What discovery in Drosophila led to the study of PEV?
An X-chromosome inversion placed the white gene (mutant phenotype) near the centromere in a heterochromatin region, leading to a mosaic eye color pattern (yet flies are genotypical wild-type).
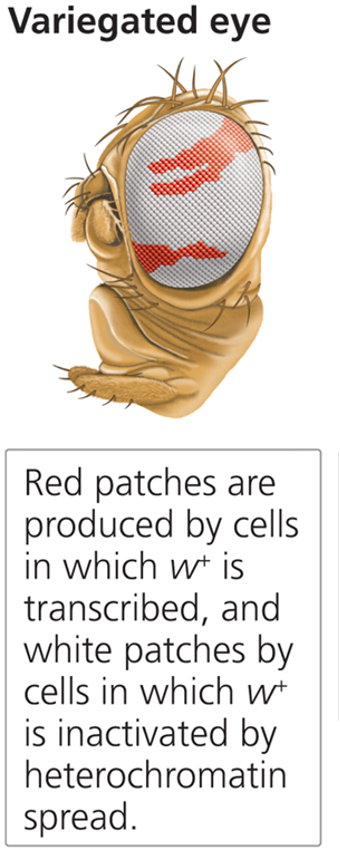
What does the white gene do in Drosophila?
Codes for a pigment transporter necessary for red eye color. When expressed in a euchromatic region, flies have red eyes. If silenced in a heterochromatic region, some cells lack pigment, leading to white patches (variegated phenotype).
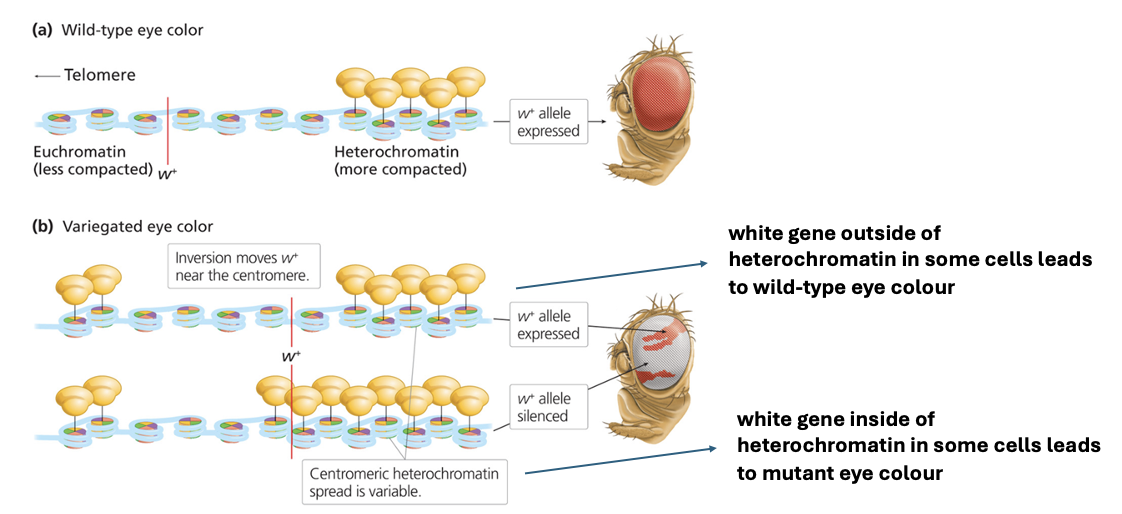
PEV and Chromatin Structure - How does epigenetics influence the variegated eye phenotype in Drosophila?
Wild-type: white gene in euchromatic region → red eye
Variegated eye phenotype: white gene in heterochromatic region → mosaic
Some cells completely silence the gene, leading to a mutant (white) eye color, while others allow partial expression, resulting in a red-and-white mosaic pattern.
This variation in gene expression due to chromatin structure is an example of Position Effect Variegation (PEV).
Mutations that affect chromatin (3)
Variegated eye
E(var) mutations
Sur(var) mutations
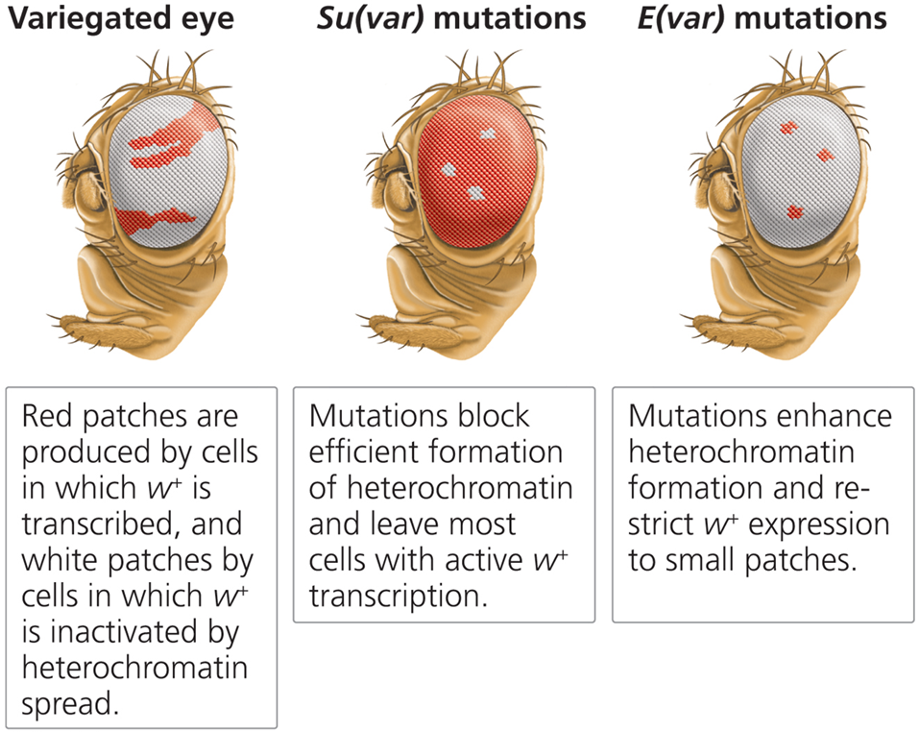
What are E(var) mutations?
Enhancers of position effect variegation that encourage heterochromatin spread, enhancing mutant phenotypes.

What are Sur(var) mutations?
Suppressors of position effect variegation that restrict heterochromatin spread, promoting wild-type phenotypes.
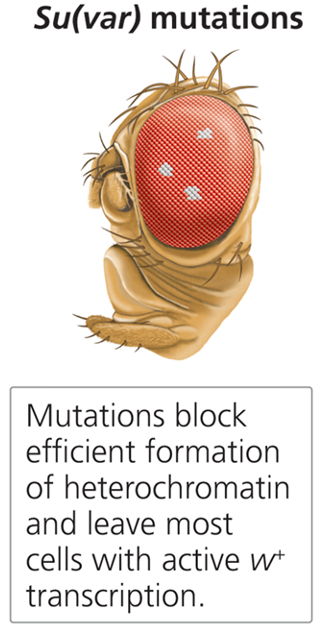
Which of the following mutations would not be an E(var) mutation?
a) A loss of function mutation in HAT genes
b) A loss of function mutation in HMT genes
c) A loss of function mutation in HDAC genes
c) A loss of function mutation in HDAC genes.
A mutation causing HDAC to not function would lead to less heterochromatin, and thus, would not be an E(var) mutation.
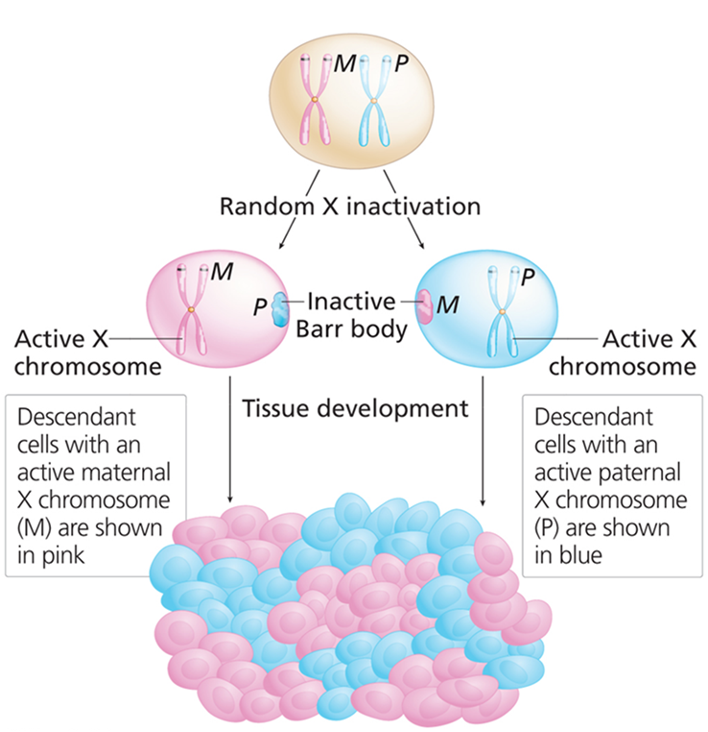
How is mosaic expression in Drosophila eyes similar to X-inactivation in female mammals?
Either the maternal or paternal X chromosome is randomly inactivated in each cell, creating a mosaic effect. Both are considered epigenetic phenomenon.
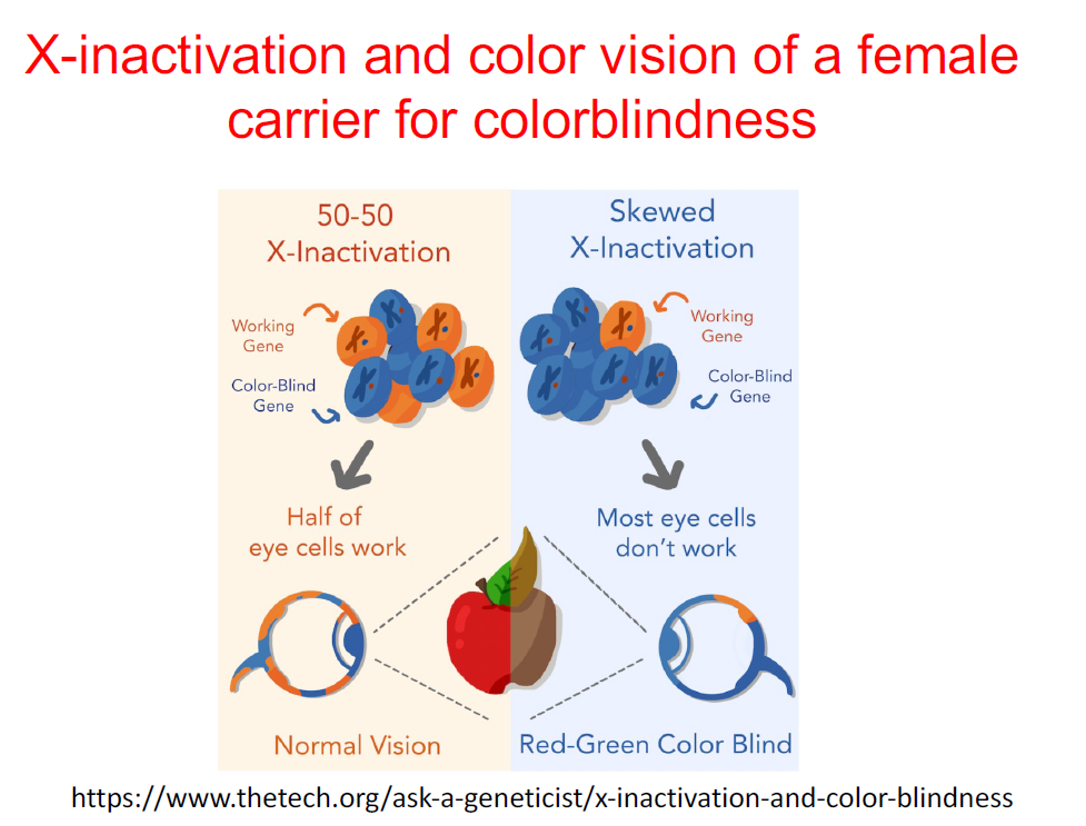
How can X-inactivation lead to partial color blindness in females?
Can randomly silence one X chromosome in each cell. If more eye cells inactivate the wild-type allele, most cells will express the color-blind allele, leading to red-green color blindness. However, this is rare since having even half of the eye cells expressing the wild-type allele is usually enough for normal vision.
How is X-inactivation controlled?
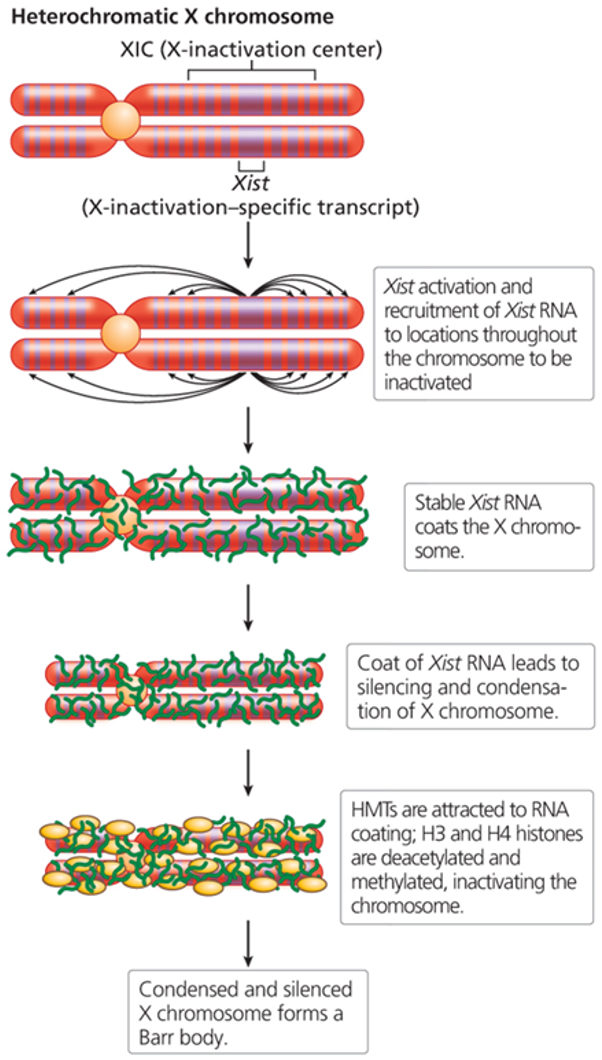
By the X-inactivation-specific transcript (Xist), a long noncoding RNA (lncRNAs).
What are long noncoding RNAs (lncRNAs)?
Long RNA molecules that lack open reading frames.
Regulate gene expression in eukaryotic cells.
Thought to act as scaffolds that link to regulatory proteins, affecting chromatin structure
How do lncRNAs relate to X-inactivation?
Play key role by spreading along the X chromosome and silencing most of its genes, turning it into heterochromatin.
Where have lncRNAs been studied?
In stem cells of mice embryos, where they help regulate gene expression and chromatin structure.
Where is the Xist gene located?
Found in the X-inactivation center (XIC) on the X chromosome.
On which type of X chromosome is Xist active? inactive?
Active on heterochromatic X chromosome
Inactive on euchromatic X chromosome
What does Xist RNA do during X-inactivation?
Produced on the X chromosome that will be inactivated. It spreads along the chromosome and silences almost all its genes. HMTs attracted to RNA coating, H3 and H4 histones are deacetylated and methylation, inactivating the chromosome.
What is genomic imprinting?
An epigenetic phenomenon where gene expression depends on whether the allele was inherited from the mother or father.
Can affect X-linked or autosomal alleles.
When offspring produce their own gametes, the previous imprinting is erased to ensure their gametes are imprinted correctly for the next generation.
What happens in maternal imprinting?
The mother’s allele/chromosome is inactivated, and only the father’s allele is expressed.
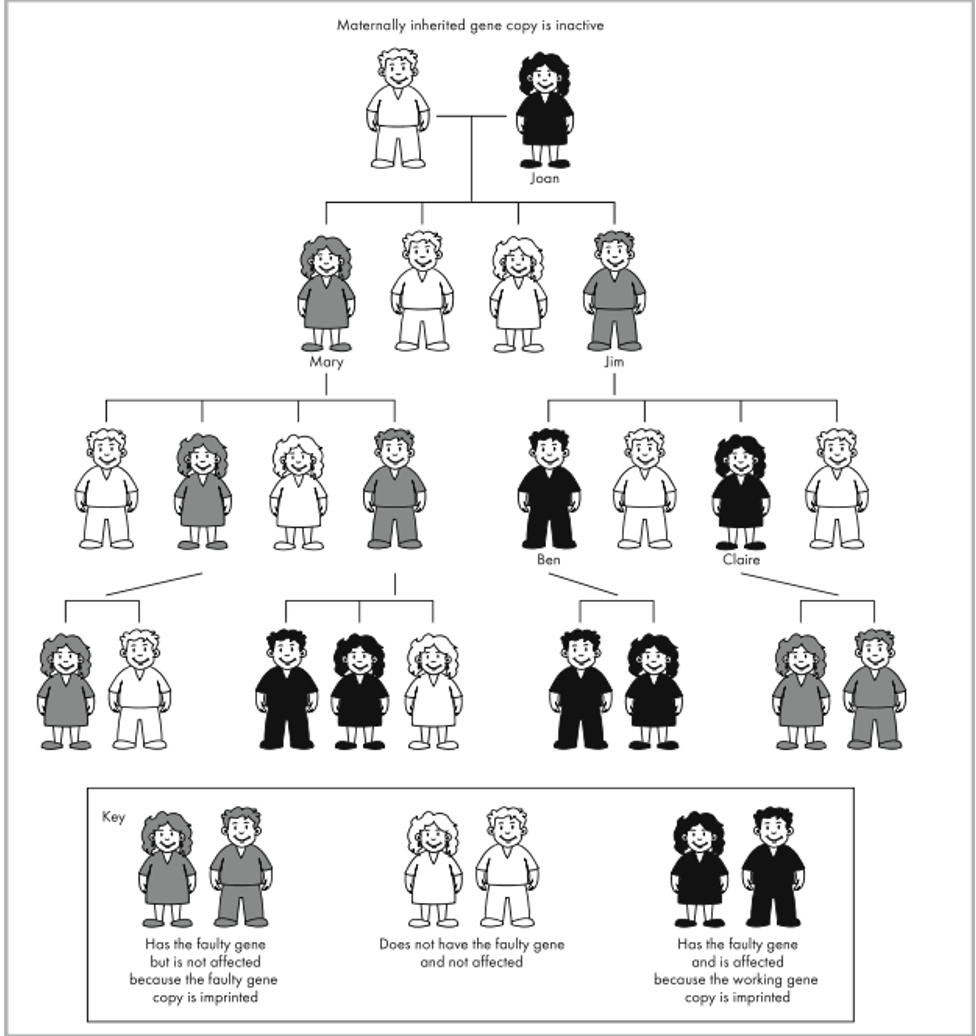
How does maternal imprinting affect disease inheritance?
Only females switch off (silence) the allele when passing it on. This means:
Affected mothers (with the disease allele) can have unaffected children if the allele is imprinted (switched off).
Males do not imprint the allele, so ONLY affected or carrier males can pass on the disease.
Example: Joan (affected) has no affected children, but her carrier son Jim can have affected children.
What happens in paternal imprinting?
The father’s allele/chromosome is inactivated, and only the mother’s allele is expressed.