Part 2.1: Transport across ER/IM Membrane
1/71
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
72 Terms
Microsomes
small vesicles produced upon breaking the ER by sonication
used to help explain the molecular aspect of ER transport
Signal Sequences in 1970s
signal sequences were discovered in the 1970s
the mRNA encoding a secreted protein was translated by ribosomes in vitro
w/o microsomes, the protein synthesized was slightly larger than the secreted protein
with microsomes the protein synthesized was slightly shorter
Blobel/Signal Hypothesis
he discovered an N-terminal signal that governs protein targeting to the ER and discovered the ER transport
signal hypothesis: there is a signal sequence that directs a protein to the ER membrane; when protein is transported, the signal sequence is then cleaved off
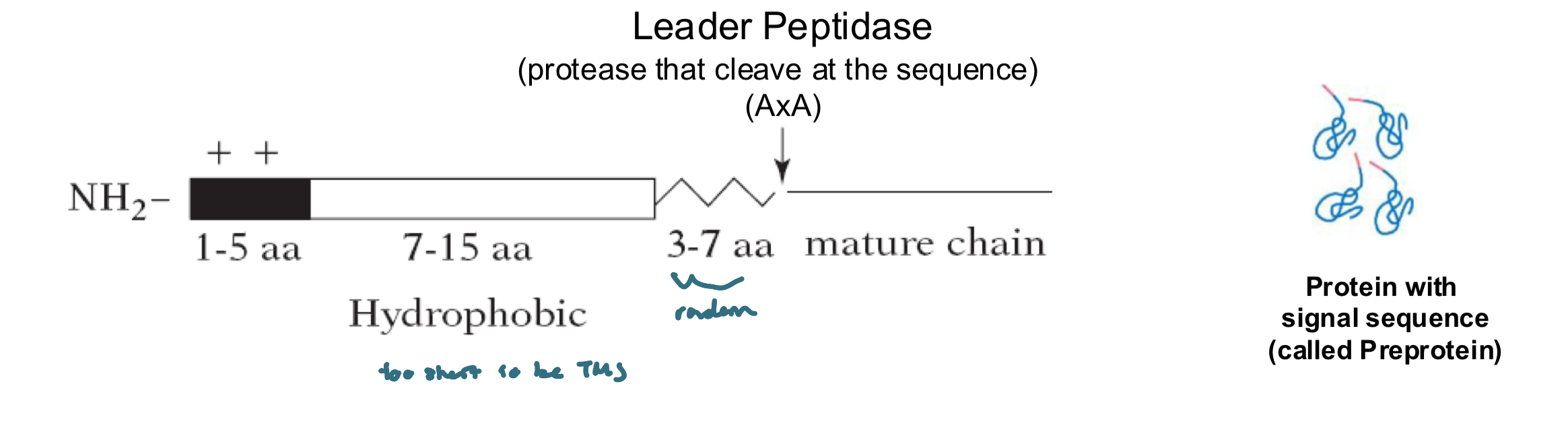
Signal Sequence
a short N-terminal a.a sequence, typically Signal Sequence~20-25 residues
placing the N-terminal ER signal on a cytosolic protein redirects the protein to the ER
the sequence varies in a.a, but each:
has 7-15 nonpolar a.a’s at its center
a + charged N-terminal
a peptidase cleavage site located 3-7 a.a after the hydrophobic sequence
Signal Sequencethere is no consensus sequence, the physical properties matter more than the exact a.a sequence
the signal sequences of all proteins having the same destination are interchageable
Signal Sequence Recognition
signal sequences are recognized by complementary receptors, called the translocons
signal peptidases remove the signal sequence during or soon after the transport process is complete
signal peptidases are located on the trans side of the membrane
Precursor vs Mature Protein
P (precursor): the full length protein with its signal peptide still attached
M (mature): the processed protein inside the microsome, and the signal peptide is removed by signal peptidase
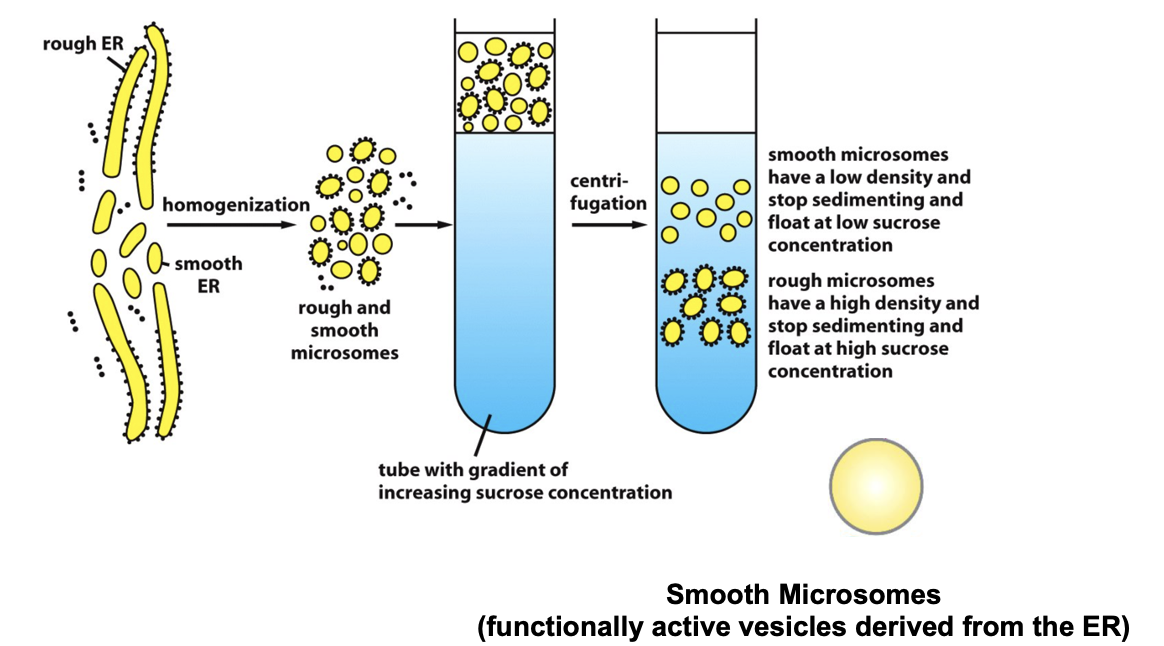
Blobel’s Experimental Goal: Recreate ER Transport in vitro
cells were broken open (homogenized)
the ER fragments reseal into microsomes (~200nm)
microsomes are relatively easy to purify by equilibrium sedimentation on sucrose gradient
microsomes behave and function like mini-ERs:
translocation, protein glycosylation, Ca2+ uptake and release, lipid synthesis
the interior of the microsome is equivalent to the internal space of ER (lumen)
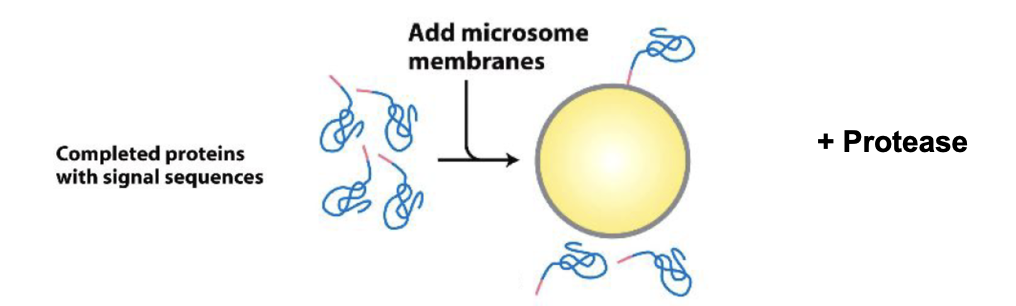
The Experiment Didn’t Work
proteins were synthesized in vitro using ribosomes and radioactive methionine (35S Met), allowing the proteins to show up on film after SDS-PAGE
then microsomes were added after the proteins were already made
Result: no transport occurred, proteins with signal sequences stayed outside the microsomes
Why? Because ER transport must occur during translation, not after
the ribosome needs to be attached to the microsome membrane while the protein is being synthesized, allowing the nascent chain to be threaded directly into the lumen
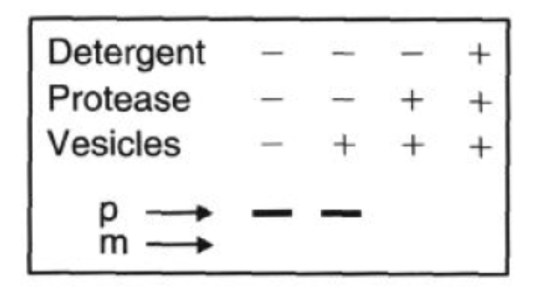
SDS PAGE of the Experiment that didn’t work
Protease: added at the end, digests proteins that are outside the microsome
Detergent: breaks open microsomes, exposing the inside. Now everything can be digested
SDS-PAGE shows which radiolabeled proteins are protected (inside) vs digested (outside)
smaller mature (M) band means the protein entered the microsome and had its signal peptide cleaved
microsomes present during translation or protease only
larger precursor (P) bands means the protein stayed outside
when microsomes added after translation
SDS PAGE of the Experiment that did work
when microsomes are added during protein synthesis, the proteins bearing the signal sequence are transported into the microsomes
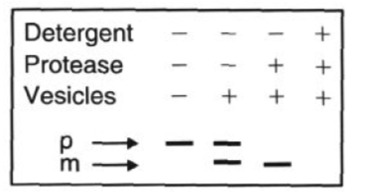
Microsomes Added during Protein Synthesis Figure
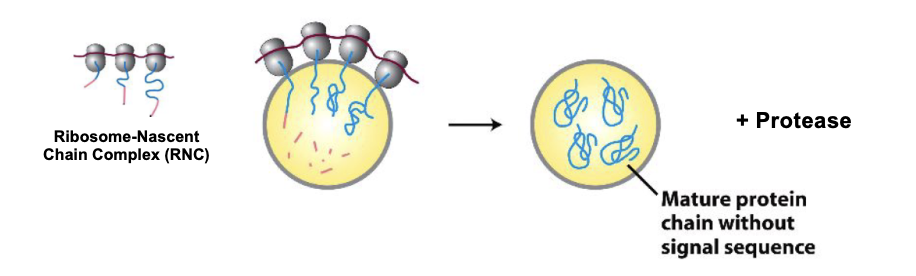
Signal Recognition Particle (SRP): Structure
an elongated particle made by 6 different polypeptides bound to a small RNA molecule (ribonucleoprotein)
the elongated RNA acts like a scaffold to organize SRP proteins
one end interacts with the ribosome a.a. entry door, the other interacts with the emerging signal sequence
the signal sequence binding site is a hydrophobic pocket lined by methionine residues
Methionine’s flexible side chains provide plasticity, allowing SRP to recognize many different hydrophobic signal sequences.
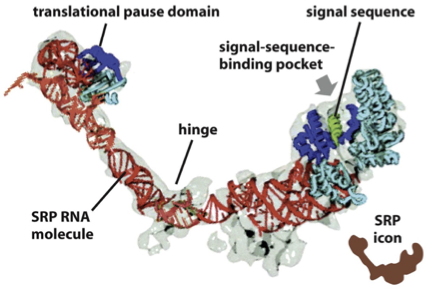
SRP: Function
SRP binds to the emerging signal sequence and blocks entry of a.a’s with their tRNA, into the ribosome
∴, SRP blocks the translation of the protein until the ribosome binds to the ER membrane
SRP brings the RNC complex to the ER membrane (spatial effect) by binding the SRP receptor on the membrane
SRP coordinates protein translocation (temporal effect)
pauses translation temporarily after binding the signal sequence, and resumes only after the ribosome binds the SRP receptor and docks to the translocon
SRP directs RNC (ribosome-nascent chain) complex to SRP membrane receptor
SRP: Function FIGURE
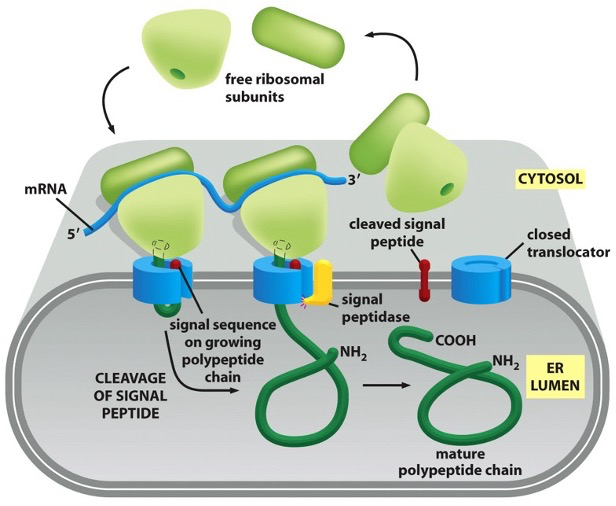
Early Model for Co-Translational Translocation
the ribosome docks on to the ER membrane, and as it makes the protein, it injects the growing chain directly into the ER lumen
the signal peptide at the start of the protein tells it to go to the ER
once inside, the signal peptide is cut off by a signal peptidase (enzyme on lumenal side)
the rest of the protein folds inside the ER lumen
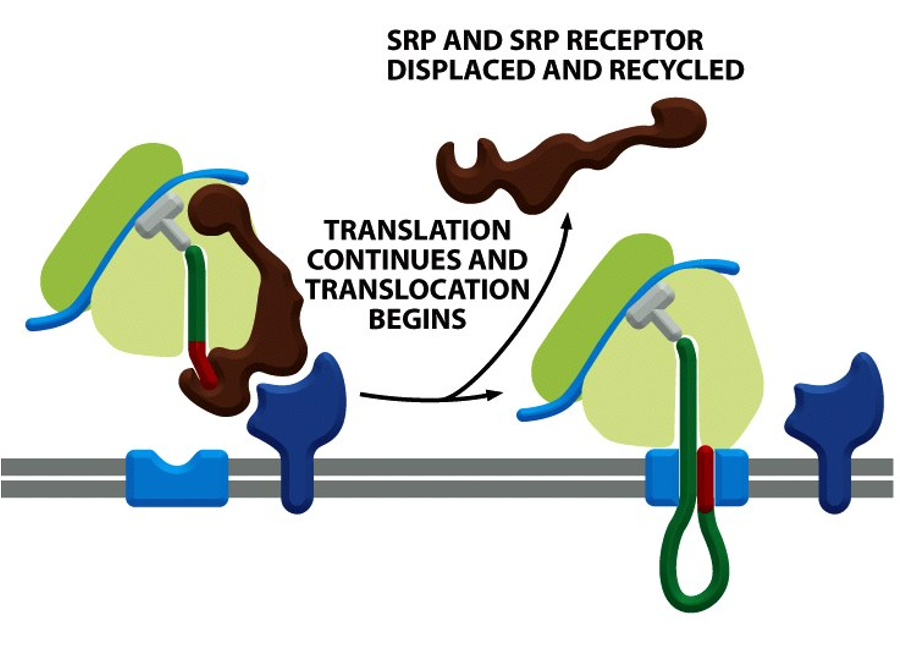
SRP: Continuation of Translation
once the SRP–RNC complex docks onto the SRP receptor on the ER membrane, SRP and SRP receptor are dissociated, releasing the ribosome to transfer to the translocon
translation resumes, and the nascent polypeptide is threaded into the ER lumen thru the translocon channel
this sequence of events is crucial to prevent protein misfolding/aggregation/activation in the cytosol
dissociation is regulated by GTP
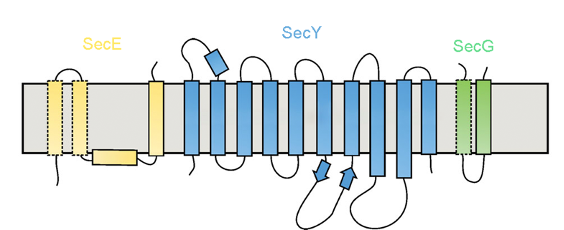
SecY/Sec61 Translocon
translocon is also called the Sec61 complex, made up of 3 subunits
the polypeptide chain is transferred thru the translocon (a membrane channel)
prokaryotes: SecY complex (Sec Y, SecE, SecG)
eukaryotes: Sec61 complex (Sec61⍺, Sec61β, Sec61γ), aka “protein conducting channel (PCC)”
archaea: SecYβ complex (SecY, Sec61β, SecG)
provides a pathway through the ER membrane for the growing chain, instead of being released into the cytosol
X-Ray Diffraction
Protein structure can be determined using X-ray diffraction
X-rays are electromagnetic radiation with a short wavelength
if a beam of X-rays is directed across a pure protein, most of the X-rays pass thru
a small fraction is scattered by the atoms in the sample
if the protein is well-ordered into a crystal, the scattered waves are well-defined
each spot in the diffraction pattern contains information about the locations of the atoms and can produce a complex 3D electron-density map
Translocon Structure
the pore is gated by a short helix(green) that keeps the channel closed
the pore opens when a polypeptide chain traverses the membrane
the pore ring is made of up 6 Ile’s (hydrophobic residues), that form a gasket around the polypeptide in transit
the structure allows for translocation and ion tight
hourglasss structure
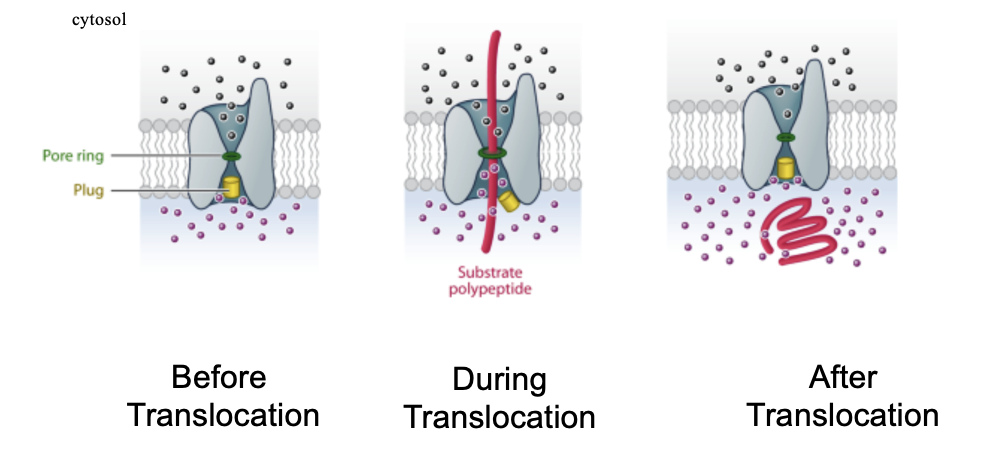
Translocon: Removal of the Plug
removal of the plug is lethal to the cell
the inner membrane loses its impermeability (molecules equilibrate across membrane) and the cell dies
Translocon Lateral Gate
SecY/Sec61α (largest subunit): forms the main pore for polypeptide translocation
SecE and Secβ/SecG (smaller subunits): stabilize the complex
Polypeptides pass through the central pore of SecY/Sec61
the plug helix blocks the pore when no polypeptide is present, preventing ion leakage.
lateral gate: SecY/61 channel can open sideways toward the lipid bilayer
allows signal sequences to enter the channel and TMS of membrane proteins to exit into the bilayer
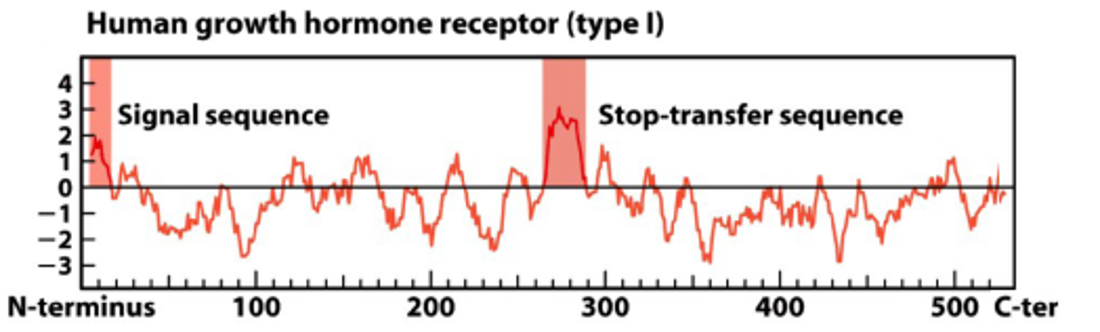
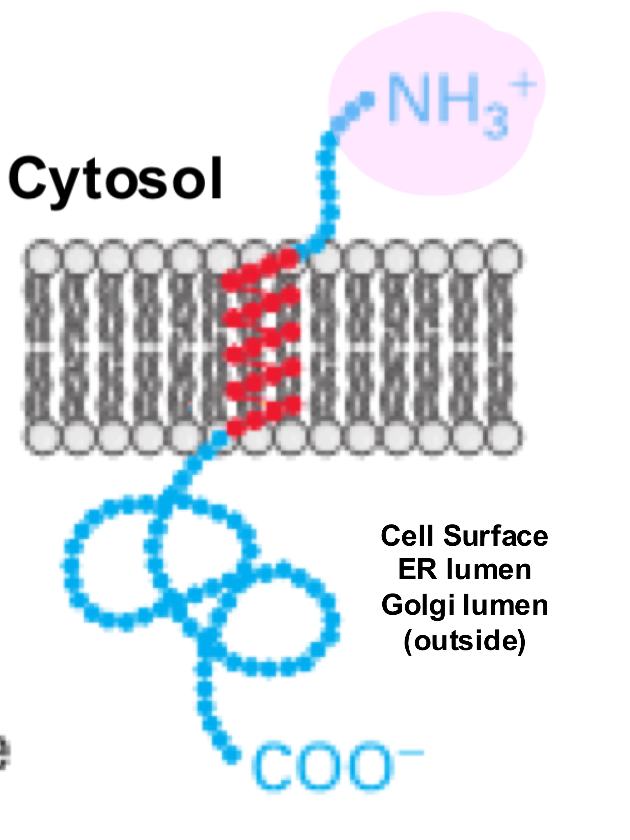
Membrane Protein Type 1
single-pass transmembrane protein with its N-terminus on the extracellular side and its C-terminus on the cytosolic side of the cell membrane
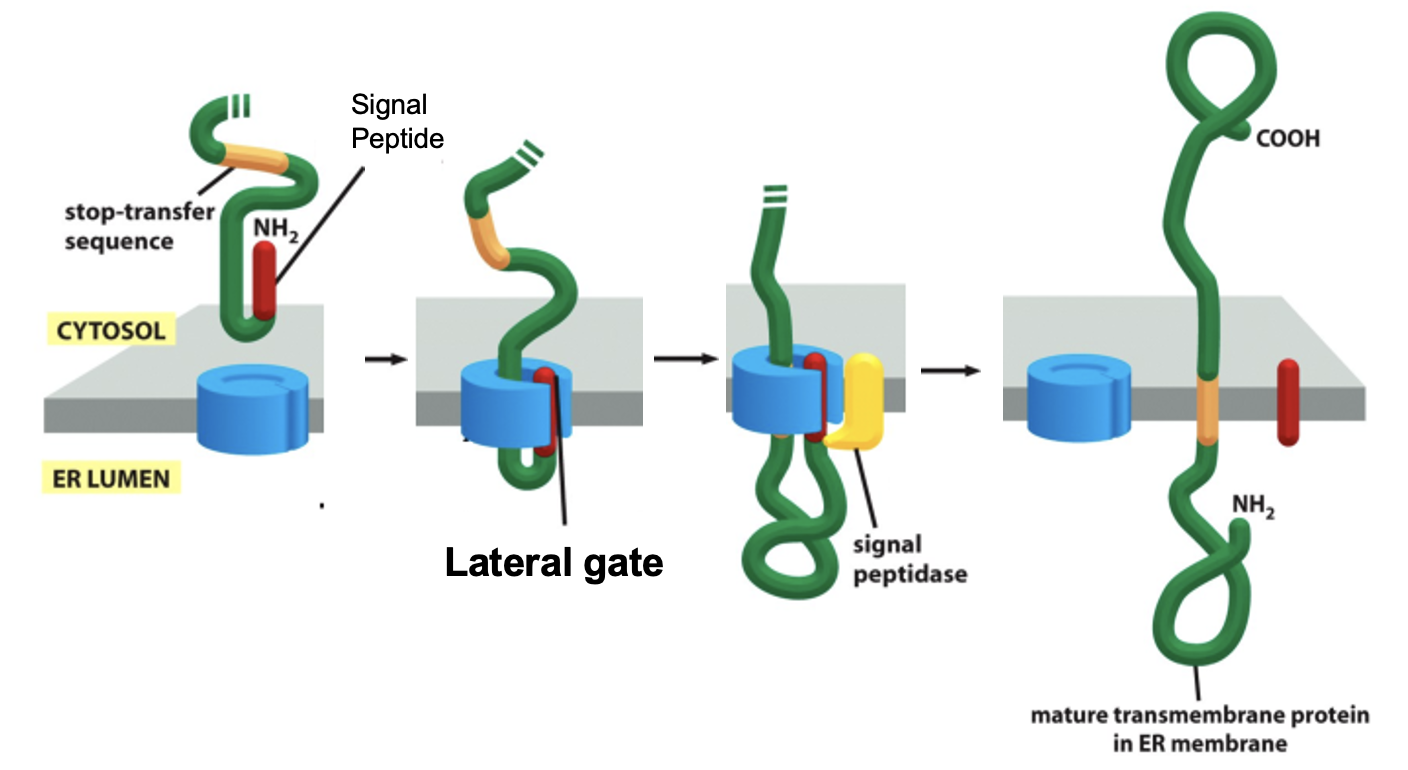
Insertion of a Single Pass Membrane Protein: Details
the hydrophobic sequence following the signal sequence (7-15 a.a’s) is the stop-transfer sequence
the stop-transfer sequence remains in the lipid bilayer as a membrane-spanning ⍺ helix (i.e becomes a TMS)
eg. glycophorin A
Insertion of a Single Pass Membrane Protein: Step 1
translation begins in the cytosol: ribosome synthesizes nascent polypeptide and the N-terminal signal sequence emerges first
Insertion of a Single Pass Membrane Protein: Step 2
SRP binds the signal sequence and pauses translation
Insertion of a Single Pass Membrane Protein: Step 3
SRP-RNC complex docks onto the SRP receptor on the ER membrane
Insertion of a Single Pass Membrane Protein: Step 4
SRP and SRP receptor dissociate and the ribosome is handed off the Sec61 (translocon), translation resumes
Insertion of a Single Pass Membrane Protein: Step 5
the N-terminal region is threaded thru the Sec61 channel via hairpin loop into the ER lumen
Insertion of a Single Pass Membrane Protein: Step 6
signal peptidase in the ER lumen cleaves the N-terminal signal sequence
Insertion of a Single Pass Membrane Protein: Step 7
the hydrophobic TMS (stop-transfer sequence) of the protein reaches the lateral gate of Sec61, halting further translocation of the polypeptide into the ER lumen
the lateral gate opens, allowing the TMS to exit sideways into the lipid bilayer
the rest of the protein is synthesizes in the cytosol
Insertion of a Single Pass Membrane Protein: Figure
Hydropathy Plot of Type 1 Membrane Protein
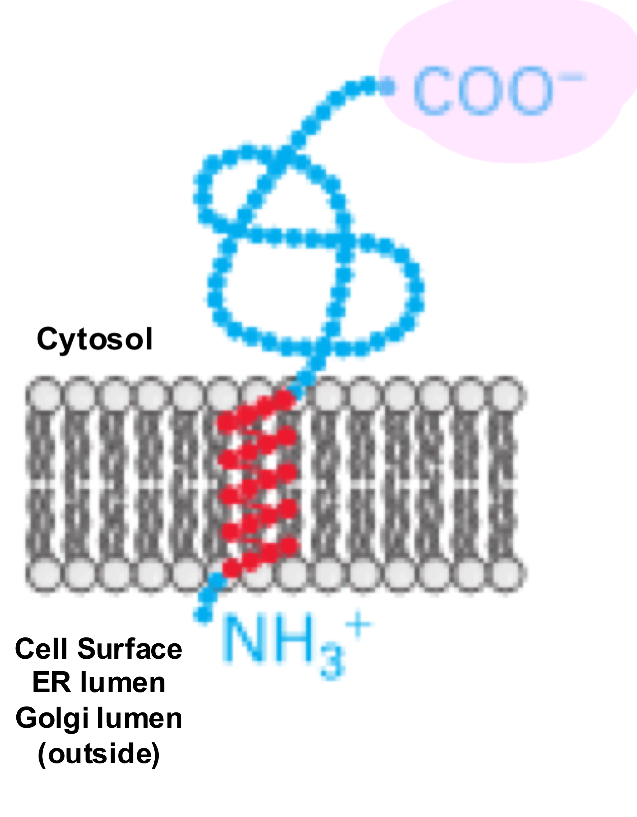
Membrane Protein Type 2
a single-pass transmembrane protein where the N-terminus is located on the cytosolic side of the membrane and the C-terminus is on the extracellular (or luminal) side
not made with a signal peptide
there is a signal anchor sequence (aka start transfer sequence) near the N-terminus of the protein, which is a TMS recognized by SRP
eg. transferrin receptor, Golgi galactosyltransferase
Insertion of Membrane Protein Type 2
translation begins in the cytosol
SRP binds to the emerging start transfer sequence and pauses translation
SRP-RNC complex docks to the SRP receptor on the ER membrane
SRP and receptor release after GTP hydrolysis and the ribosome is handed off to Sec61, translation resumes
the sequence is inserted into the channel as a hairpin loop
the charge distribution around the signal anchor determines orientation
positive-inside rule
therefore N-terminus stays in cytosol
the start-transfer sequence exits the translocon laterally thru the gate into the bilayer
the rest of the protein continues to be synthesized in the ER lumen
Insertion of Membrane Protein Type 2: Figure
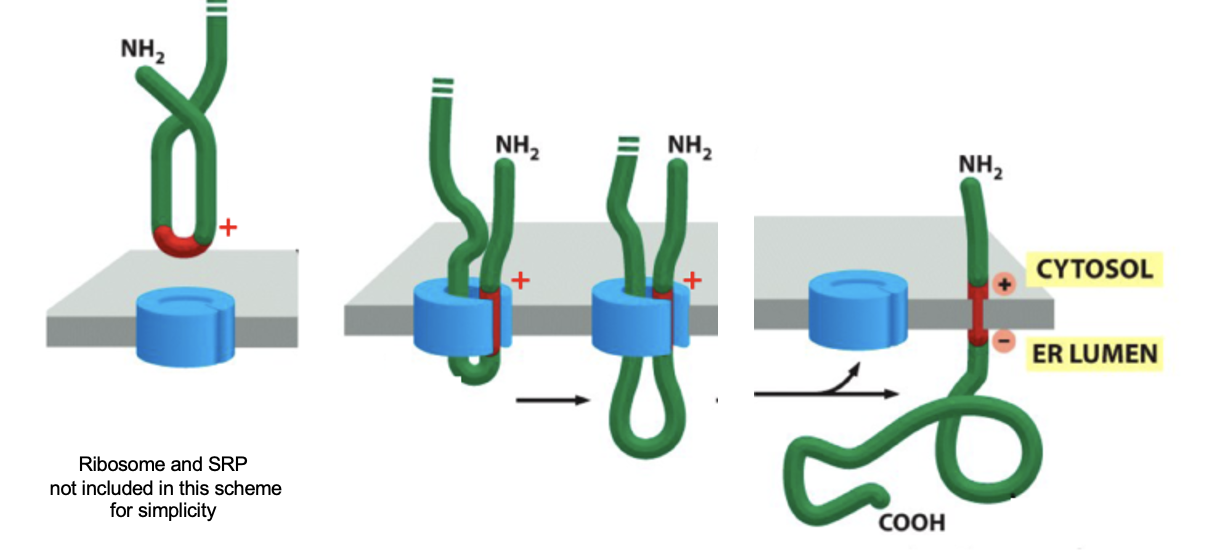
Positive-Inside Rule
+ charged residues (eg. Lys, Arg) on the cytosolic side interact with - charged phospholipid head groups of cytosolic leaflet of ER membrane
Possible explanations:
effect of - charged phospholipids located in the inner leaflet
in bacteria, the effect of the proton gradient (charge separation creating dipole across the membrane)
Membrane Protein Type 3
inserted using a "signal-anchor" sequence that results in the N-terminus on the extracellular side and the C-terminus on the cytosolic side,
lacks the cleavable N-terminal signal peptide
there is a signal anchor sequence (aka start transfer sequence) near the N-terminus of the protein, which is a TMS recognized by SRP
eg. cytochrome P450
Insertion of Membrane Protein Type 3: Details
two modes of insertion are possible: NH2 in (Type 2) or NH2 out (Type 3)
orientation depends on positive/negative a.a charge distribution around the start transfer sequence
+ charges tend to remain on the cytosolic side
- charges are present around the translocation channel
Insertion of Membrane Protein Type 3: Steps
translation begins in the cytosol and an internal, hydrophobic signal anchor sequence emerges. It acts as an ER targeting signal and membrane anchor
SRP recognizes and binds this hydrophobic region, translation is paused
SRP-RNC complex docks at SRP receptor on ER membrane
GTP hydrolysis releases SRP, translation resums
the C terminal of the start transfer sequence contains + charged residues, so this side stays in the cytosol
the hydrophobic signal anchor (TMS) enters the Sec61 channel then exits laterally into the bilayer via lateral gate
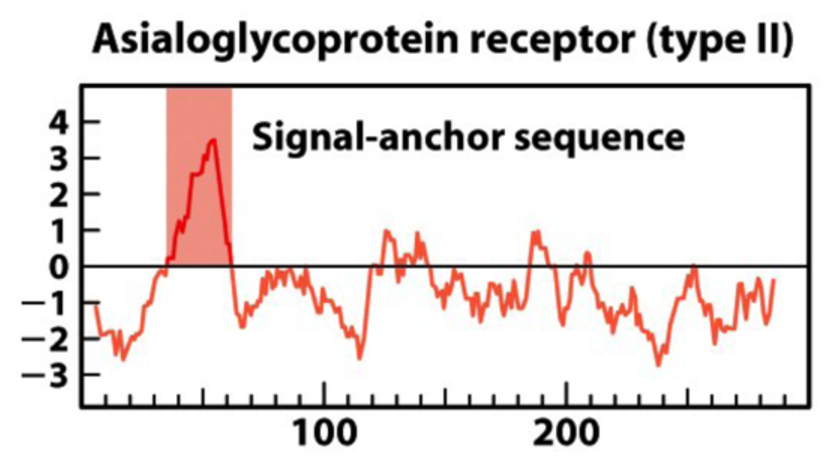
Hydropathy Plot of Membrane Type 2/3
Membrane Protein Type 4
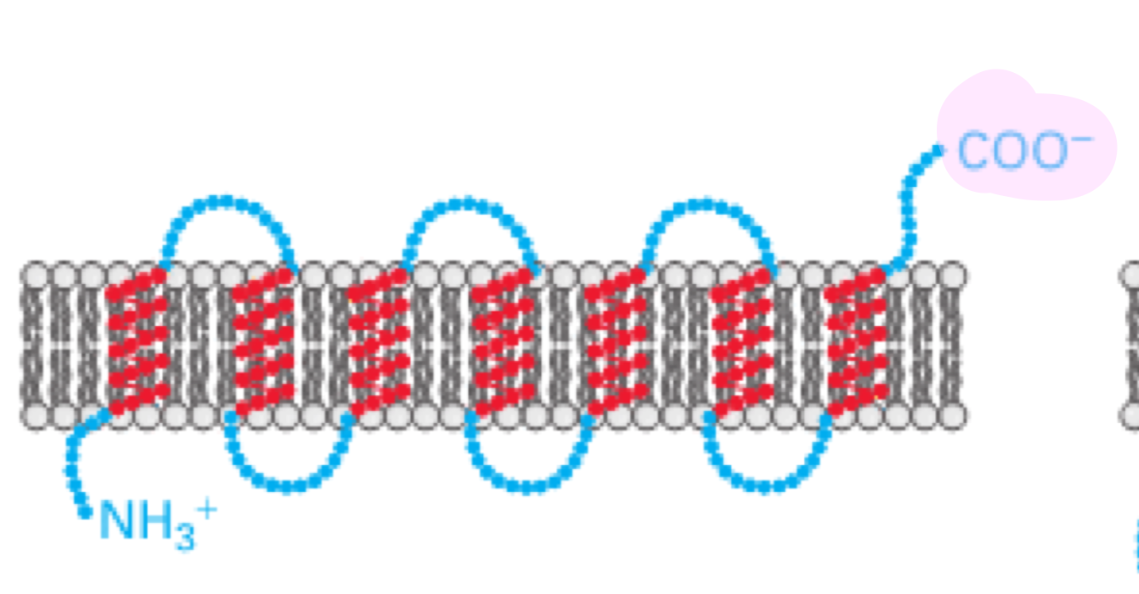
multi-pass transmembrane proteins
use several internal signal-anchor (start-transfer sequences) and stop-transfer sequences
the N-terminus and C-terminus orientation depend on the number of transmembrane segments
even # of TMS: N and C on same sides
odd # of TMS: N and C on opposite sides
the biogenesis of multi-pass membrane proteins depend on start-transfer and stop transfer sequences
start-transfer signals initiate translocation, which continues until reaching a stop-transfer sequence
subsequent start transfer sequences reinitiate translocation
SRP scans for the first hydrophobic segment that emerges from the ribosome
a similar scanning process continues until all the hydrophobic regions are inserted
Reporter Enzyme
fused to different parts of the protein being studied, typically the N-terminus or C-terminus
these enzymes have activity only if there are in a specific cellular compartment (eg. cytosolic vs. lumenal), so by seeing where the enzyme is active, researchers can tell which side of the membrane a part of the protein ended up on
Reporter Enzyme: Alkaline Phosphatase
alkaline phosphatase hydrolyzes a substrate called XP
XP is membrane impermeable
when alkaline phosphatase is transported to the periplasm, XP is hydrolyzed and bacterial colonies turns blue
when alkaline phosphatase remains in the cytosol, bacterial colonies are white
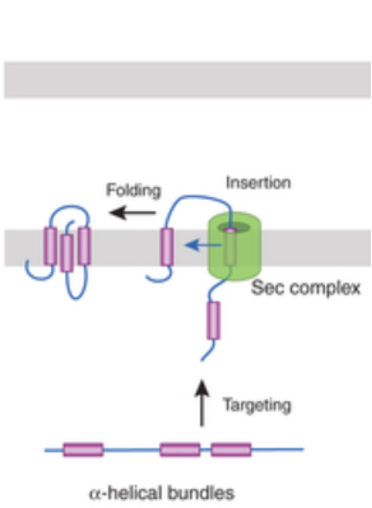
Membrane Protein Insertion in Bacteria: Step 1 - Targeting by SRP
Bacteria do not have ER, but still use SRP + Sec machinery to insert proteins into their inner membrane (IM)
proteins destined for the inner membrane usually contain hydrophobic ⍺-helical TMS
as these hydrophobic sequences emerge from the ribosome, bacterial SRP recognize them, pausing translation and directing the ribosome-nascent chain complex to the membrane
Membrane Protein Insertion in Bacteria: Step 2 - Docking at the Membrane
SRP brings the ribosome to its receptor in the bacterial inner membrane
this positions the growing polypeptide above the SecYEG translocon
Membrane Protein Insertion in Bacteria: Step 3 - Insertion and Folding
the ribosome continues translation
the nascent chain is threaded into the Sec translocon
hydrophobic helices exit laterally from SecYEG into the inner membrane, where they fold into their proper arrangement
final structure: ⍺-helical integral membrane protein
Membrane Protein Insertion in Bacteria Figure
Can a β-Strand serve as a Stop Transfer Sequence
no a single β-strand is not hydrophobic enough (alternating polar/non-polar residues), not long enough to span the membrane and not helical
a membrane spanning β-barrel required 8 - 22 β strands assembled together
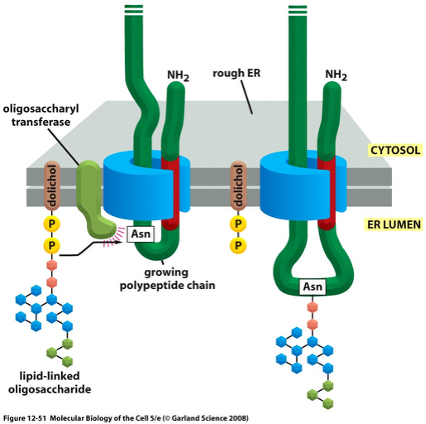
OST Complex
Oligosaccharide Transferase
nascent proteins enter the ER lumen unfolded, allowing lumenal chaperones (eg. BiP) to assist folding and assembly
BiP functions both as a chaperone and, in post translation translocation, as an ATP-driven pulling motor
about half of all eukaryotic proteins are glycosylated, but very few proteins in the cytosol are glycosylated because the OST is located in the ER lumen
OST transfers a preassembled 14-sugar oligosaccharide onto an Asn in an N-X/S motif (N-linked glycosylation)
a dolichol (lipid molecule) holds the preformed oligosaccharide in the ER, linked by high-energy pyrophosphate bond
10% of oligosaccharides are linked to the OH group of Ser or Thr
these O-linked oligosaccarides are formed in the Golgi apparatus
OST Complex Figure
Uses of Glycosylation
the carbohydrate layer protects cells against mechanical and chemical damage
the oligosaccharide side chains are enormously diverse in their arrangement of sugars
the diversity makes them important in specific cell-recognition processes
oligosaccharides are used as tags to mark the state of protein folding
glycosylation is required for proper folding
Calnexin/Calreticulin
two ER chaperons (calnexin/calreticulin) are carbohydrate binding proteins (aka lectins)
bind to oligosaccharides on incompletely folded proteins and retain them in the ER until the protein is properly folded
a protein enters the ER with 14-sugar N-linked oligosaccharide, this glycan has 3 terminal glucose residues
two glucoses are trimmed, calnexin/calreticulin recognize the one glucose form
calnexin binds the protein, holding it in the ER and preventing folding when bound
a β-glucosidase removes the final glucose, releasing the protein from calnexin
checkpoint: if the protein is correctly folded, it moves forward (export from ER), if it isn’t, it reenters the cycle
if unfolded, a new glucose is added
the cycle repeats until the protein folds correctly
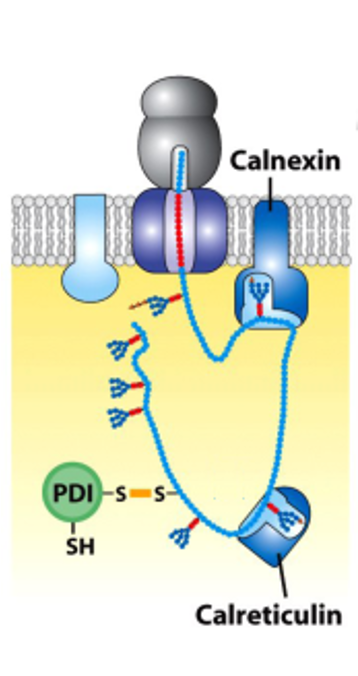
Protein Disulfide Isomerase (PDI)
an enzyme that catalyzes the formation, breakage and rearrangement of disulfide bonds in proteins (crucial for proper protein folding and quality control)
functions in the ER, but also has roles at the cell surface and in the cytosol
acts as both an enzyme and a chaperone (preventing misfolded proteins from aggregating and can assist in the folding process of nascent proteins)
Molecular Chaperones
Proteins that direct the correct folding and assembly of polypeptides w/o being components of the final structure
chaperones can maintain protein in an unfolded state (for transport into organelles or insertion into membranes, or prevention from aggregating while they fold later)
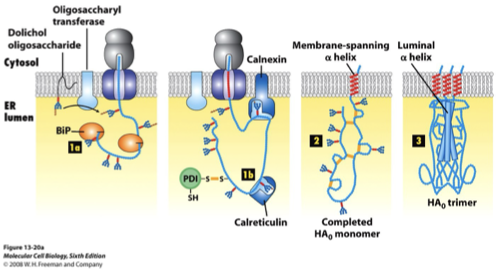
Protein Modifications during Transport FIGURE
glycosylation
disulphide bond formation
folding and oligomerization
Influenza Hemagglutinin (HA)
type of hemagglutinin found on the surface of influenza viruses
antigenic glycoprotein (able to trigger an immune response in the body)
responsible for binding the virus to the cell that is being infected
“hemagglutinin” comes from the protein’s ability to cause RBC’s to clump together in vitro
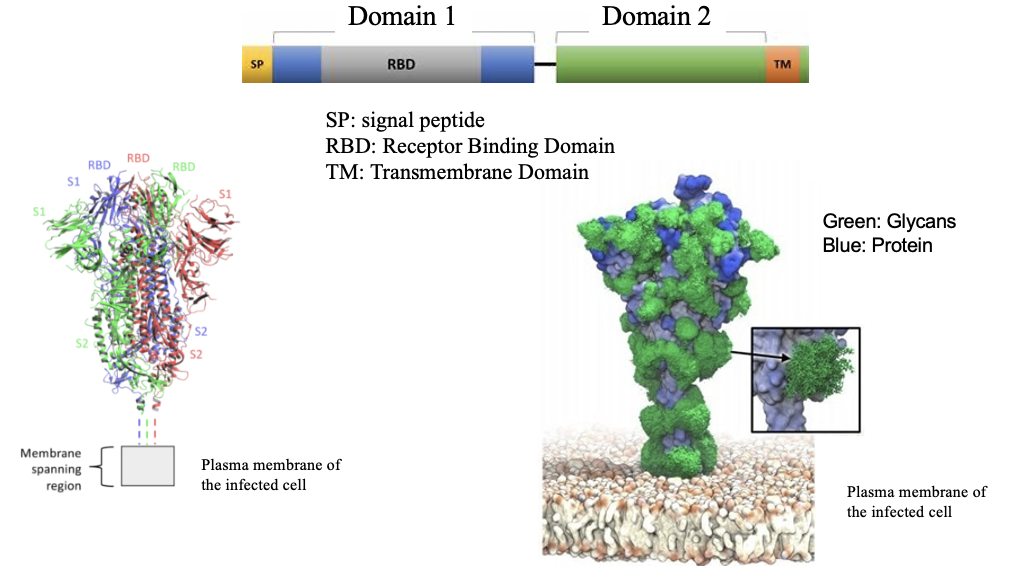
Sars-CoV-2 Spike Protein
a glycosylated membrane protein trimer processed in ER/Golgi
~22 N-linked glycans per monomer → ~66 per trimer
the glycans help Spike fold and shield it from antibodies by forming a glycan shield
are odded by OST in the ER, and then modified in the Golgi
made on ER membrane-bound ribosomes using:
an N-terminal signal peptide (targets it to the ER)
transmembrane domain (anchors it in the viral envelope)
glycosylation machinery (OST + Golgi enzymes)
trimerization requires proper folding, calnexin/calreticulin folding cycle, ER chaperones and correct disulfide bonds
Sars-CoV-2 Spike Protein Figure
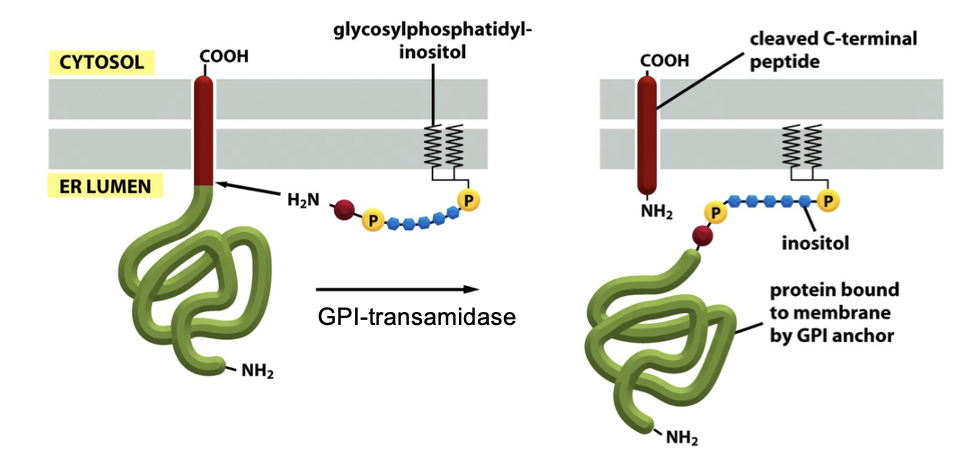
GPI-Linked Protein: GPI Anchor
some membrane proteins acquire a glycosylphosphatidylinositol (GPI) anchor
protein is initially made as a type 1 membrane protein, has an N-terminal signal peptide that targets it to the ER, and has a C-terminal transmembrane helix that would normally anchor it
GPI transamidase (ER protein) attaches a GPI anchor to the C-terminus of some membrane proteins that exits the translocon (occurs when the C-terminal TMS is cleaved off)
GPI-anchored proteins face outside the cell once delivered to the PM, and is anchored thru a lipid rather than a TMS
Example: trypanosome shed their coat of GPI-anchored surface proteins when attacked by the immune system
GPI-Linked Protein: GPI Anchor Advantages
release of the protein from the membrane is easy
eg. in response to signals that activate a membrane phospholipase
phosphatase easily cleaves this anchor
faster diffusion on the lipid bilayer than TMS
preferential localization of PM proteins into lipid rafts along with other proteins
the raft doesn’t anchor them, it’s just where they tend to cluster
GPI Anchor FIGURE
GPI Protein: Example - VSG Protein
Variable Surface Glycoprotein
the VSG protein forms a coating layer at the surface of the parasite
the parasite escapes the immune system by rapidly changing the VSG coat
a phosphatidylinositol-phospholipase C (PI-PLC) rapidly removes the VSG coat from the cell surface (hydrolyze the P-bond of the GPI anchor)
GPI Protein: Trypanosoma Brucei
Trypanosoma Brucei is covered with a dense coat of VSGs, hiding most of the parasite’s surface proteins (ion channels, transporters, receptors)
this means the host immune system can only target the exposed extracellular loops of VSG, but because VSG is the only “visible” antigen, the immune system attacks
the parasite escapes being killed by the immune system, by changing the VSG molecules it expresses (antigenic variation); enabled by:
genetic switching: the parasite has hundreds of VSG genes, and host antibodies specific to the old VSG become useless
rapid removal (shedding) of old coat, allowed by the GPI anchor which is cleaved quickly
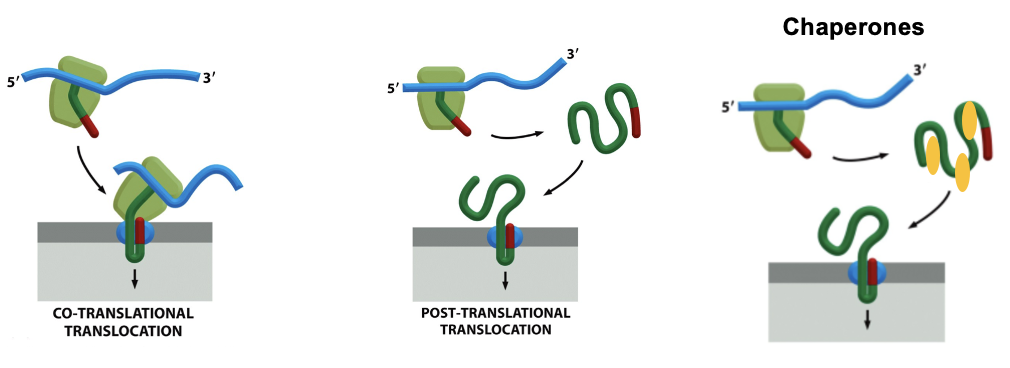
Two Modes of Protein Translocation
Proteins that need to enter the ER (eukaryotes) or periplasm (bacteria) must cross a membrane thru a translocon and there are 2 major ways they get thru that pore
Co-Translation Translocation: the protein is threaded into the translocon as it is being synthesized by the ribosome, the ribosome docks on the ER membrane (via SRP and SRP receptor) and the growing polypeptide enters the translocon immediately
used for secreted, membrane proteins and proteins targeted to ER lumen
Post-translational Translocation: the protein is first completely made in the cytosol and then targeted to the translocon. The entire finished chain must pass thru the channel
fully folded proteins cannot fit thru the translocation pore (too big)
cells use chaperones to keep the protein loosely folded or unfolded enough to pass
very common in yeast and bacteria
Two Modes of Protein Translocation FIGURE
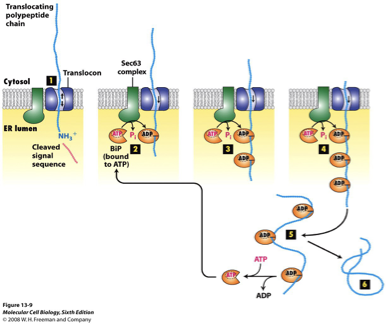
BiP
in post-translocation, translation is finished and there’s no ribosome pushing the chain through the ER
BiP is a molecular chaperone, grabbing the chain so it can move forward into the lumen and preventing it from sliding back out
BiP + Sec62/Sec63 is the engine that drives post-translational import into the ER
BiP Step 1
Targeting the finished polypeptide to Sec61/Sec62/Sec63
the polypeptide is synthesized in the cytosol, cytosolic chaperones keep it loosely unfolded
the polypeptide binds the Sec61/Sec62/Sec63 translocon at the ER membrane
cytosolic chaperones are released as the proteins begins entering the channel
BiP Step 2
BiP binds polypeptide (ATP → ADP switch)
BiP-ATP binds Sec63’s J-domain
following ATP hydrolysis, BiP tightly binds to the polypeptide substrate in its ADP state, preventing the polypeptide from sliding back into the cytosol
BiP Step 3
Repeated cycles pull the chain through
as the chain moves forward a little, another BiP-ATP binds, hydrolyzes ATP and grabs the new section
several BiP molecules bind sequentially, each grabbing further down the chain
this process is repeated until the polypeptide has completely traversed the channel
BiP Step 4
Nucleotide Exchange Release BiP
when the chain is fully inside the ER, bIP releases the chain when ADP is replaced with ATP
protein can now fold normally
BiP FIGURE
SecA
a processive enzyme, it catalyzes multiple consecutive reactions on a single substrate molecule without releasing it after each catalytic cycle (e.g. DNA/RNA polymerase)
in bacteria, the SecA ATPase pushes the protein across the SecY complex