BISC 202 DNA replication and transcription and translation (W11-12)
1/114
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
115 Terms
DNA structure
DNA is a double helix.
It consists of two antiparallel strands: one running 5' to 3' and the other 3' to 5'.
5' end vs 3' end of DNA
The 5' end has a phosphate group attached to the 5th carbon of the sugar.
The 3' end has a hydroxyl group (-OH) attached to the 3rd carbon of the sugar.
complementary base pairing in DNA
Adenine (A) pairs with Thymine (T) via 2 hydrogen bonds.
Cytosine (C) pairs with Guanine (G) via 3 hydrogen bonds.
DNA vs RNA
DNA contains deoxyribose sugar, RNA contains ribose.
DNA is double-stranded, RNA is single-stranded.
DNA uses thymine (T), RNA uses uracil (U) instead of thymine.
origin of replication
short stretch of DNA with a specific sequence of nucleotides that forms a "bubble"; where DNA replication begins
leading strand
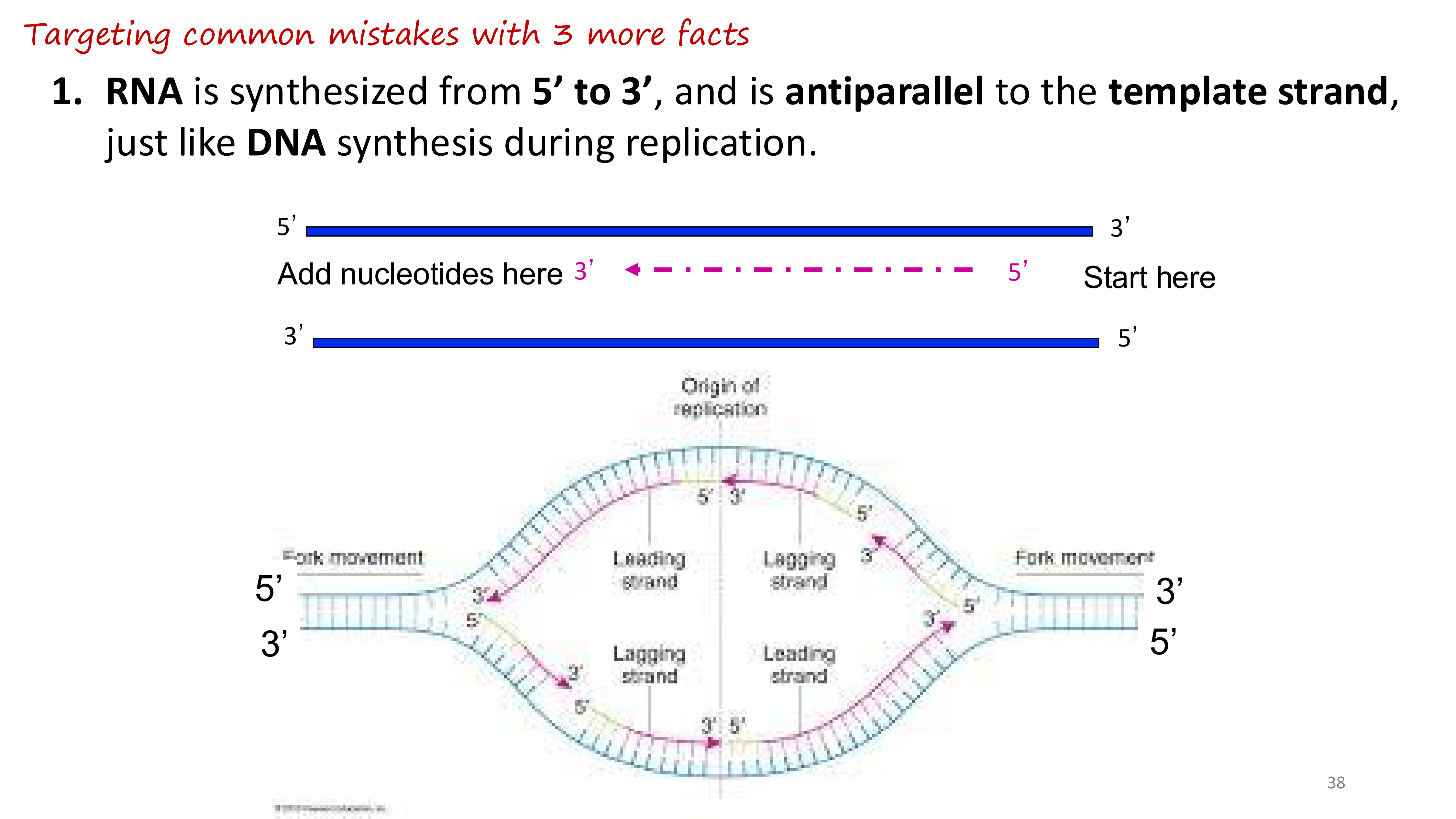
The leading strand is synthesized continuously in the 5' to 3' direction.
It occurs at the replication fork in the nucleus of eukaryotic cells.
lagging strand
The lagging strand is synthesized in short fragments called Okazaki fragments.
It is called discontinuous because these fragments are later joined together.
enzymes in DNA replication
• Helicase- unwinds parental double helix
• Single-strand binding proteins- stabilize separate strands and keep from re-pairing
• Primase- Makes RNA primer
• DNA polymerase I- replaces RNA primer
• DNA polymerase III- adds nucleotides in the 5' to 3' direction
• Ligase- joins Okazaki fragments and seals other nicks in sugar phosphate backbone
• Topoisomerase- relieves the strain
• Sliding clamp- holds DNA polymerase
what is the complementary DNA strand for 5'-ATGCGT-3'
3'-TACGCA-5'
how does DNA polymerase III proofread during replication?
DNA Polymerase III checks each newly added base for mistakes and removes incorrect nucleotides, ensuring accuracy in DNA replication
nuclease
an enzyme that can cut out segments left behind by DNA polymerase III
opening the helix
helicase, SSBPs, topoisomerase
helicase
catalyzes the breaking of hydrogen bonds between base pairs to open the double helix
single-strand DNA-binding proteins
stabilize single-stranded DNA
topoisomerase
breaks and rejoins the DNA double helix to relieve twisting forces caused by the opening of the helix
leading-strand synthesis
primase, DNA polymerase III, sliding clamp, DNA polymerase I
primase
catalyzes the synthesis of the RNA primer
DNA polymerase III (leading)
extends the leading strand
sliding clamp
holes DNA polymerase III and moves it in the required direction
DNA polymerase I
remove the RNA primer and replaces it with DNA
lagging-strand synthesis
primase, DNA polymerase III, sliding clamp, DNA polymerase I, DNA ligase
DNA polymerase III (lagging)
extends the lagging strand
DNA ligase
catalyzes the joining of Okazaki fragments into a continuous strand
DNA replication steps
1) Helicase- unwinds the parental double helix
2) DNA topoisomerase - breaks and rejoins the DNA double helix to relieve twisting forces caused by the opening of the helix
3) Single-strand binding proteins (SSBP) stabilize unwound DNA, aided by DNA gyrase.
4) Primase synthesizes a short RNA primer for DNA polymerase to bind to in the 5' to 3' direction to start replication on each strand.
5) DNA polymerase synthesizes the leading strand in 5' to 3' direction while the lagging strand is made discontinuously by primase making short pieces and then DNA polymerase extending these to make Okazaki fragments.
6) DNA ligase joins the Okazaki fragments together
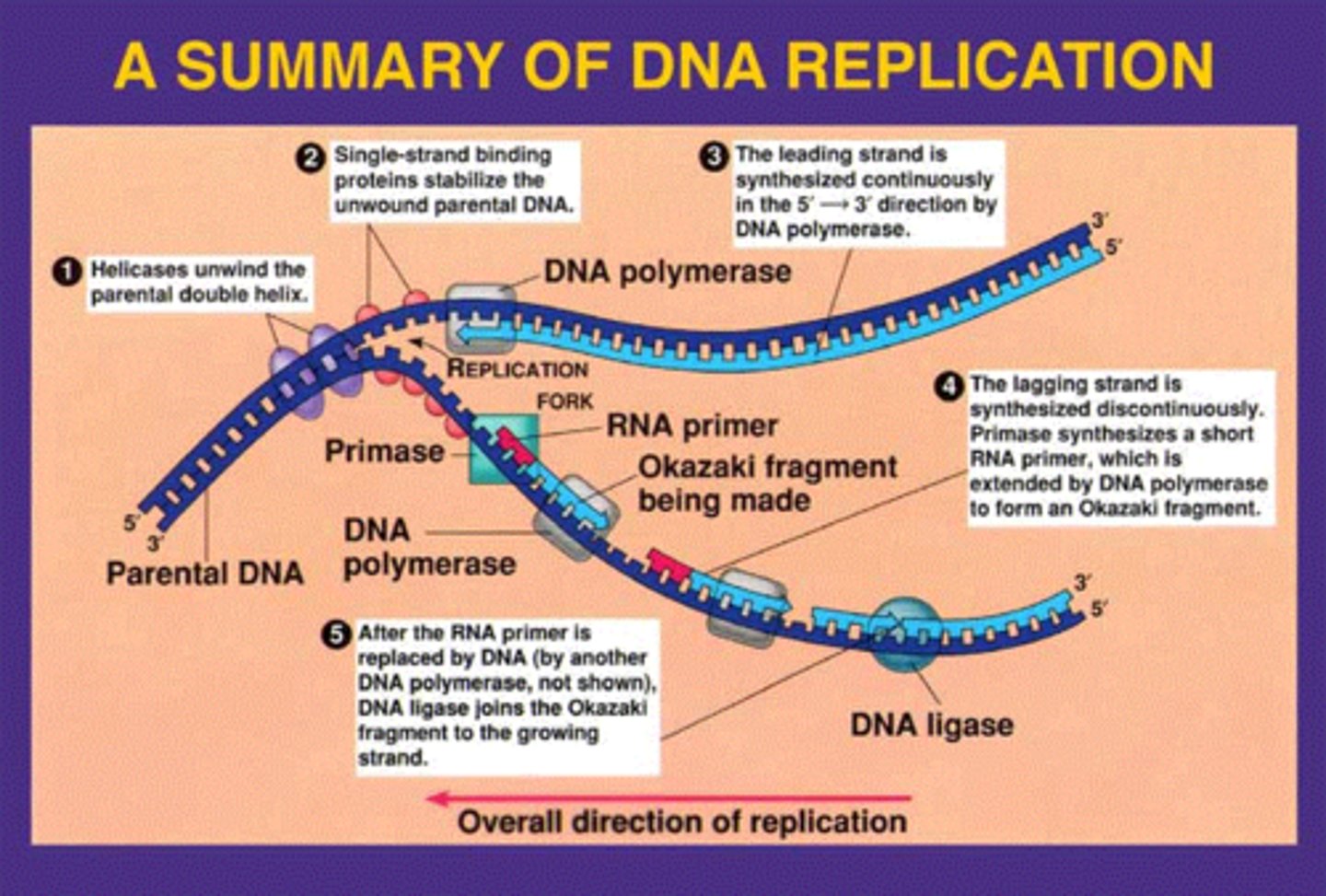
leading vs lagging strand
leading: one RNA primer, no Okazaki fragments, no DNA ligase
lagging: many RNA primers, Okazaki fragments, DNA ligase
both: helicase, ssbp's, topoisomerase, primase, DNA pol I and III, sliding clamp
what is the importance of the antiparallel nature of DNA?
In an antiparallel double helix, the two strands run in opposite directions (5' to 3' and 3' to 5'), which is important for replication and complementary base pairing.
for which type(s) of cells is DNA ligase needed, to join the leading strands to other DNA
both prokaryotic and eukaryotic cells
where will prokaryotes lose DNA during replication?
it will not lose any DNA
where will eukaryotes lose DNA during replication
at the 5’ ends of each new strand only
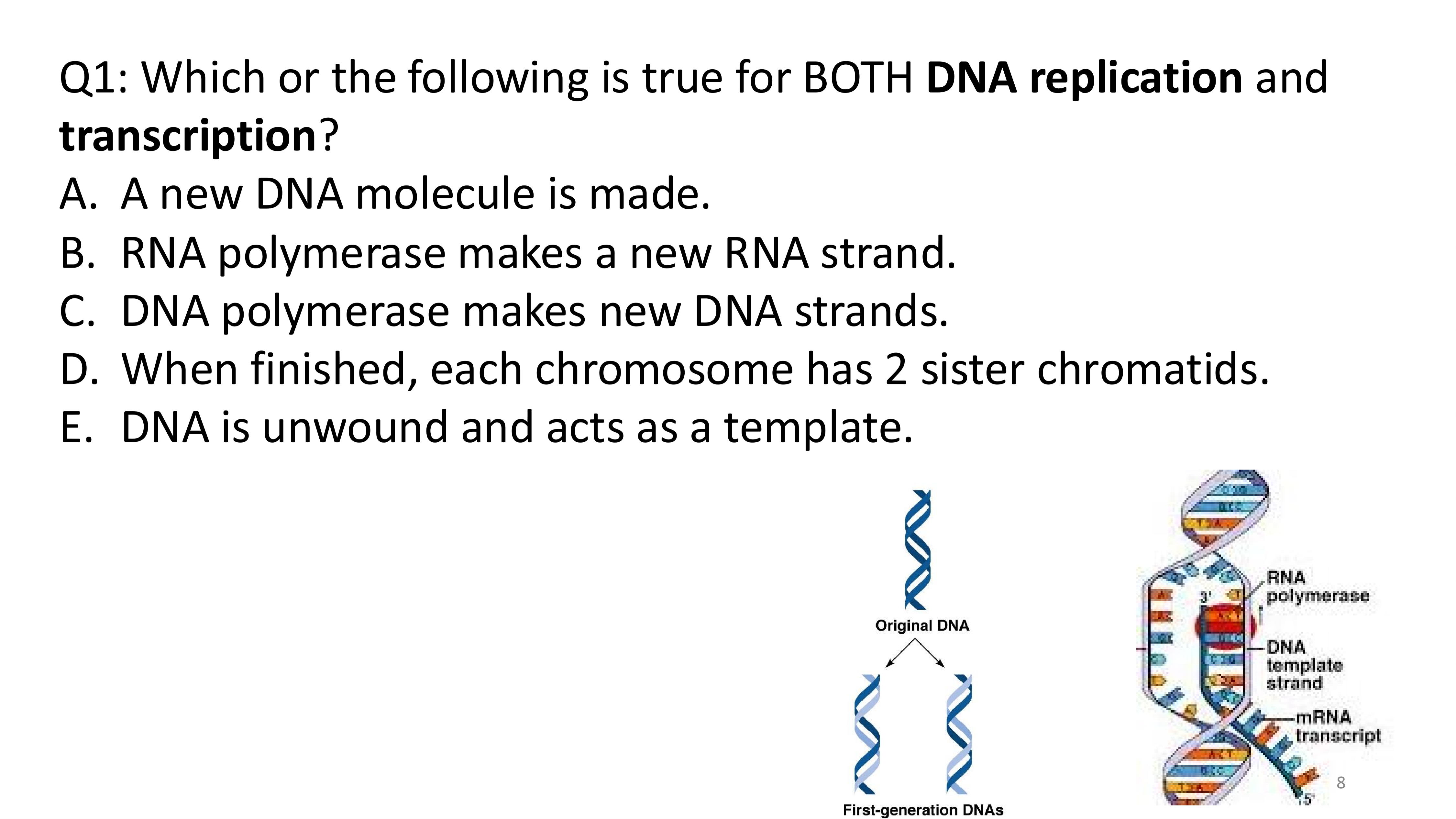
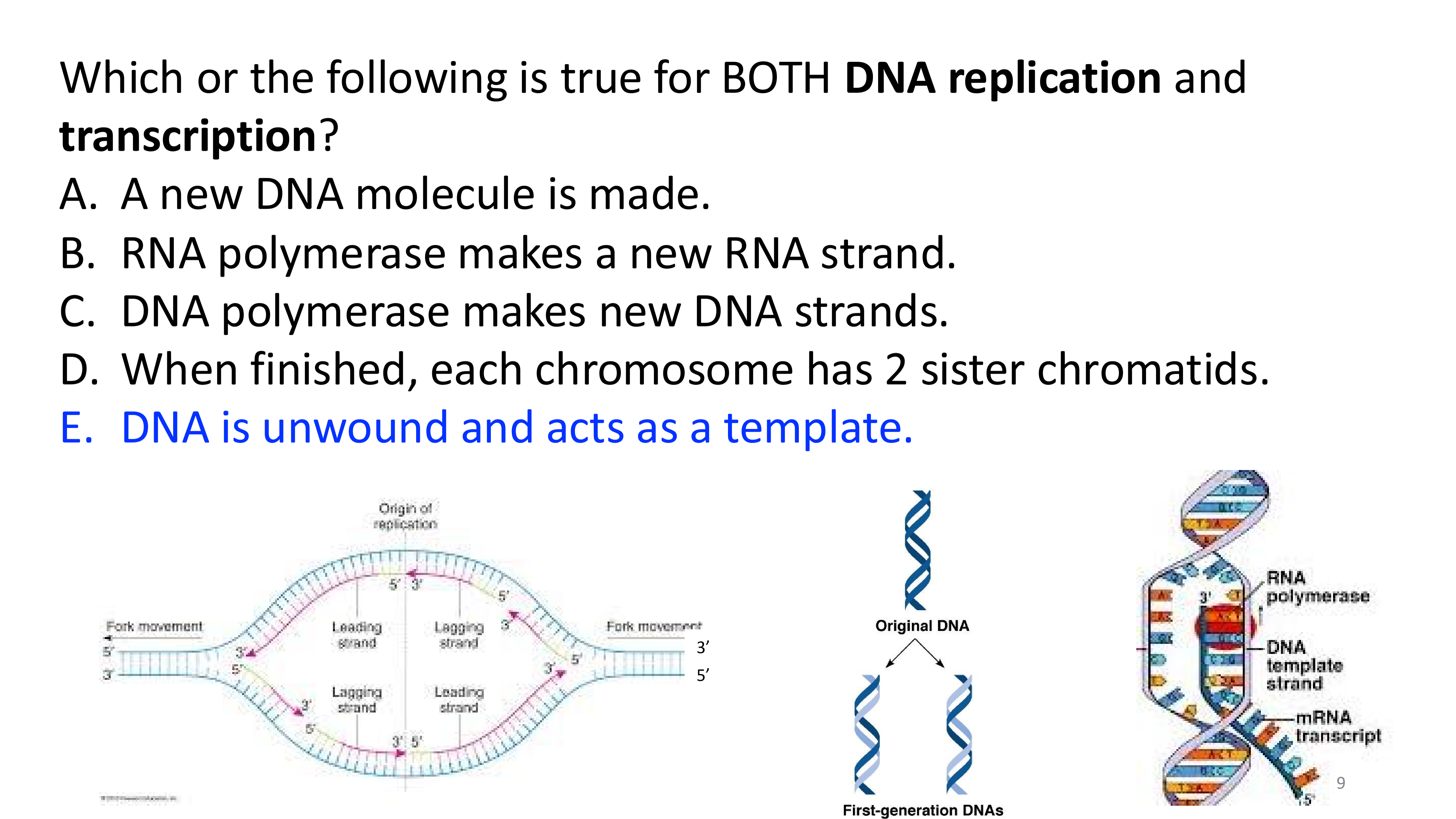
transcription big picture

key players in transcription
DNA, RNA, RNA polymerase
DNA vs RNA
DNA is double stranded, RNA is single stranded
DNA has deoxyribose sugar, RNA has ribose sugar
RNA has uracil instead of thymine, to bind to A
RNA polymerase
made up of 5 subunits plus the transcription factor
sigma factor (s70) is only there during initiation of transcription
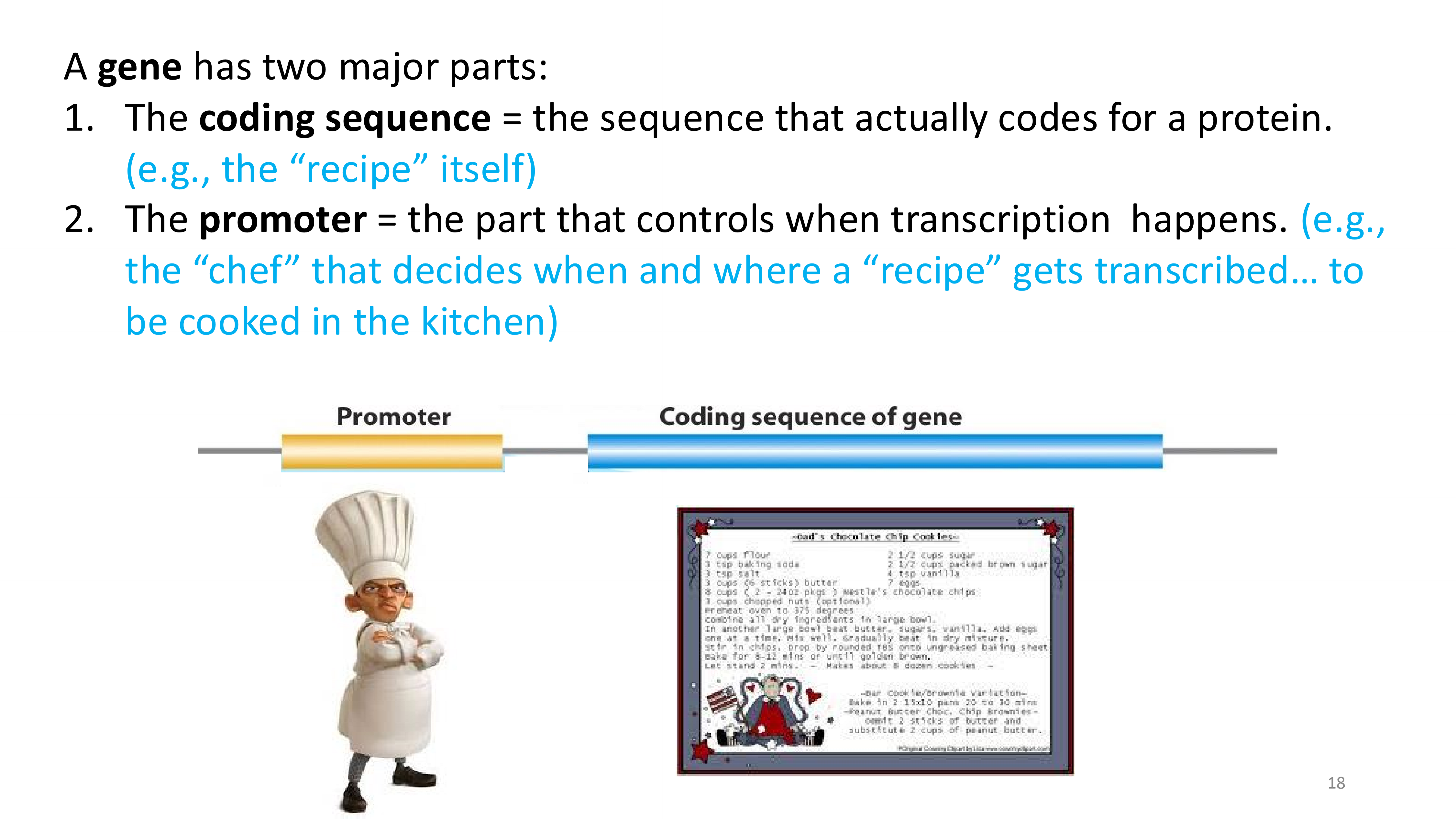
major parts of a gene
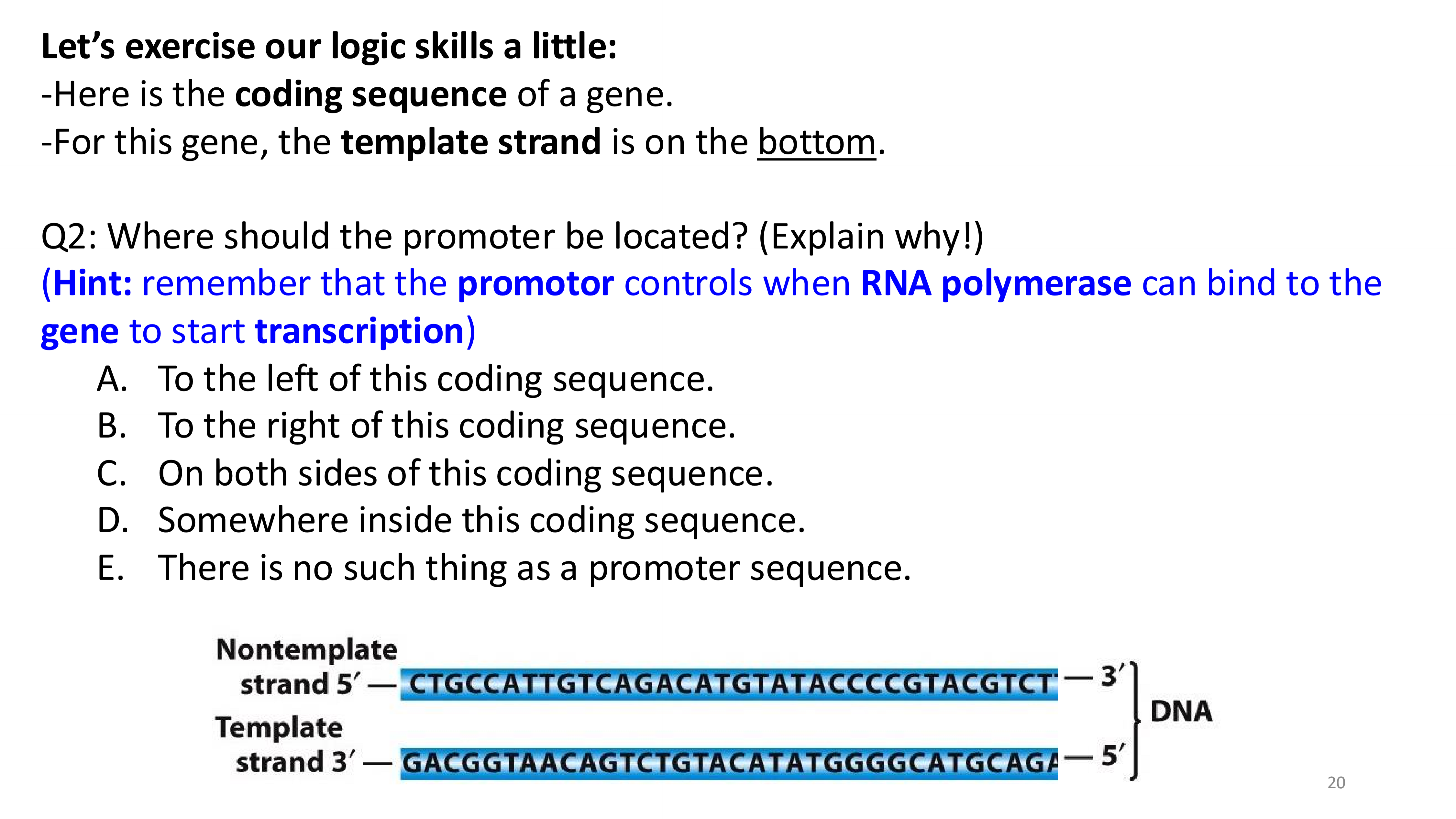
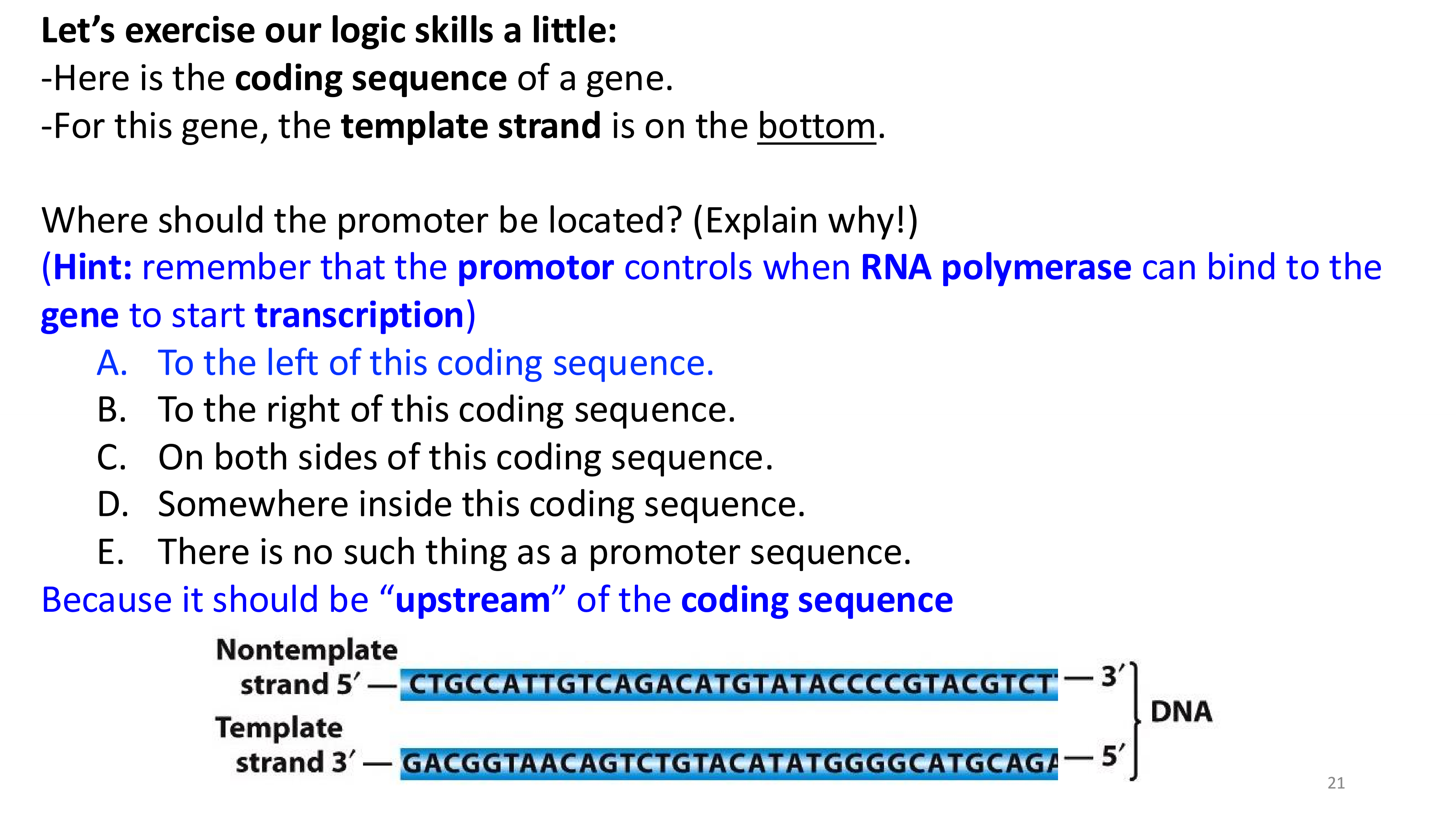
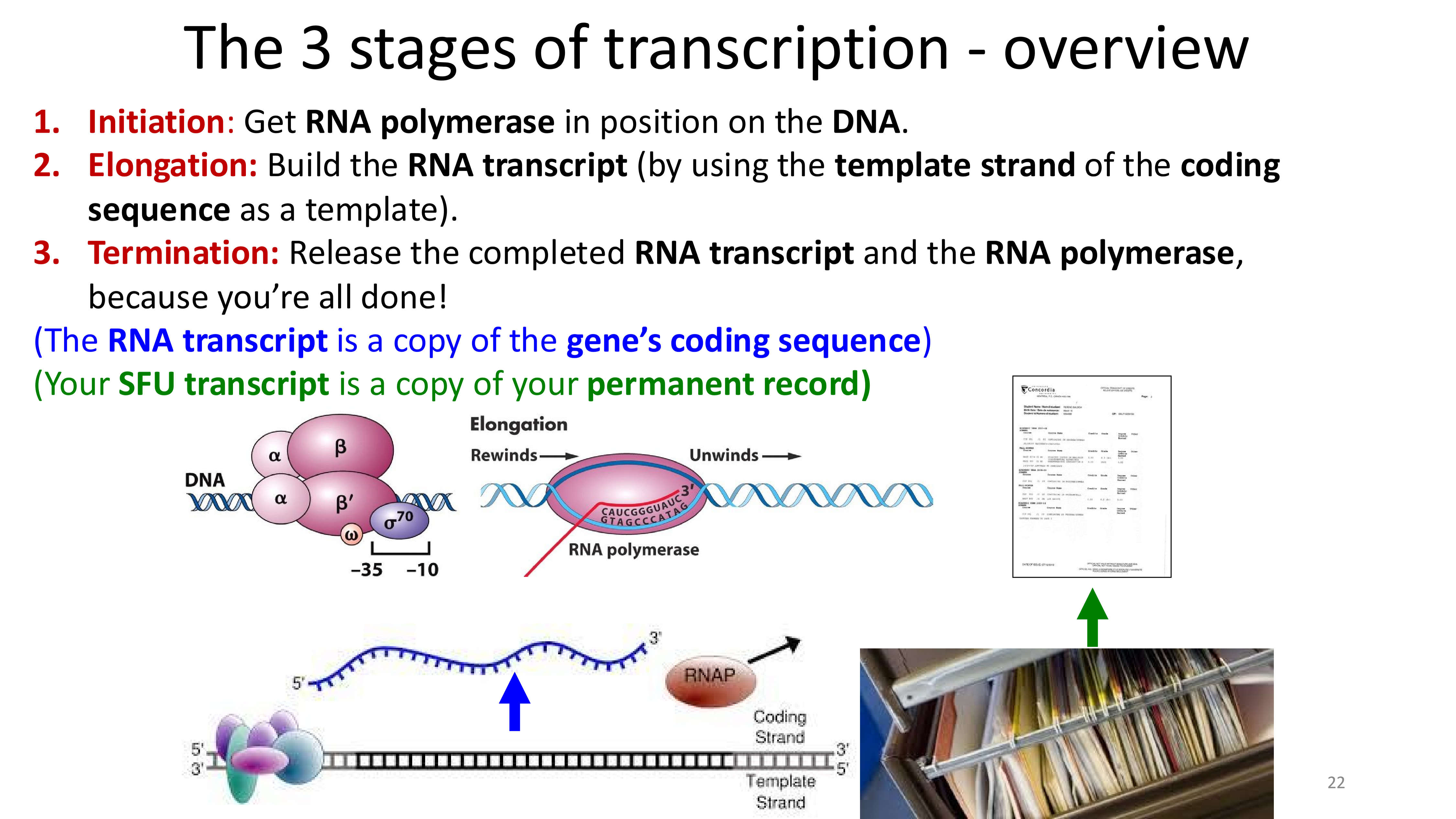
transcription stages
transcription initiation important locations
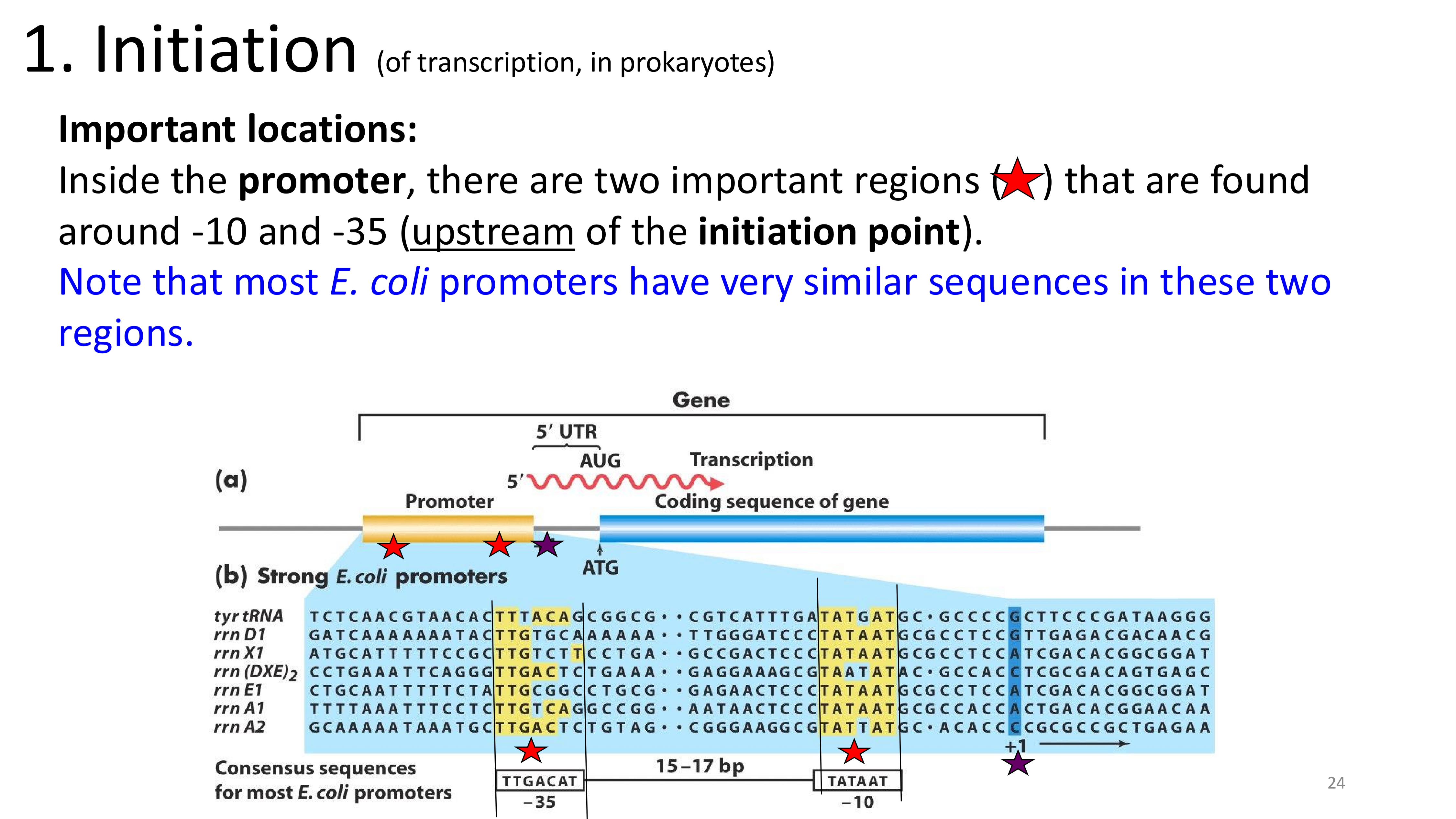
transcription initiation which regions are important and why?
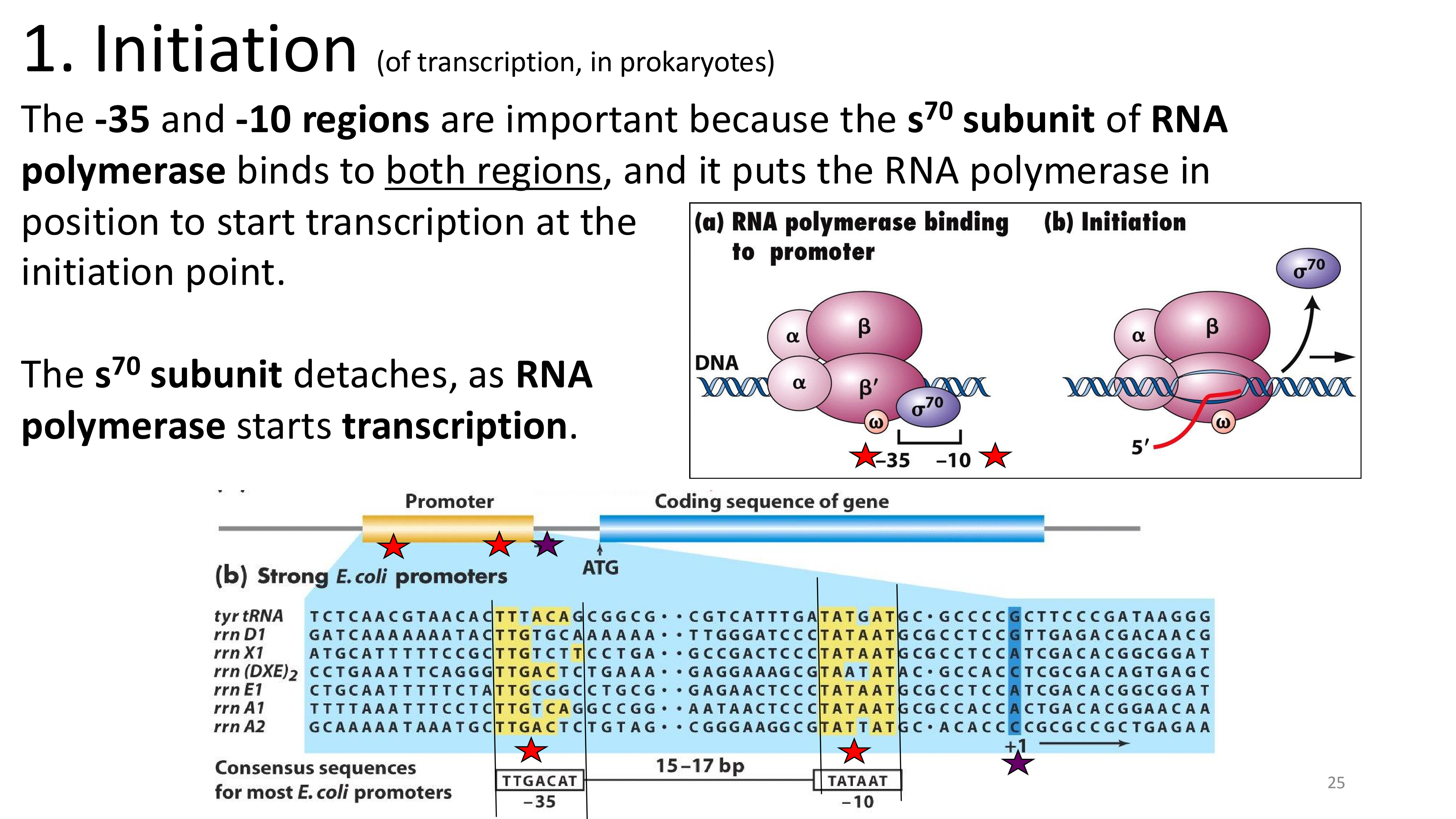
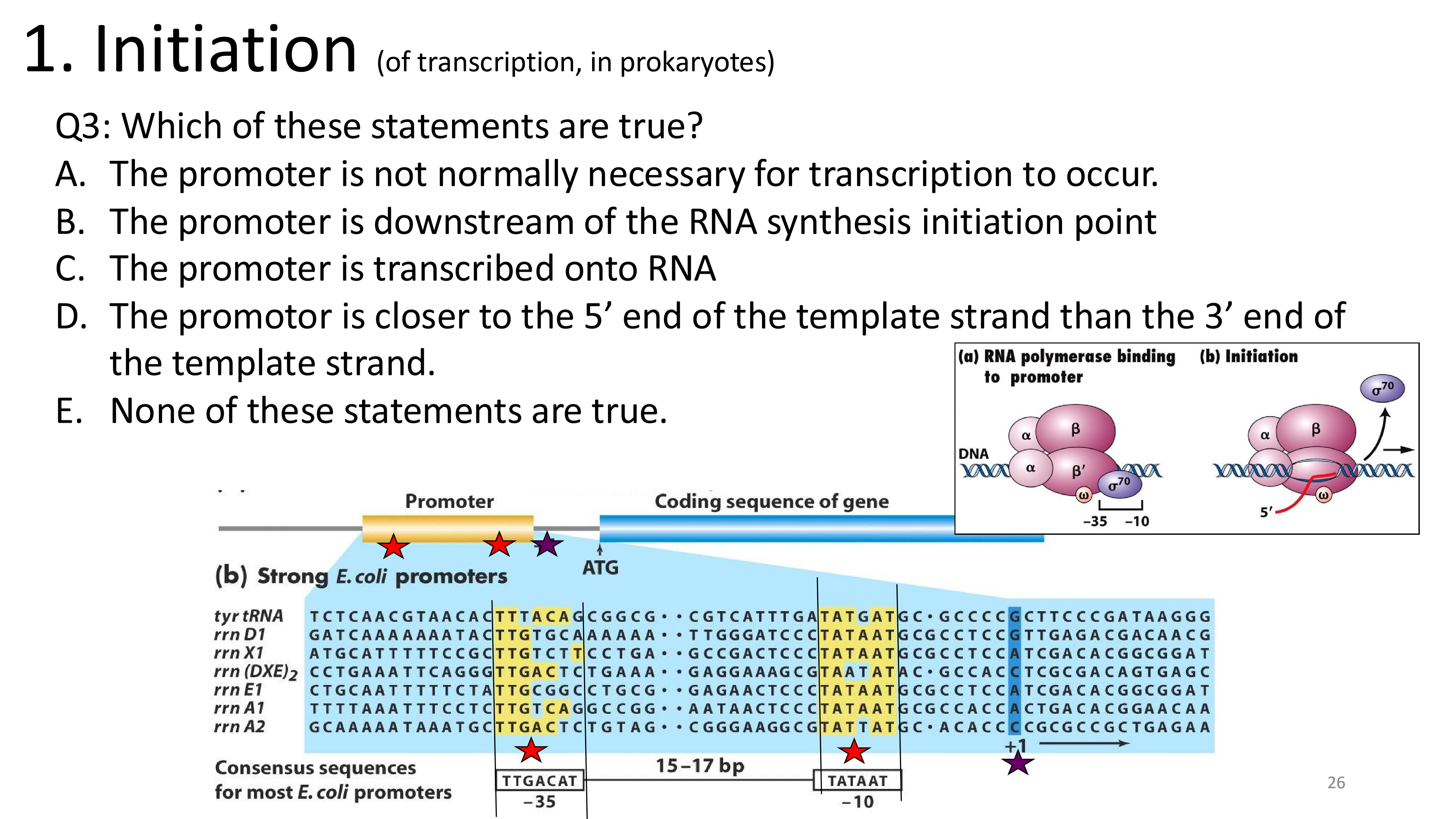
E. None
transcription elongation
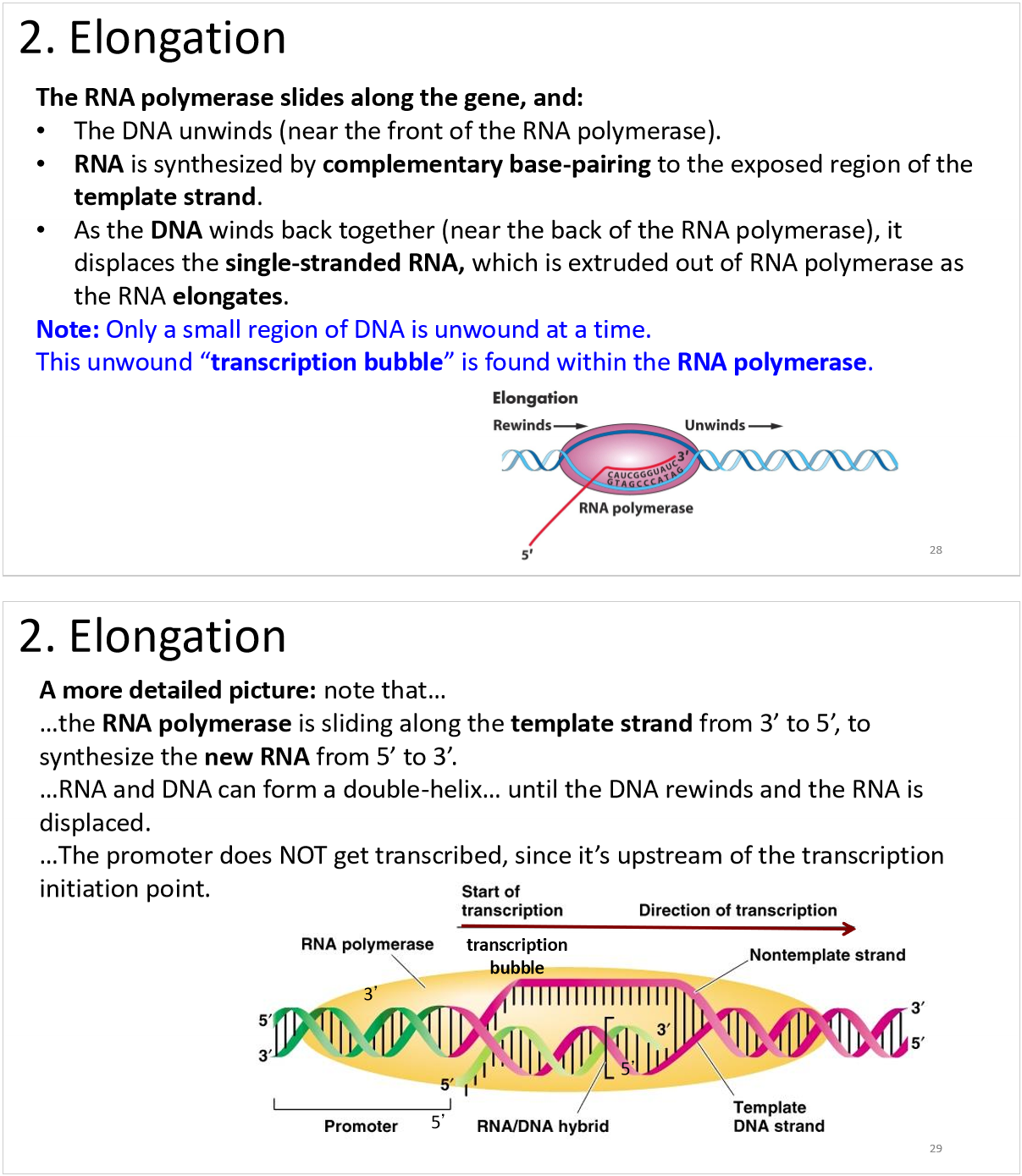
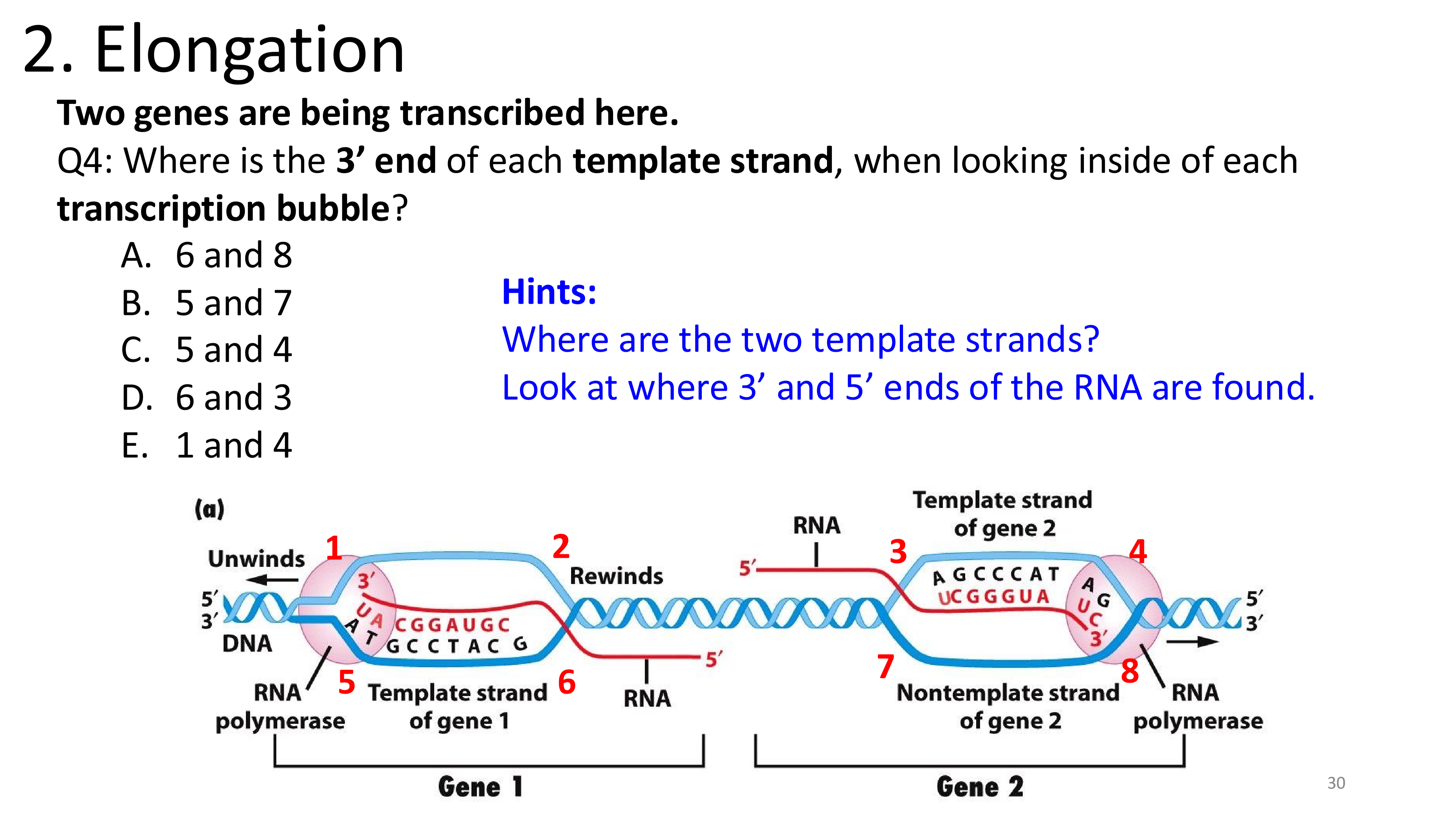
D. 6 and 3
how many hydrogen bonds does each base pair have?
A-T and A-U have 2 hydrogen bonds and are less stable
G-C pairs have 3 hydrogen bonds and are more stable
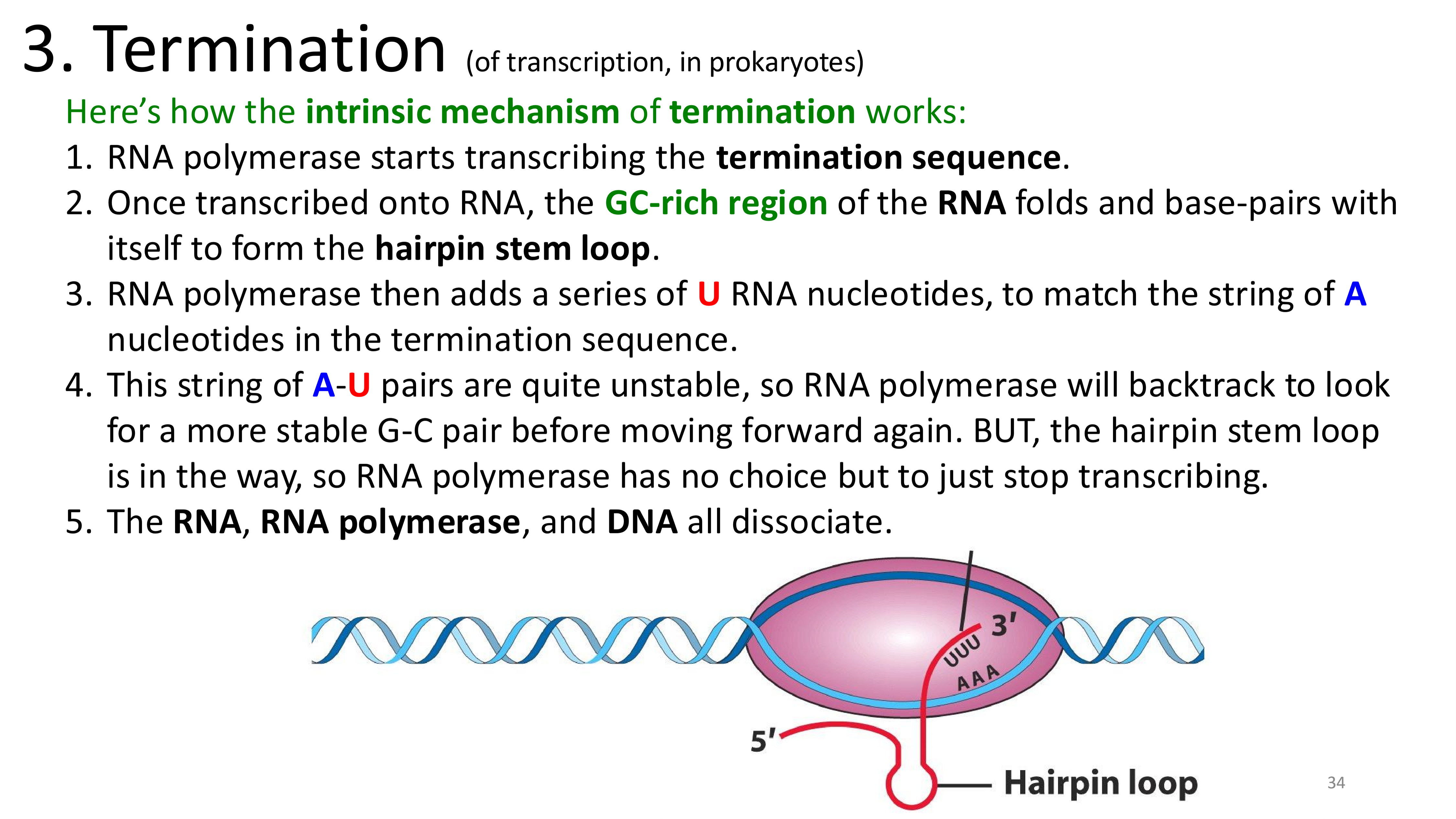
transcription termination
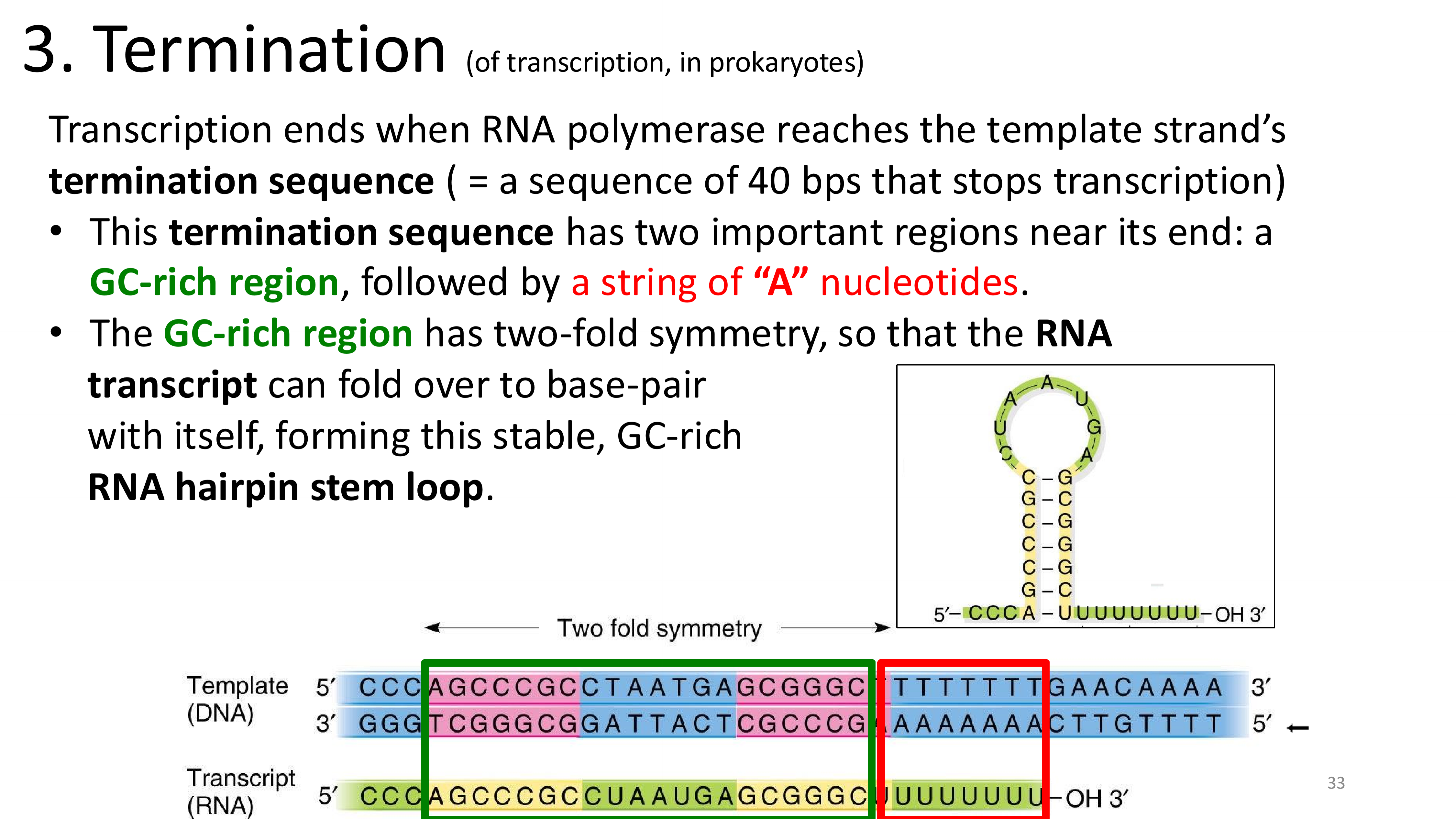
transcription termination intrinsic mechanism
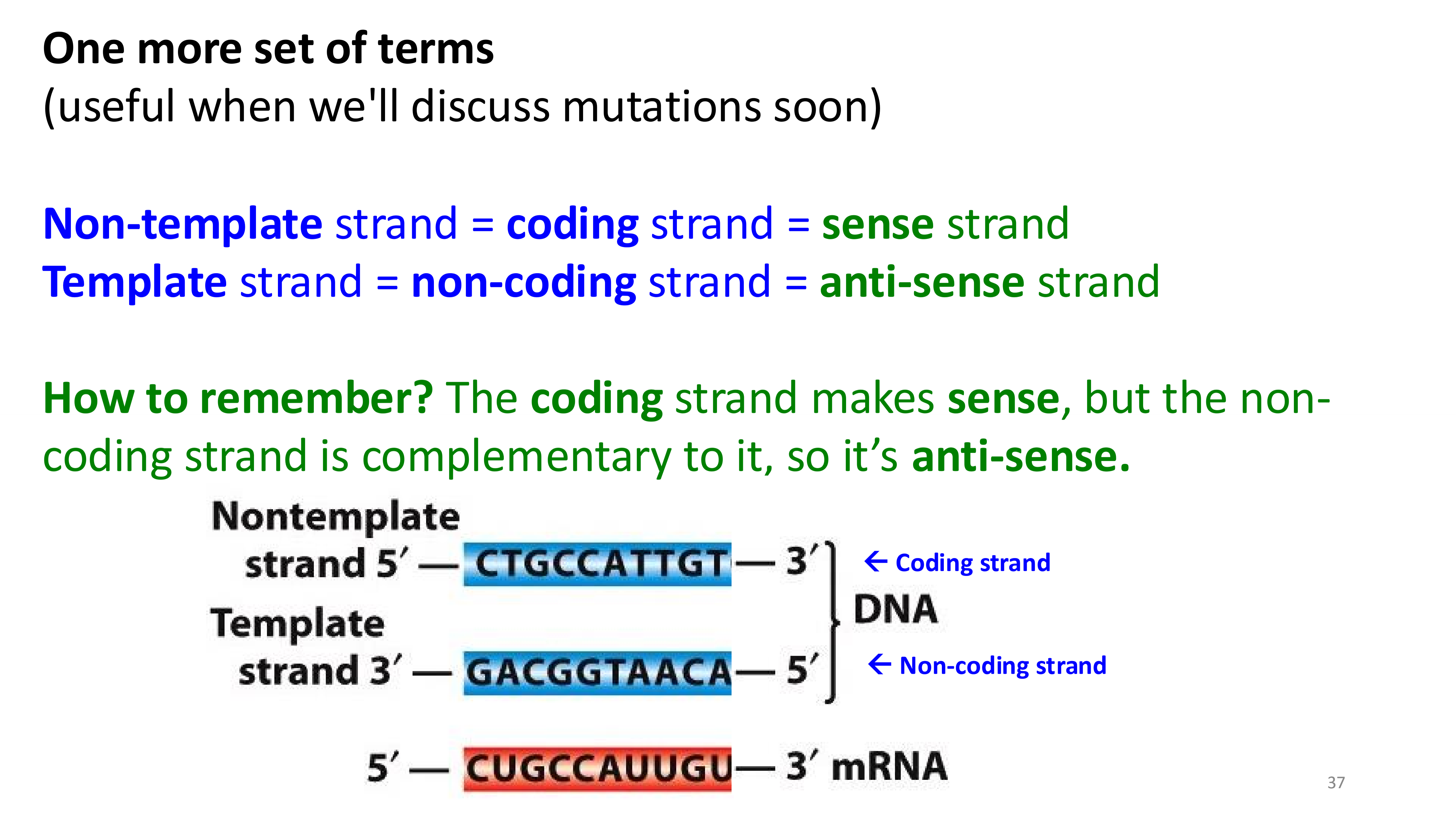
non-template strand vs template strand
what direction is RNA synthesized in transcription
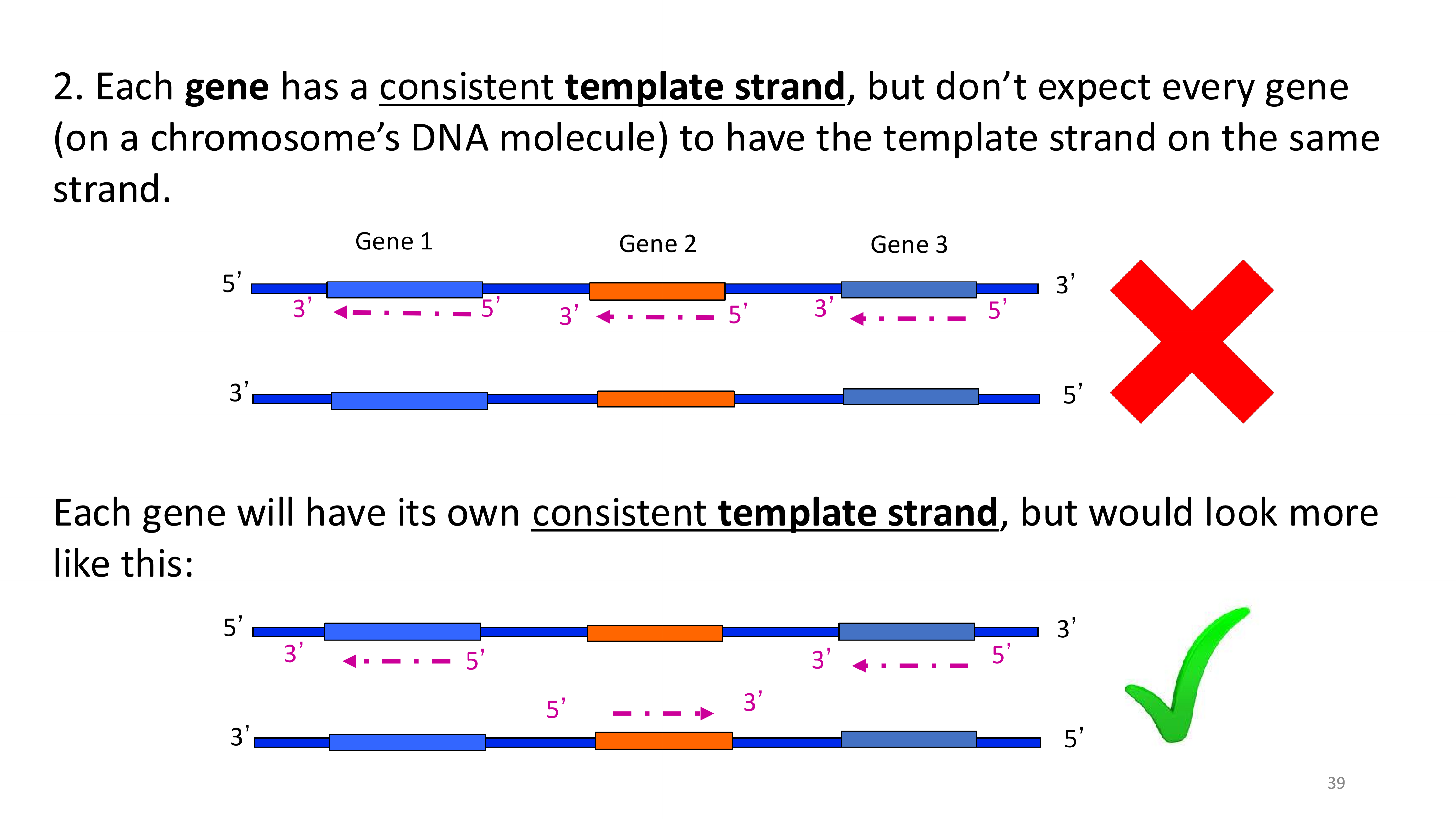
does every gene have the template strand on the same strand?
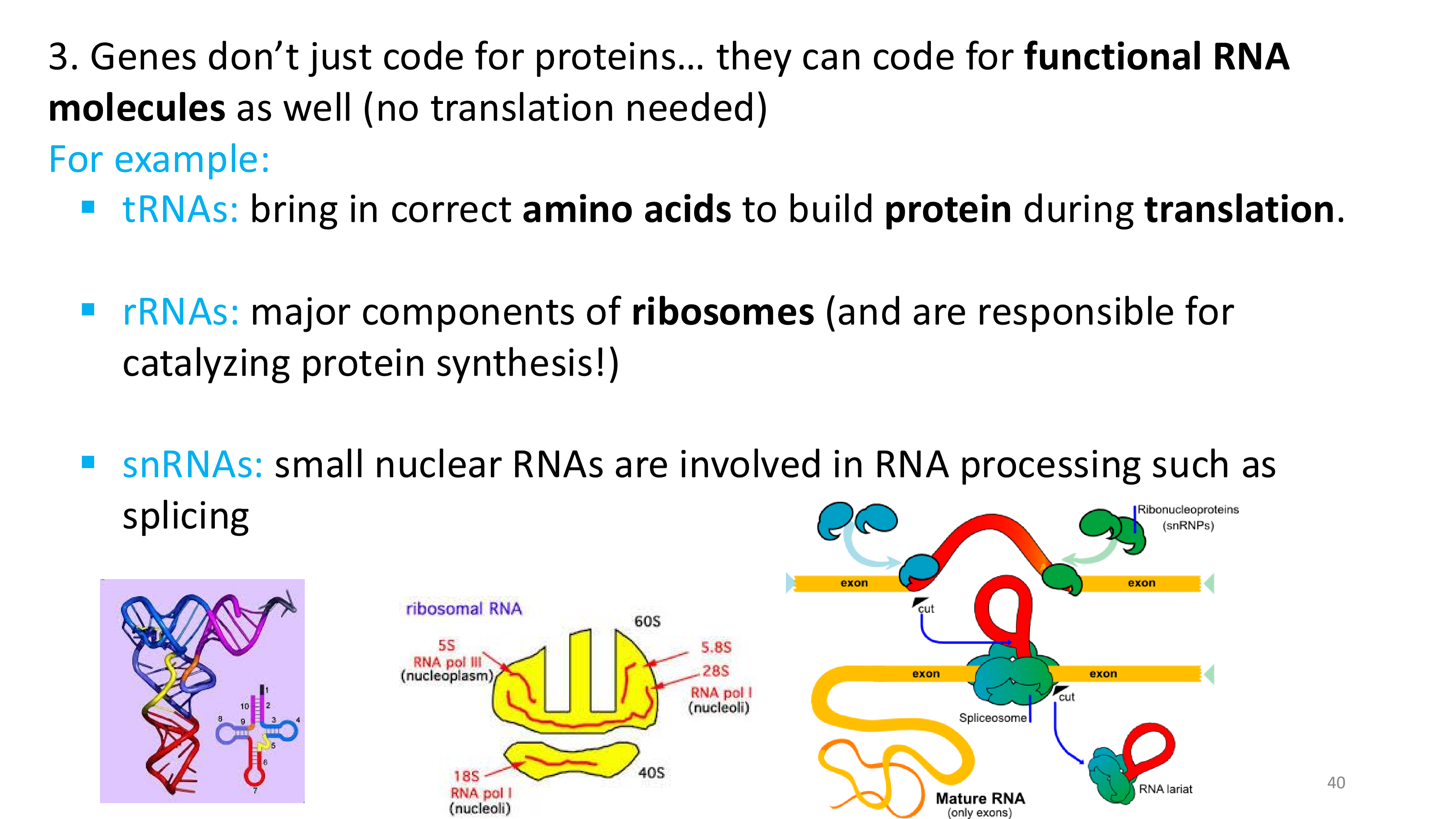
what do genes code for other than proteins?
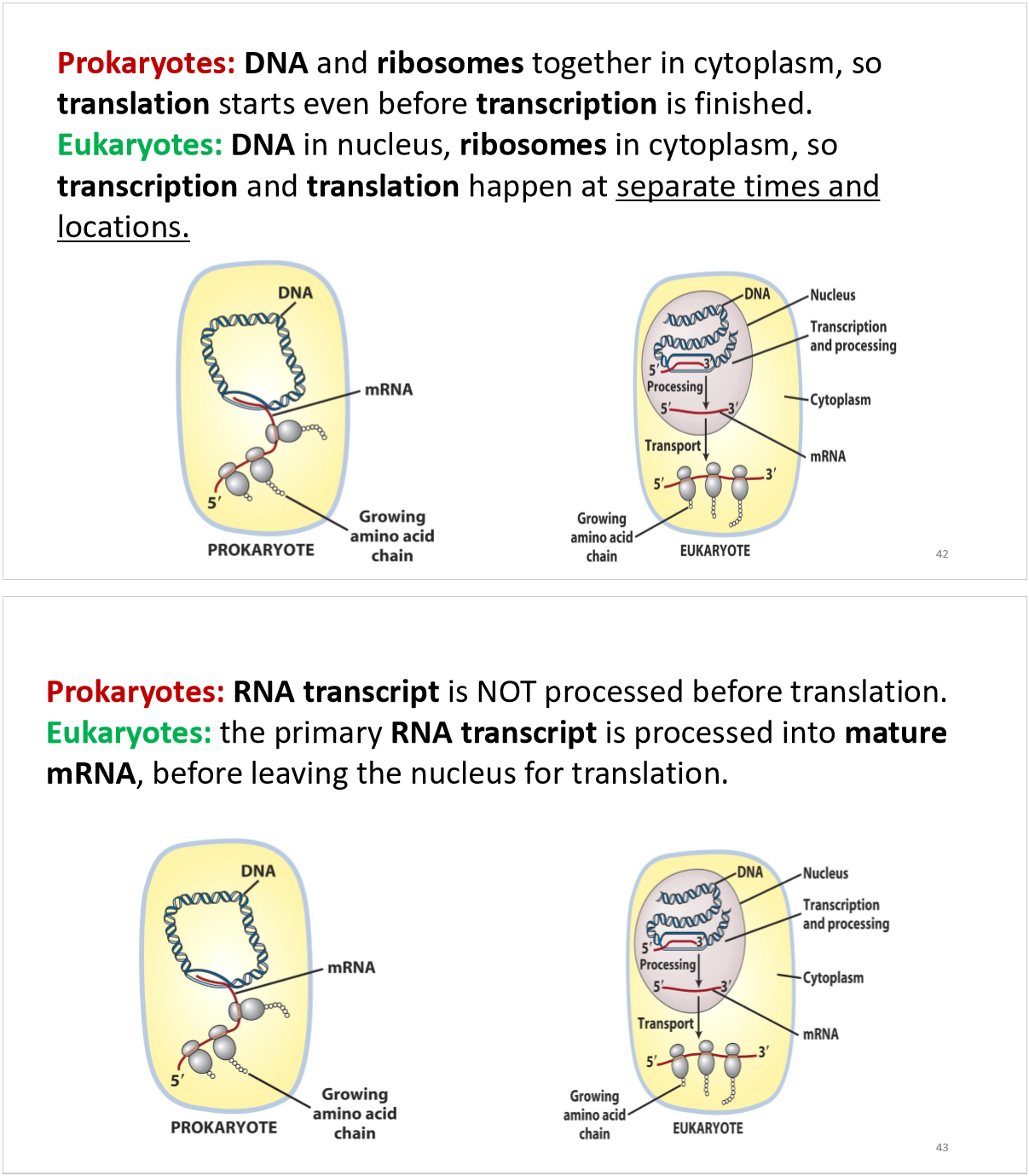
transcription in prokaryotes vs eukaryotes
RNA processing
only happens in eukaryotes
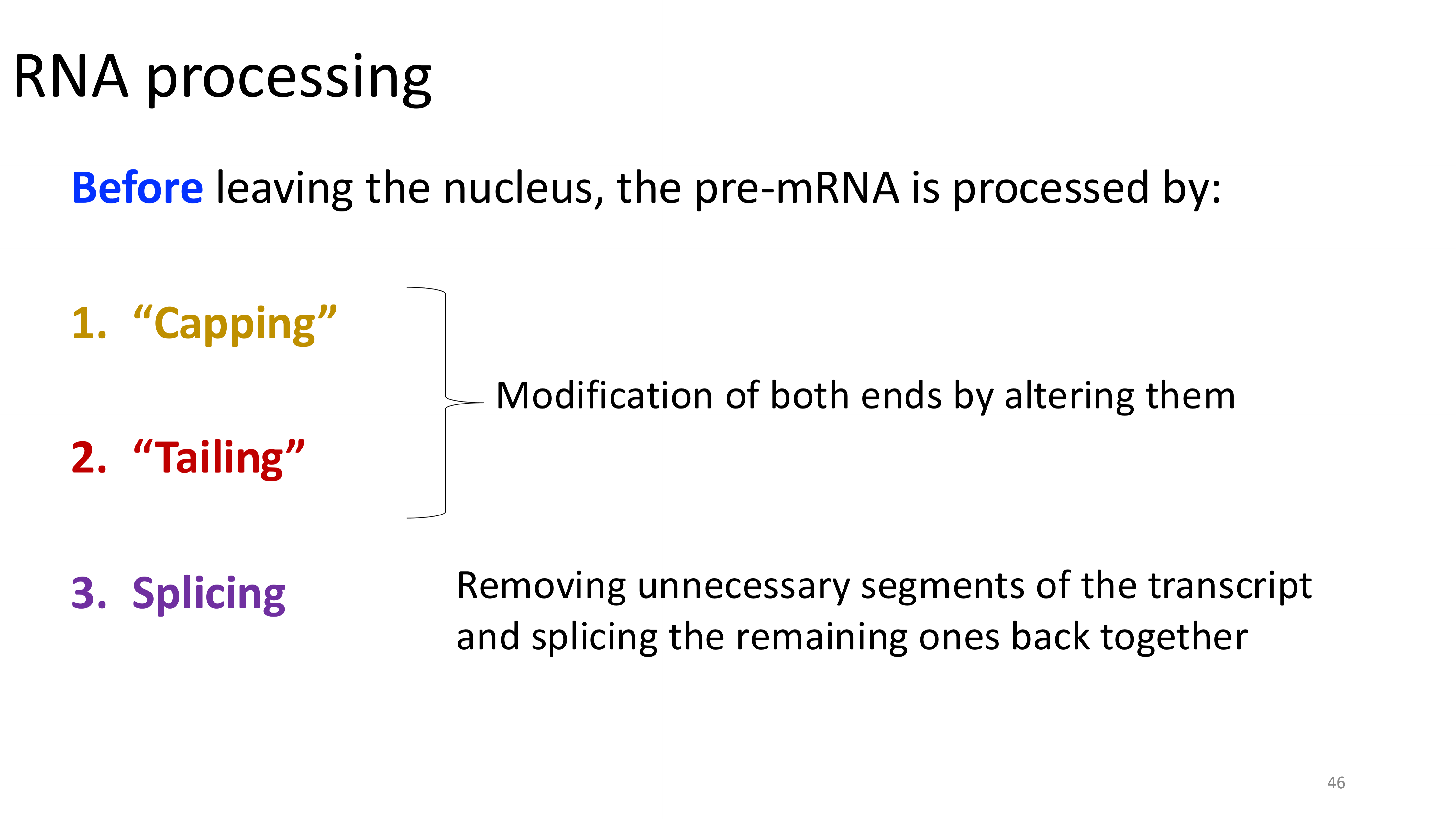
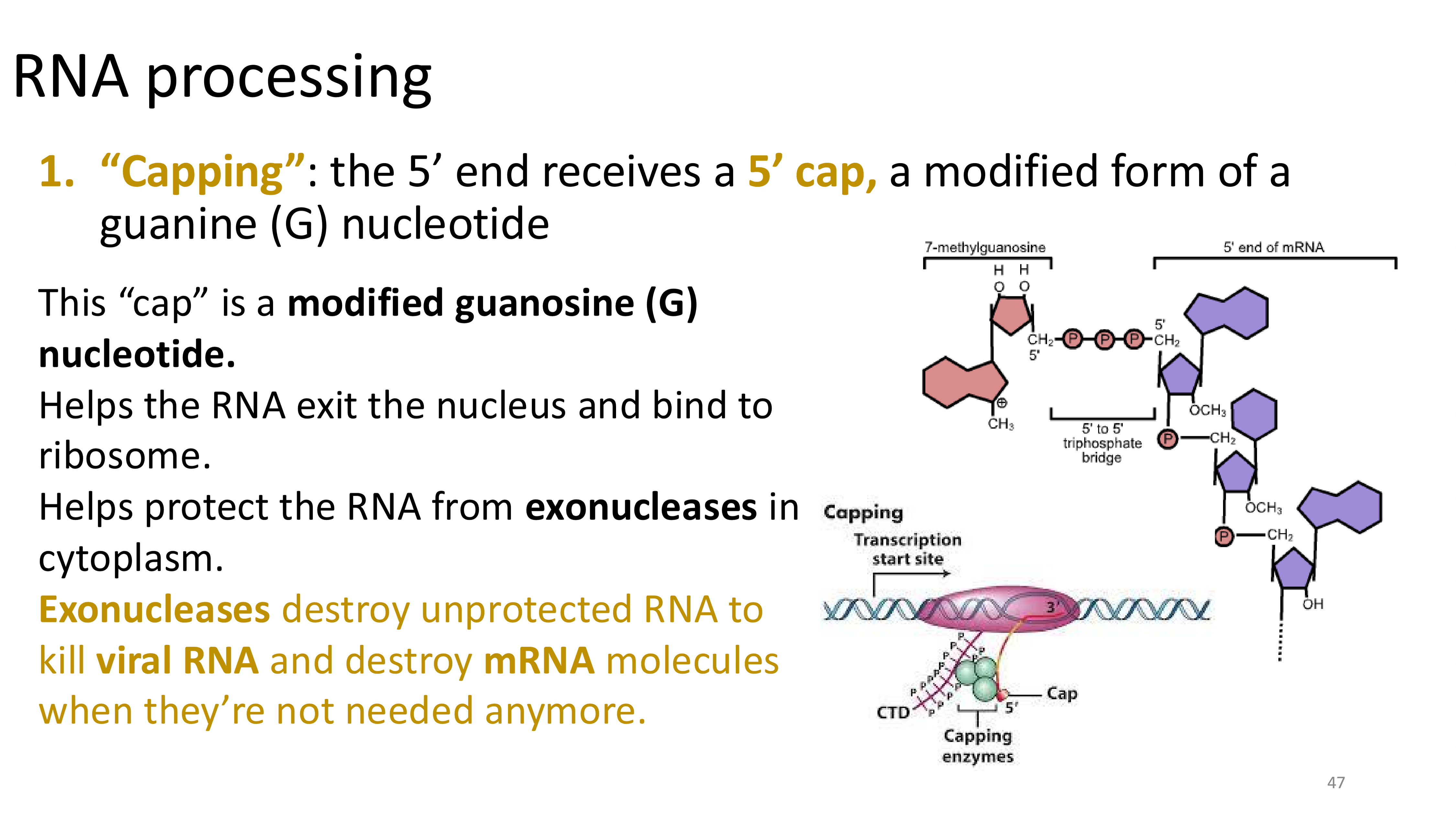
capping
only happens in eukaryotes
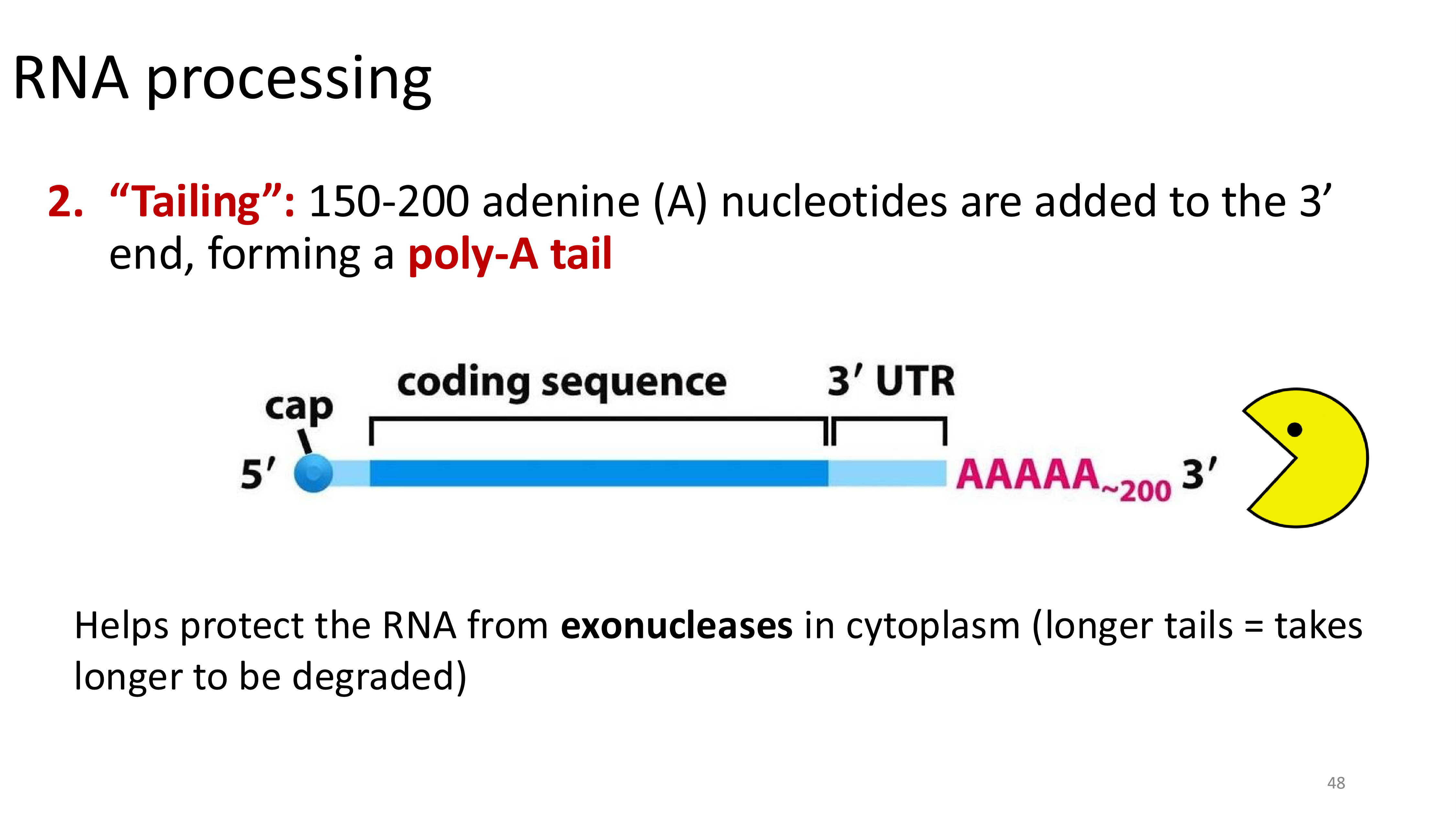
tailing
only happens in eukaryotes
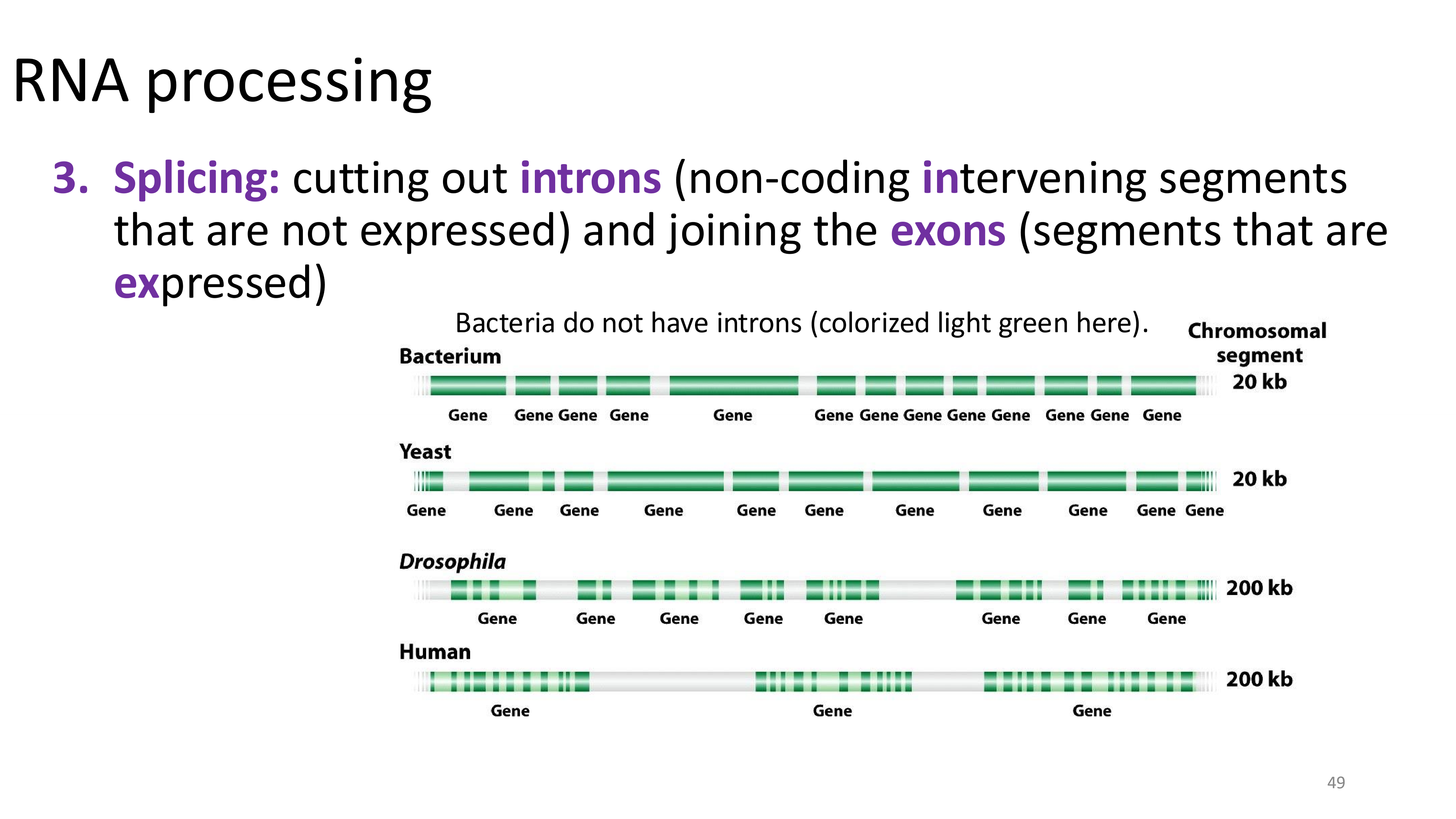
splicing
only happens in eukaryotes
what carries out splicing?
splicing is carried out by spliceosomes: protein/RNA complexes
snRNP
lariat structure
cut-out intron is also called ‘lariat’ structure (it gets degraded)
process of splicing
The spliceosome forms on pre-mRNA, bringing together snRNPs and other proteins.
The spliceosome removes introns by cutting them out.
The cut intron forms a lariat structure and gets degraded.
The exons are joined together to produce mature mRNA.
The RNA itself helps catalyze the excision process.
DNA replication vs transcription
replication: copies both DNA strands; entire genome is replicated
transcription: copies one strand of DNA (template strand); only a segment is transcribed to mRNA
initiation in transcription
RNA polymerase recognizes and binds to a promoter region on double stranded DNA, and DNA unwinds to allow transcription to begin
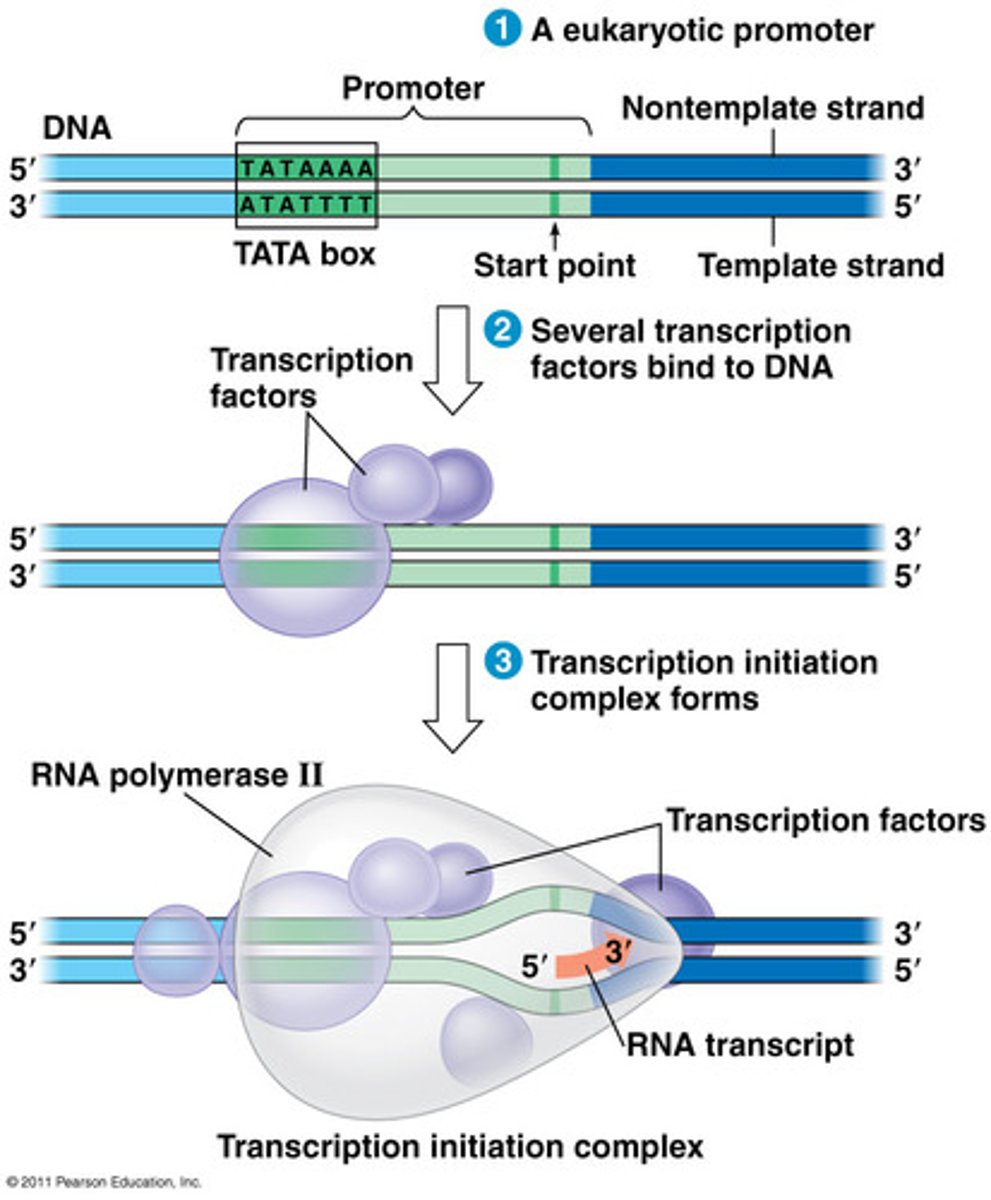
elongation in transcription
-RNA polymerase II unwinds the double helix
-RNA polymerase II adds complementary base pairs to DNA template strand
-As elongation occurs, completed portions of mRNA are released
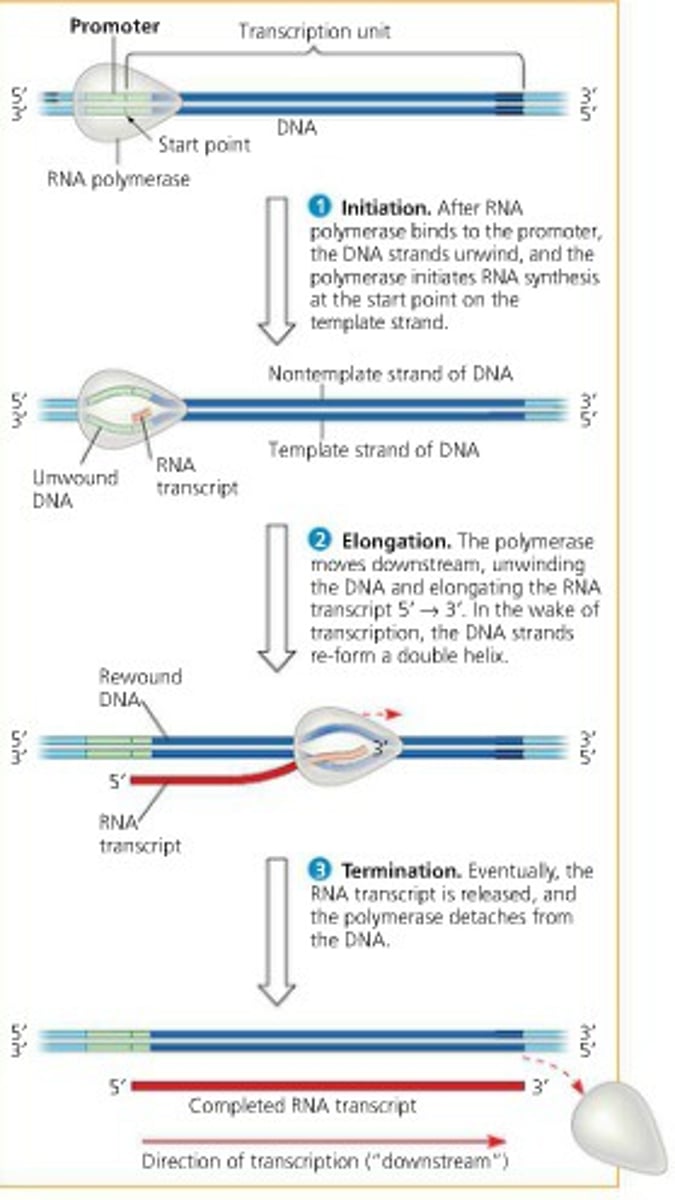
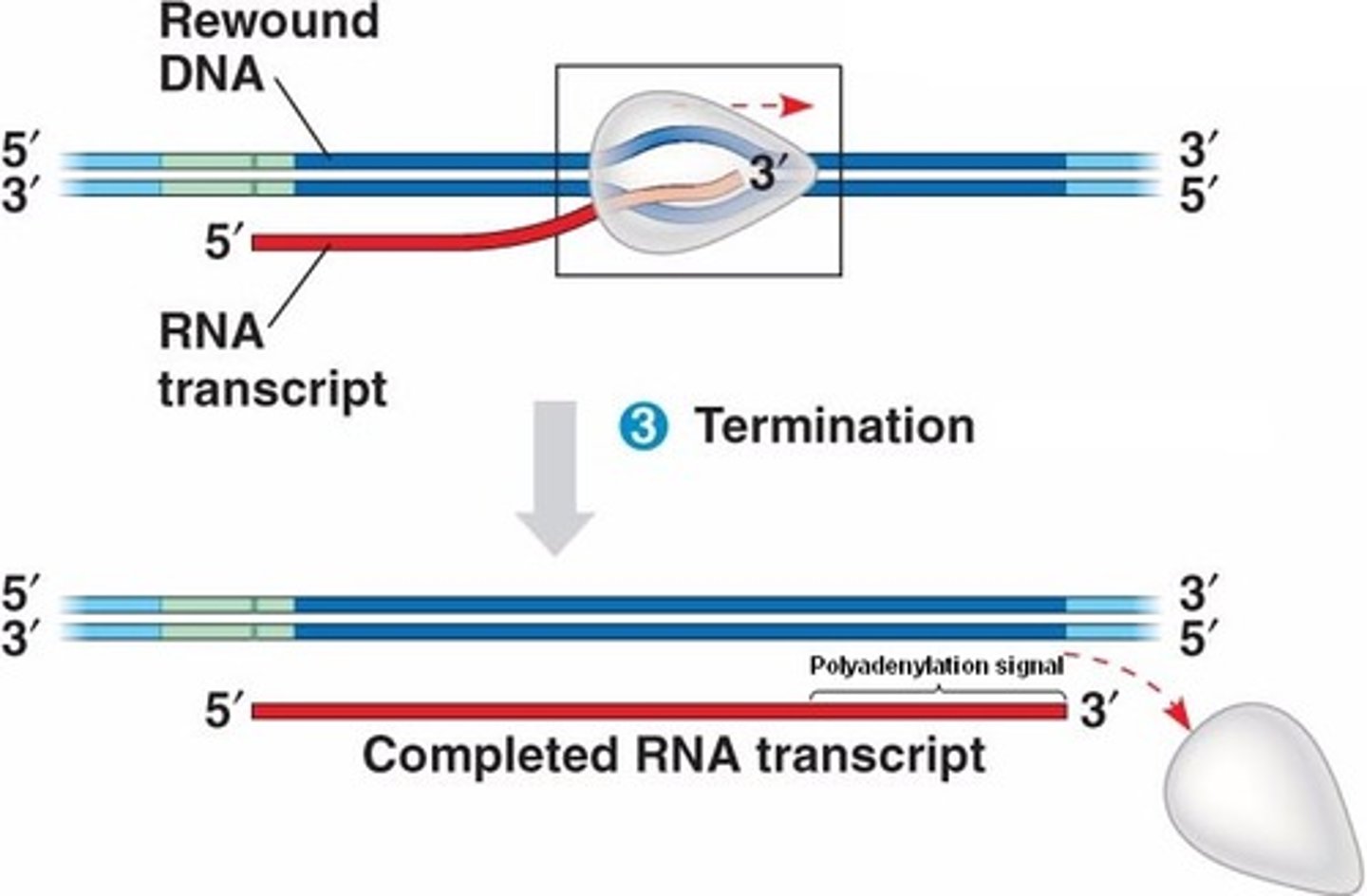
termination in transcription
- mRNA synthesis seizes when RNA polymerase reaches the termination sequence
- mRNA and RNA polymerase are released
- the newly formed RNA strand detaches from the DNA
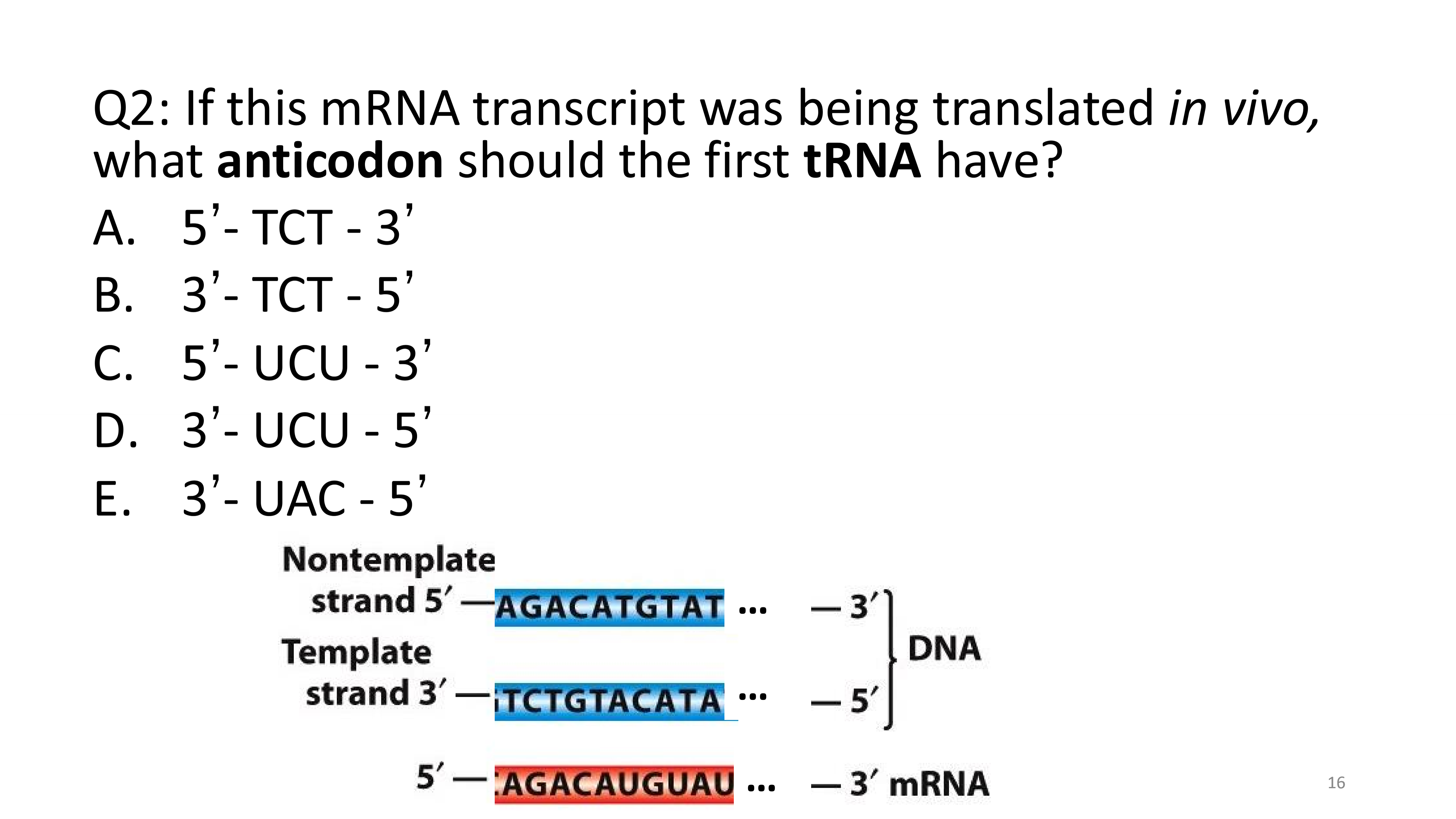
3’ - UAC - 5’
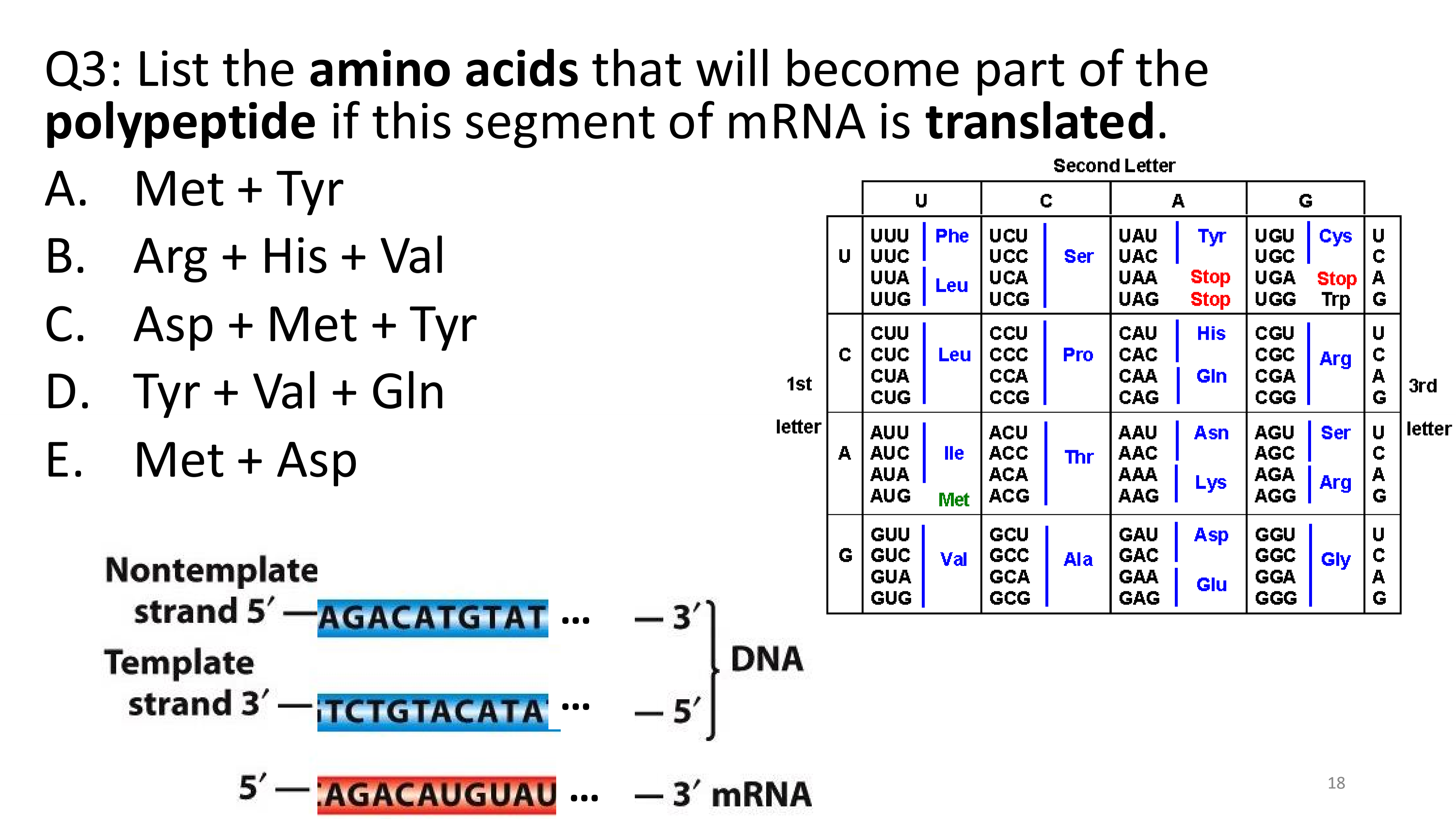
Met + Tyr
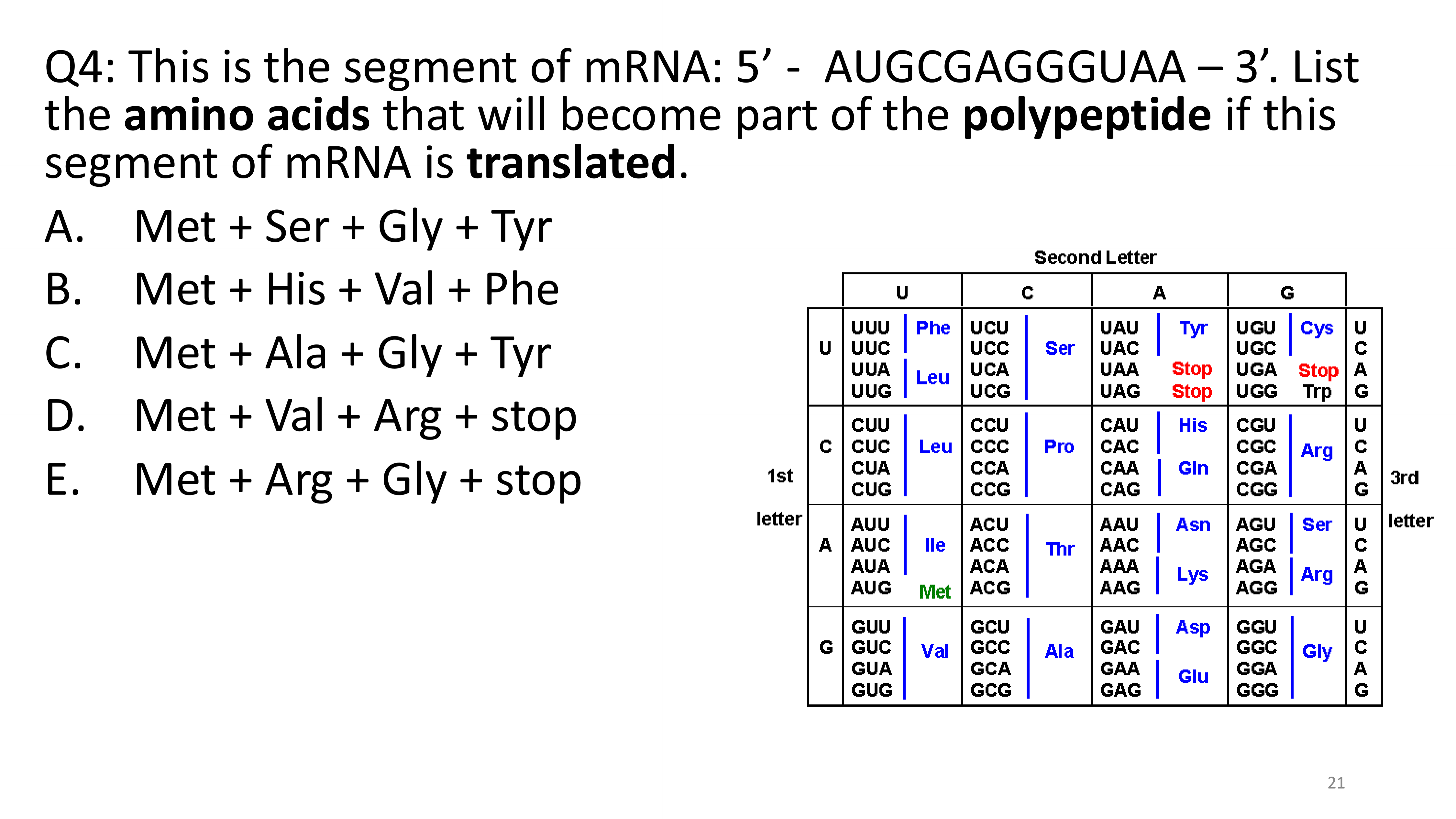
E. Met + Arg + Gly + stop
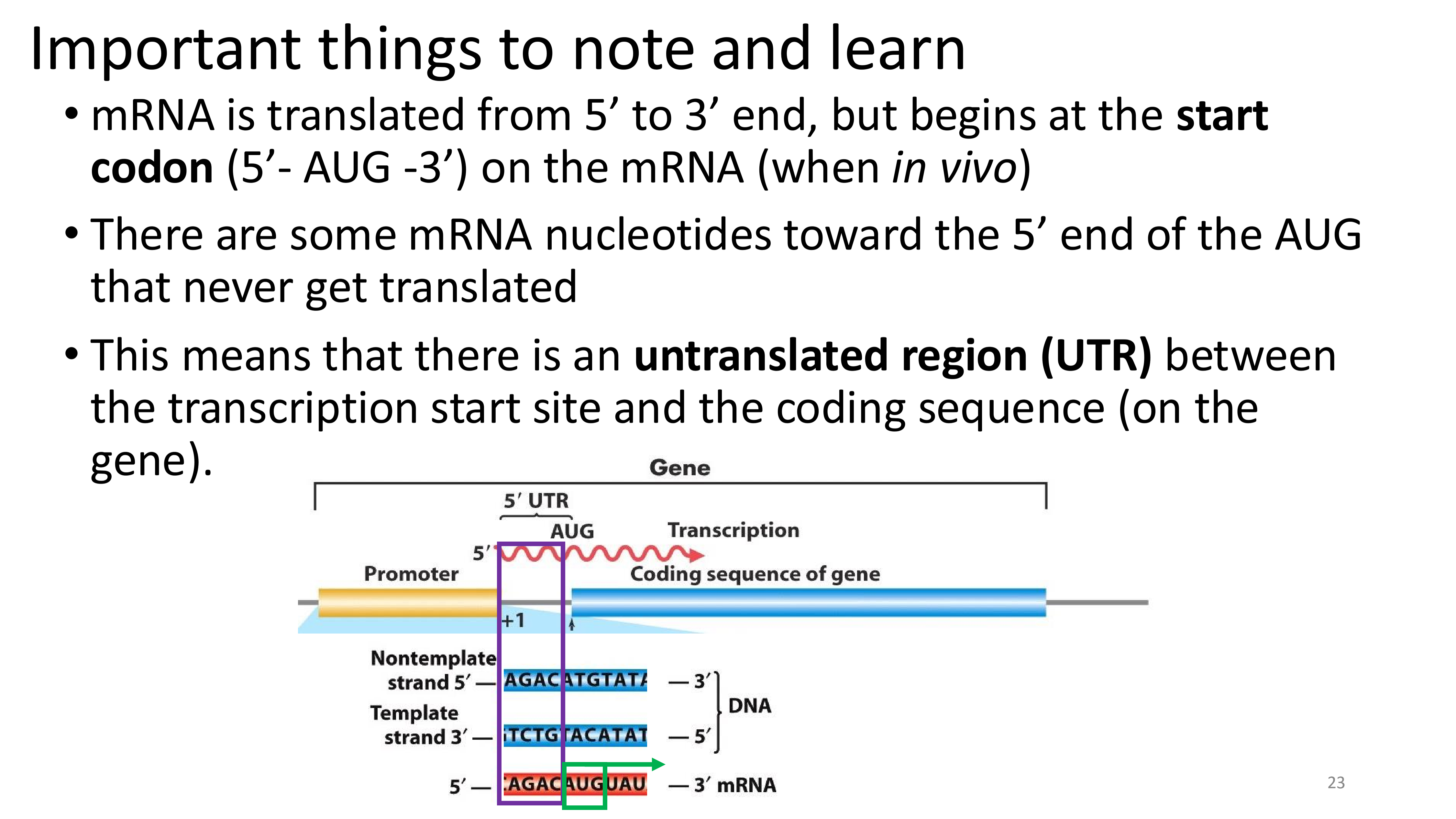
important things to note about translation
parts needed for translation
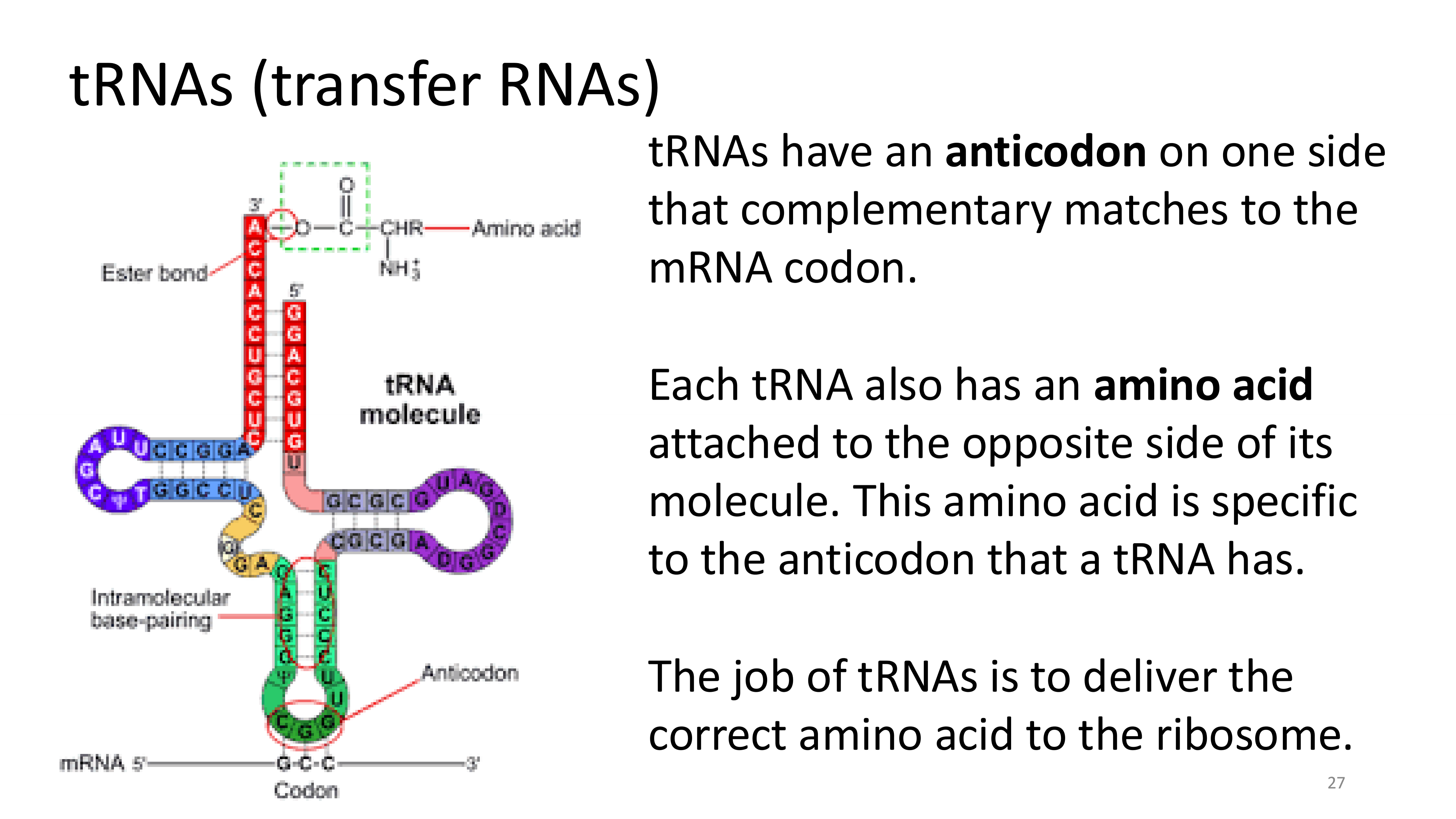
mRNA, amino acids, tRNA, ribosomes
mRNA
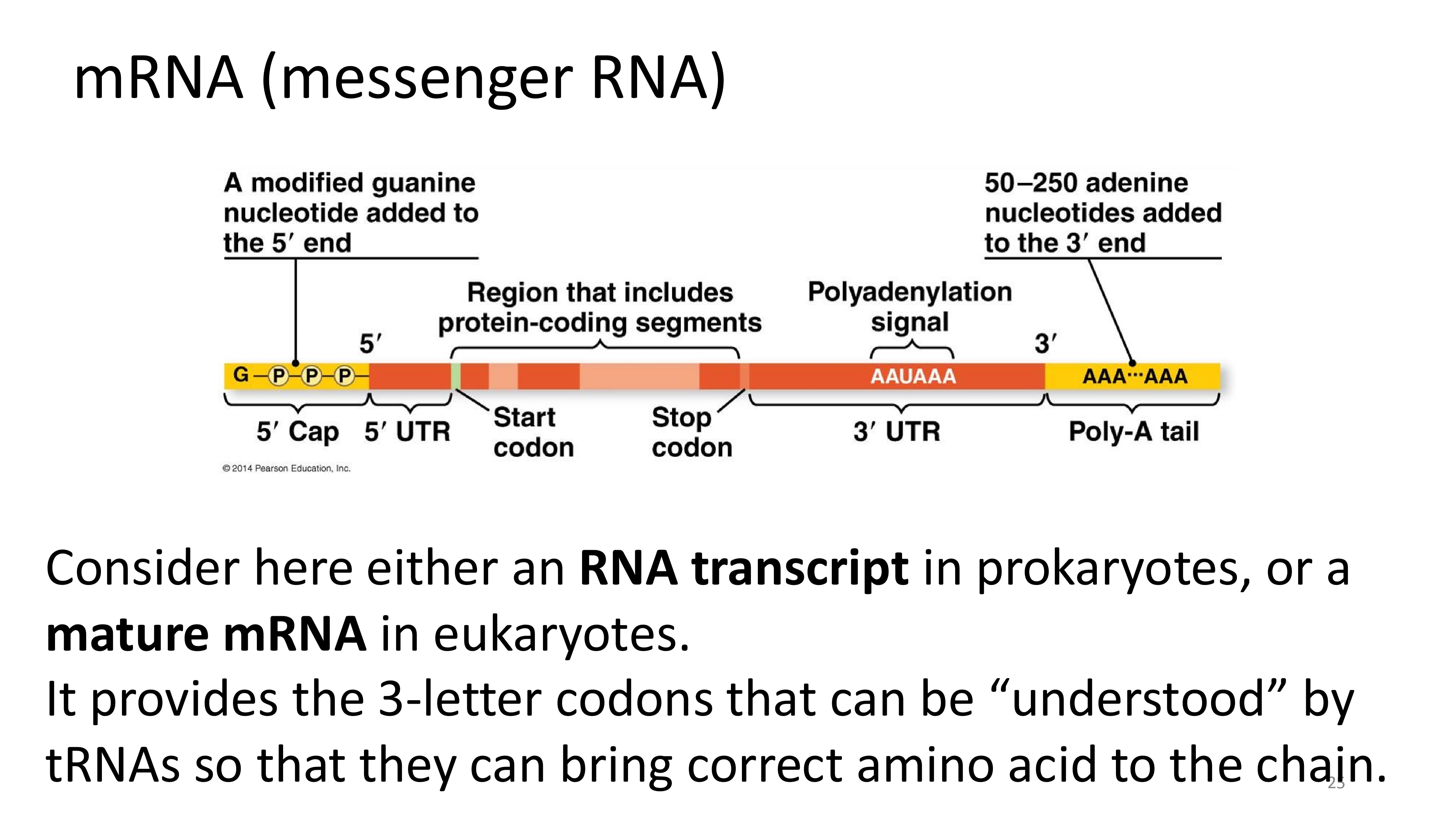
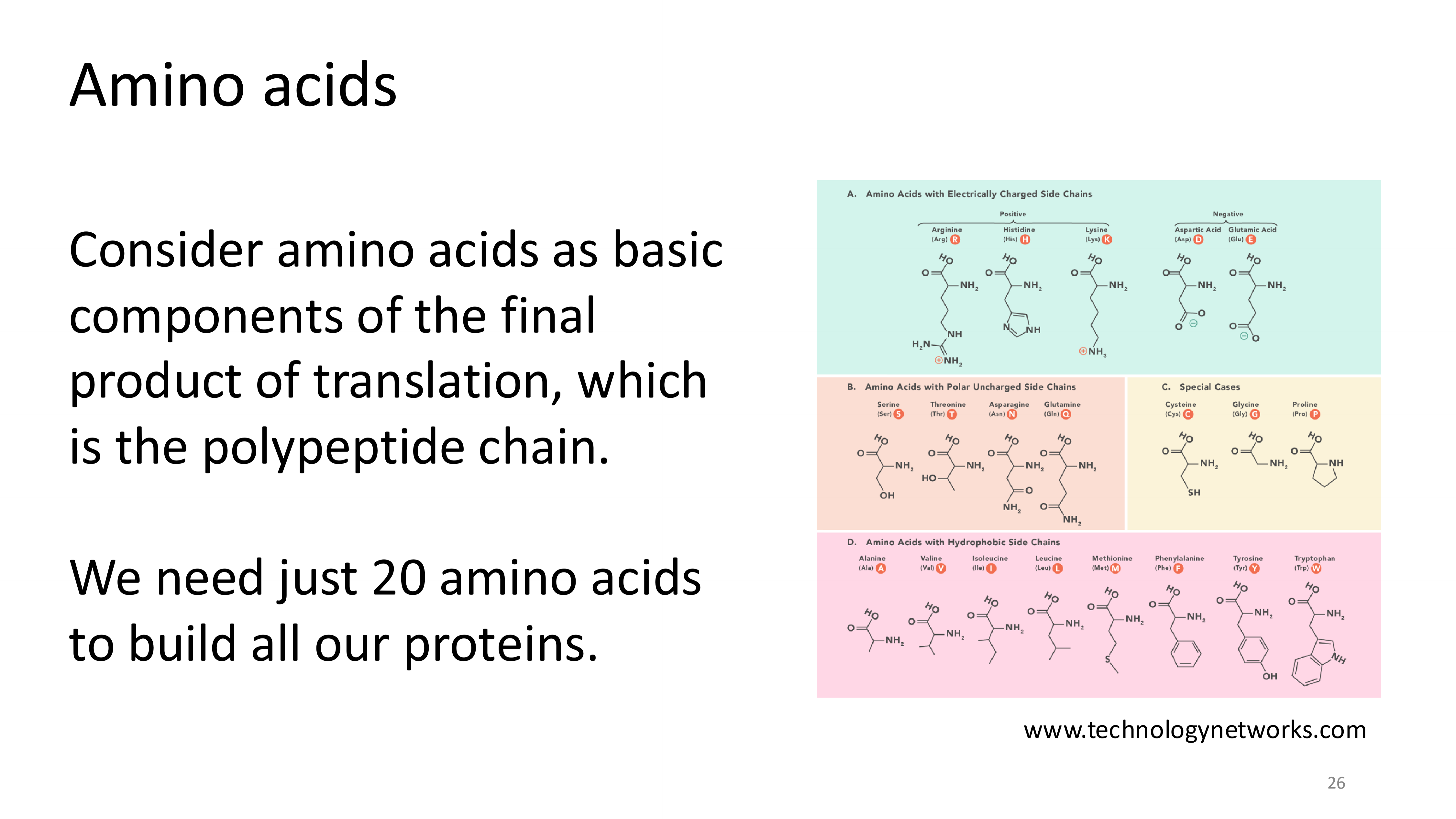
amino acids
tRNAs
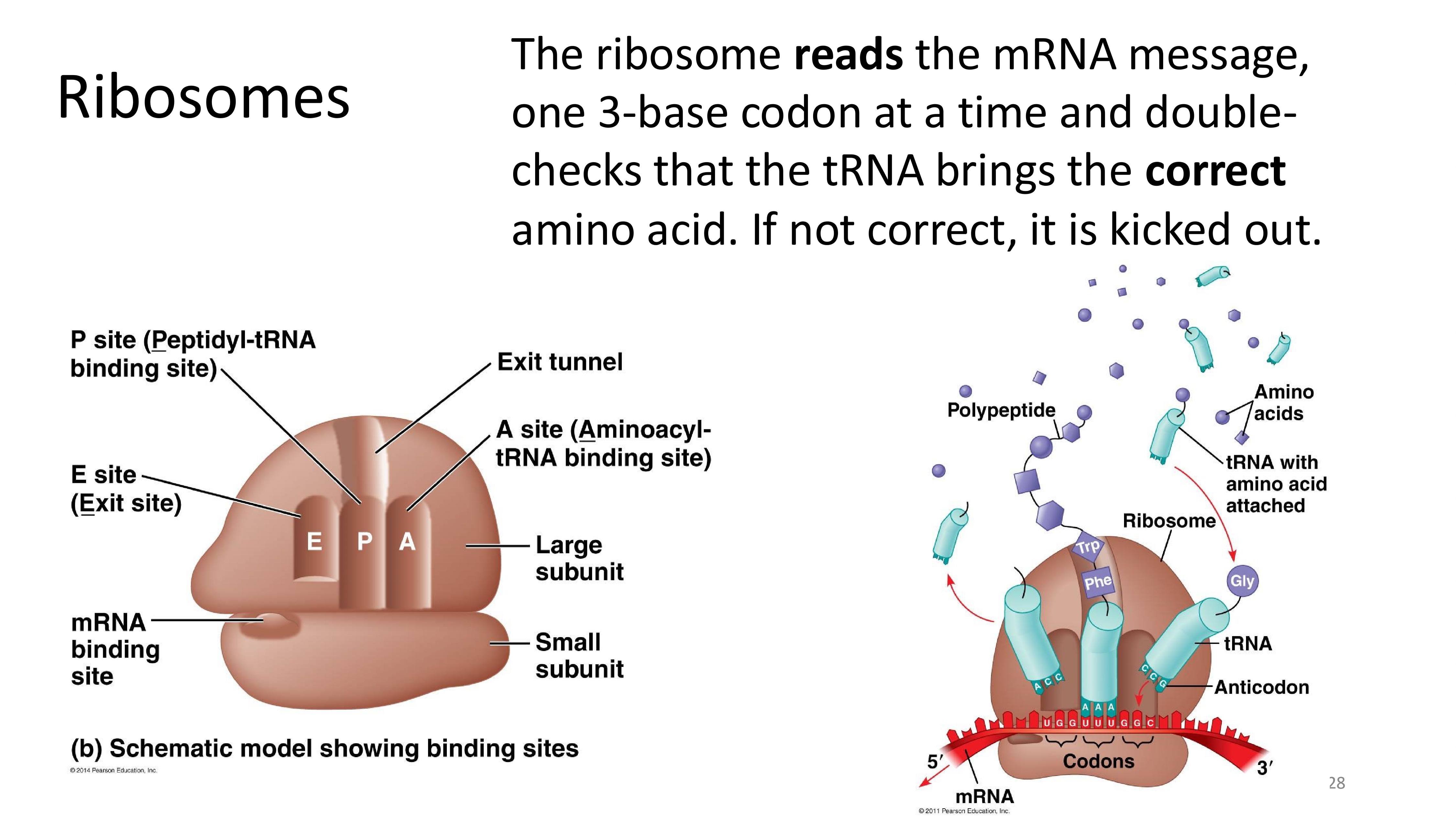
ribosomes
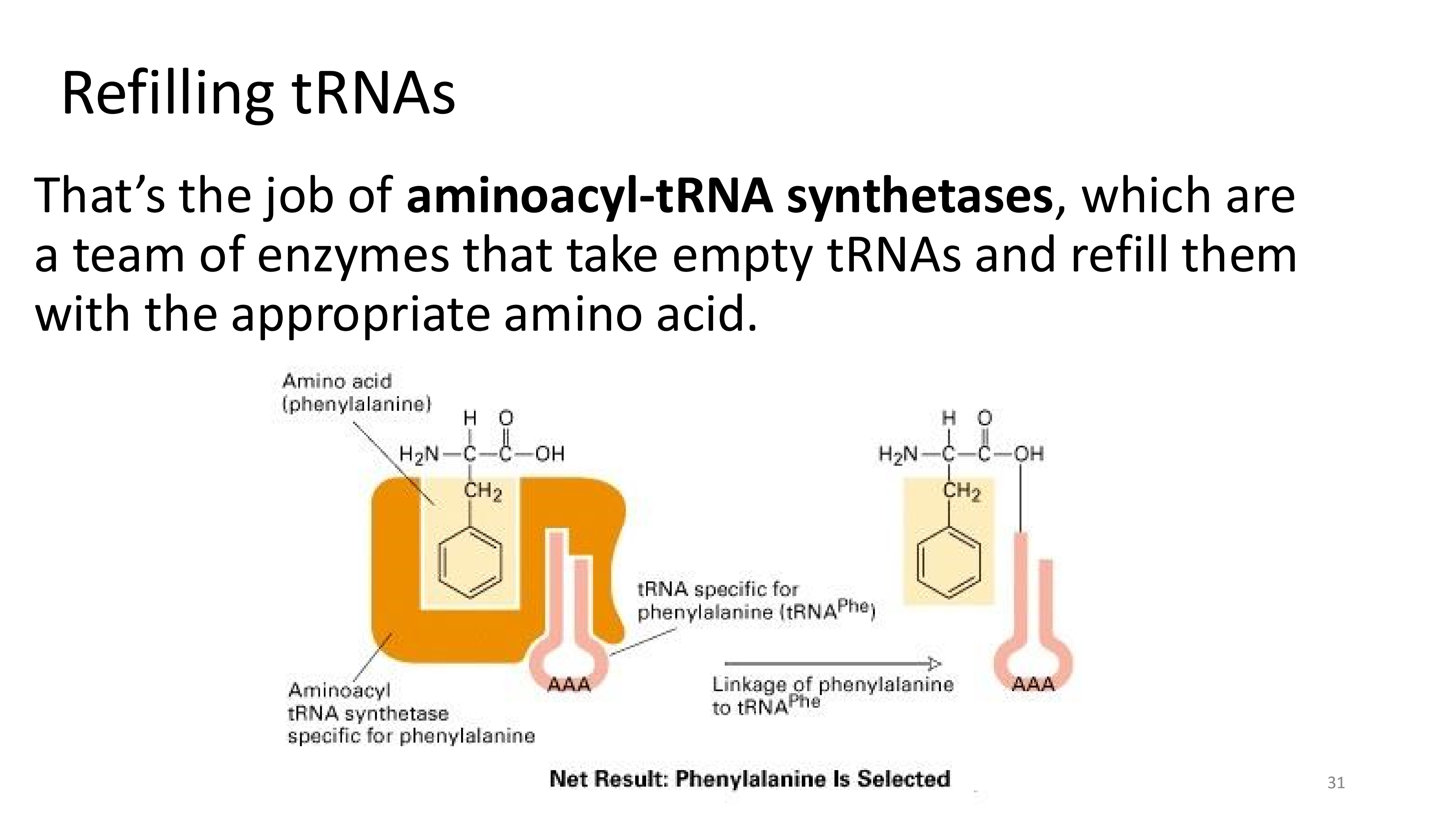
how are tRNAs refilled?
proteins vs RNA
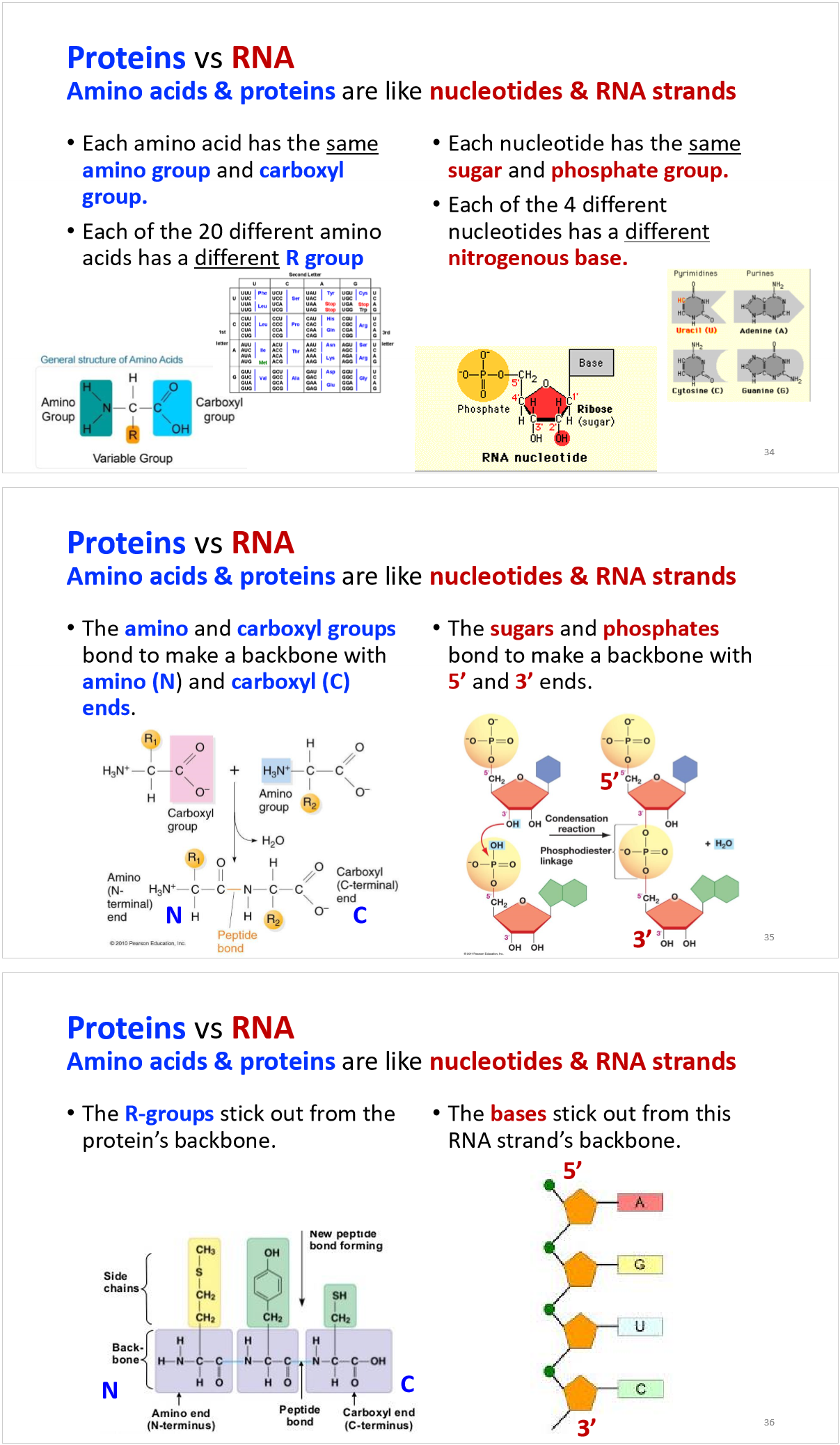
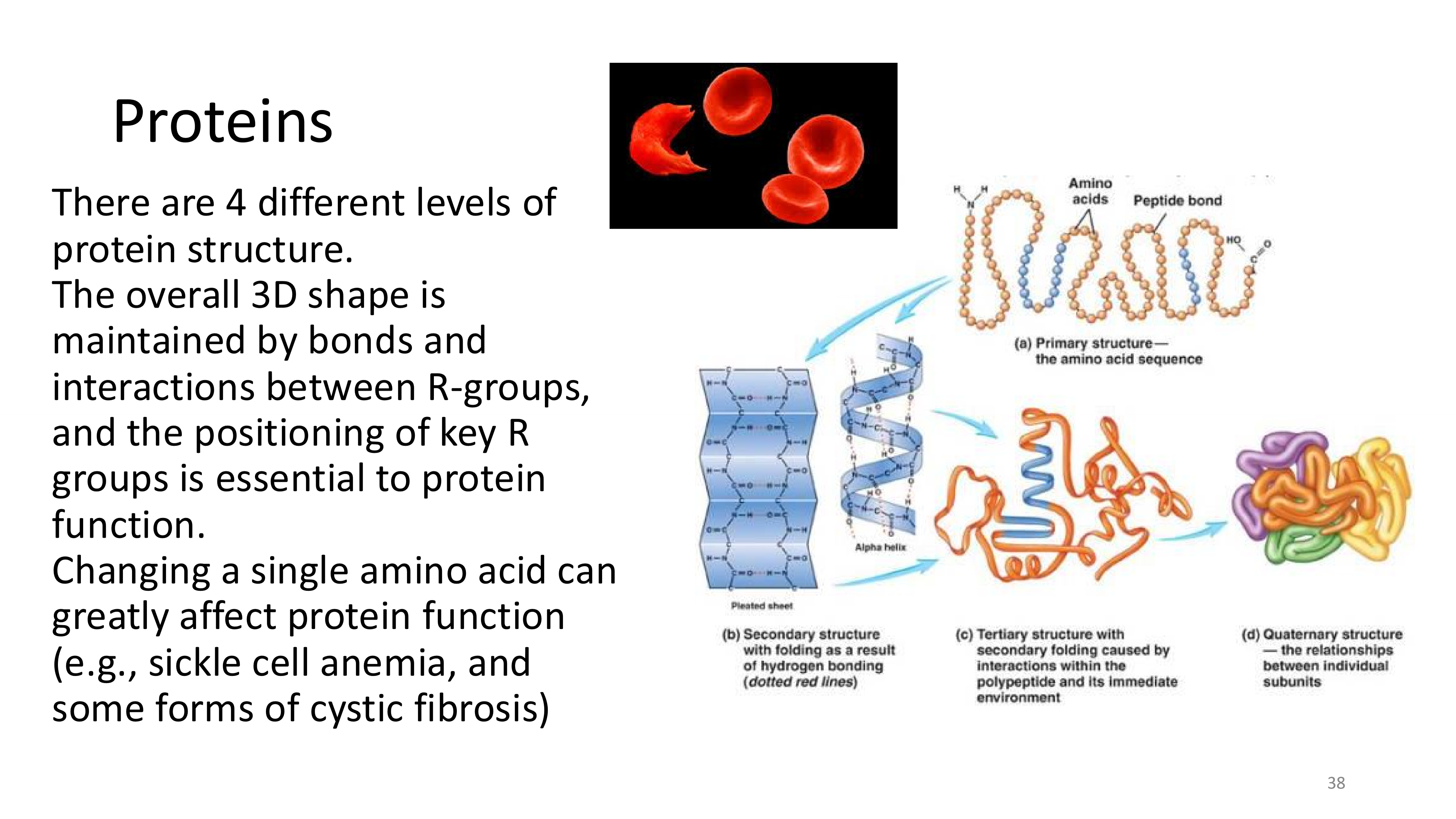
how are the amino acids determined in translation?
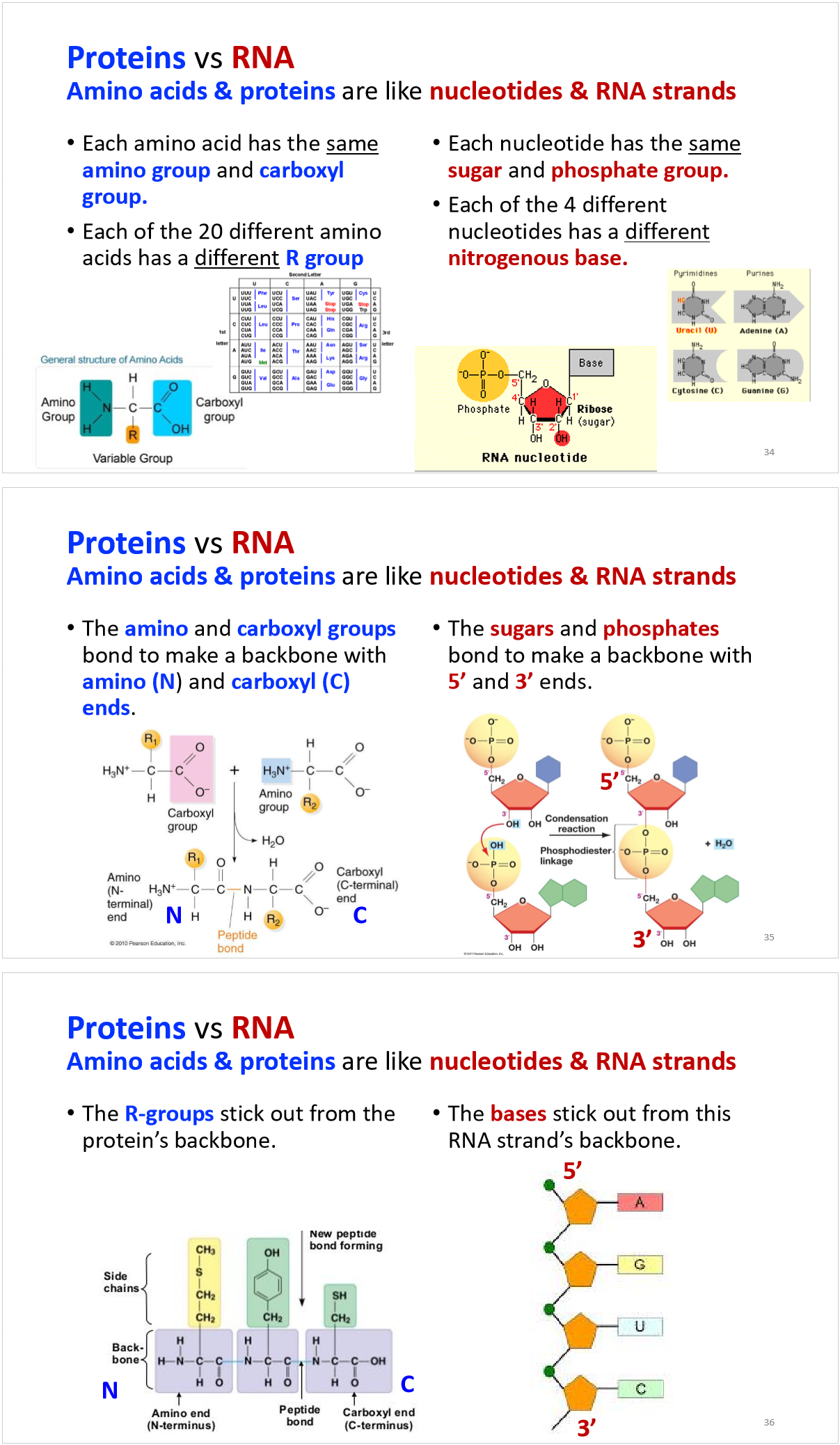
proteins

E. all of the above can be affected
translation main steps

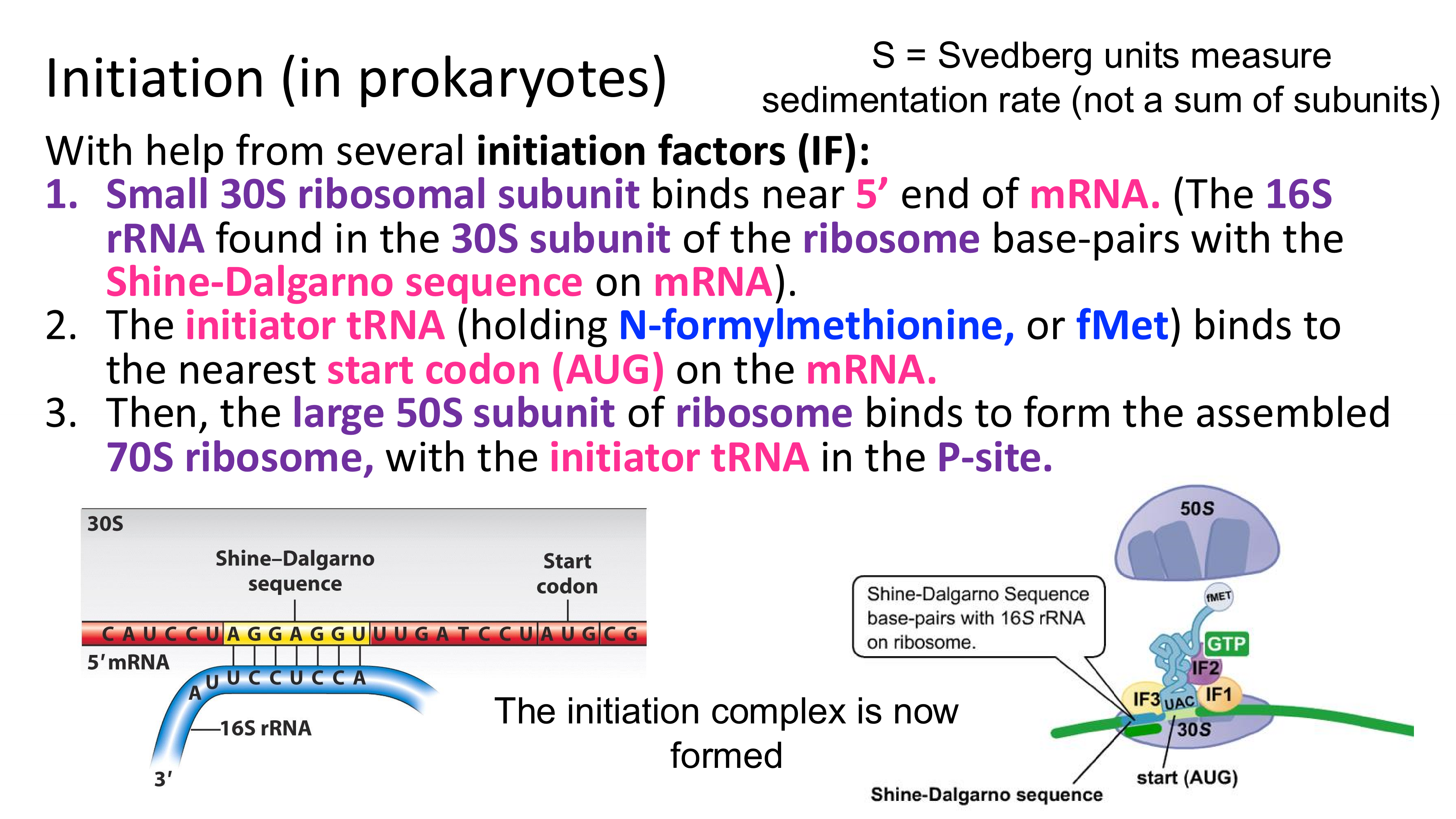
translation initiation in prokaryotes
translation initiation in eukaryotes
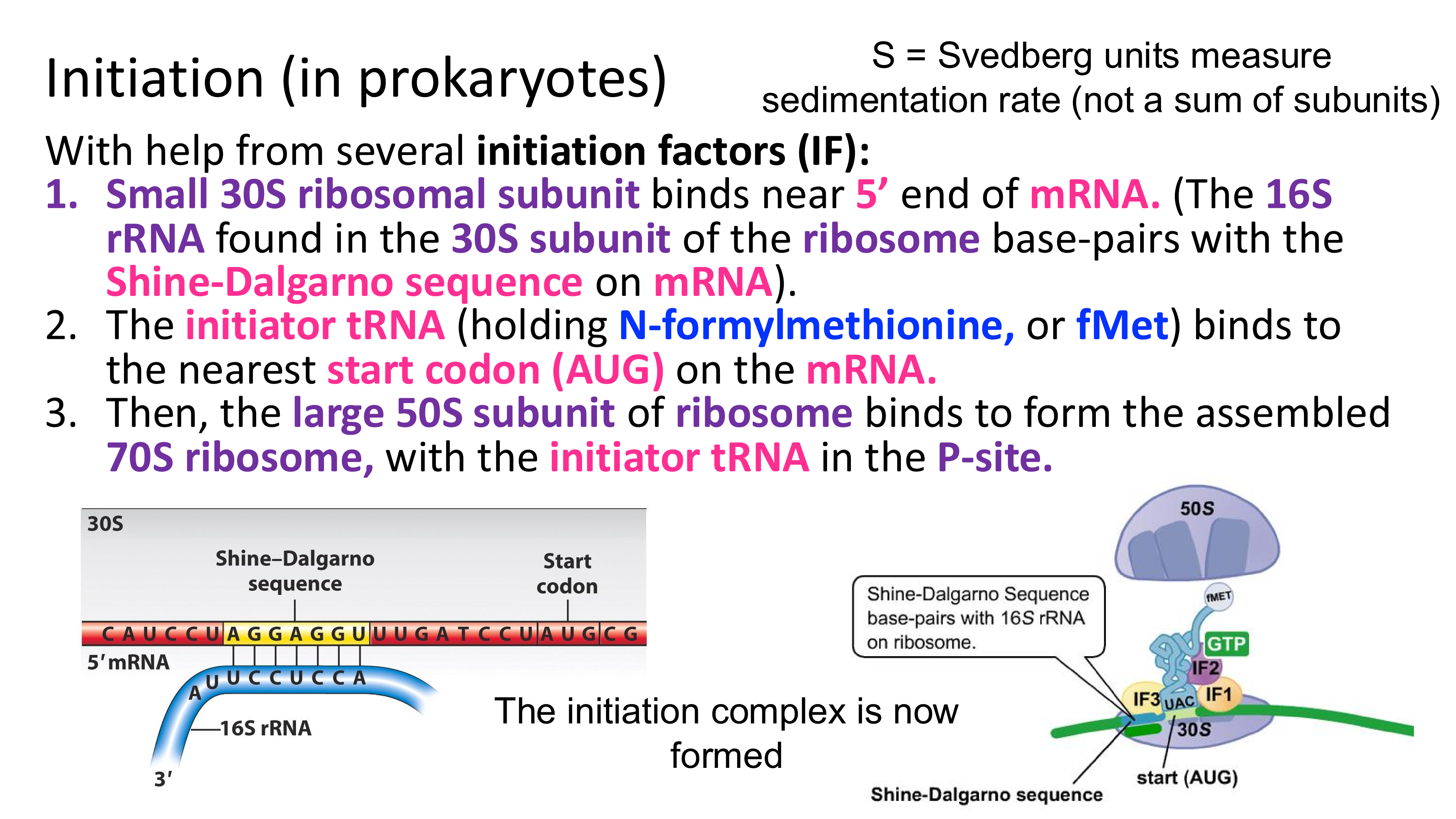
in prokaryotes, how does the small ribosomal subunit “find” the first AUG on RNA (so that it can set the reading frame at this start codon)?
it binds to the Shine-Dalgarno sequence (the start codon is slightly downstream)
in eukaryotes, how does the small ribosomal subunit “find” the first AUG on RNA (so that it can set the reading frame at this start codon)?
it joins tRNAMet and “scans” the mRNA from 5’ to 3’
once the small ribosomal subunit is bound to mRNA and initiator tRNA is bound to the start codon (AUG) what happens next?
the large ribosomal subunits binds, and initiation is complete
translation elongation in prokaryotes
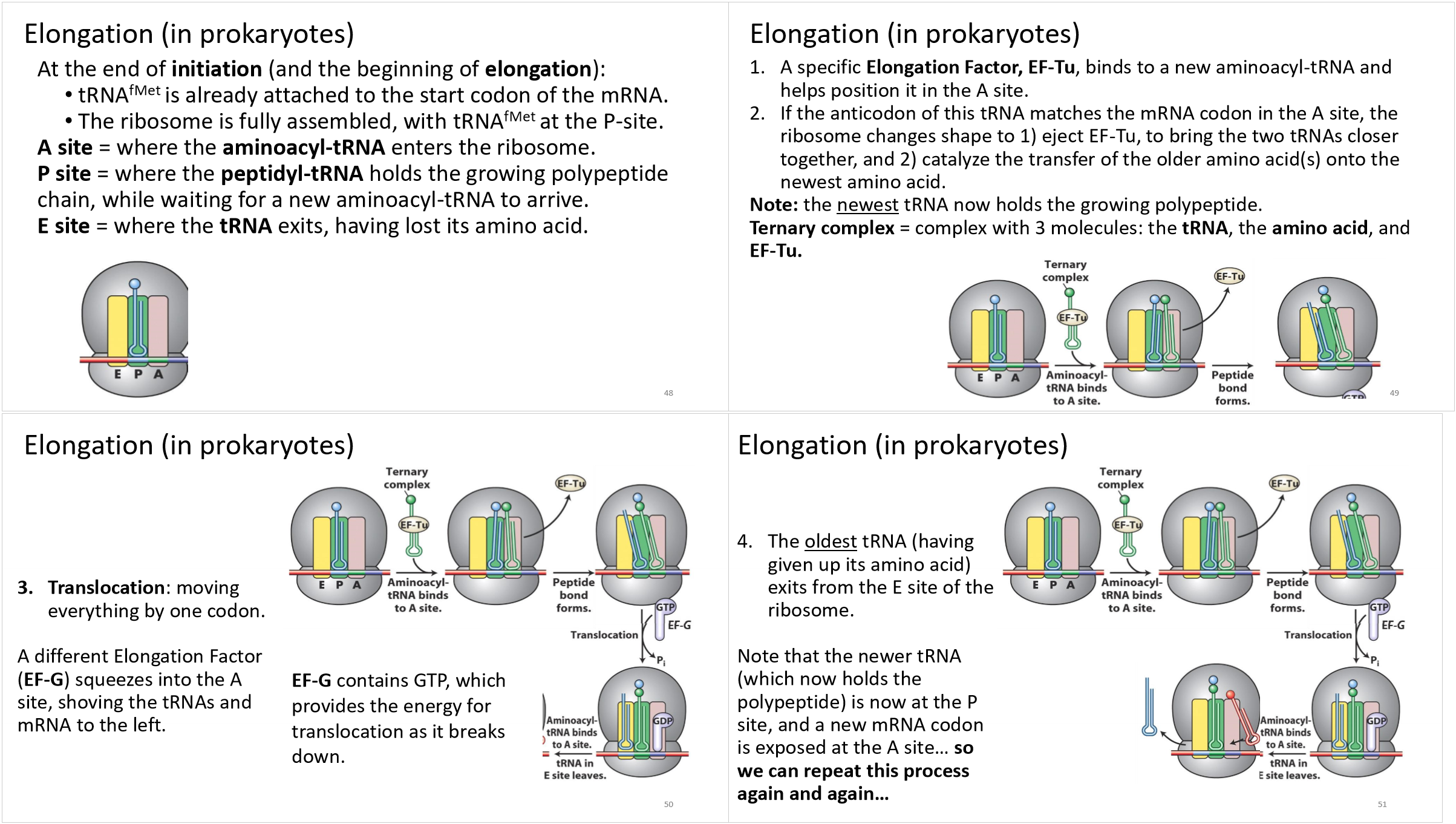
when will translational elongation end?
when the stop codon reaches the A site
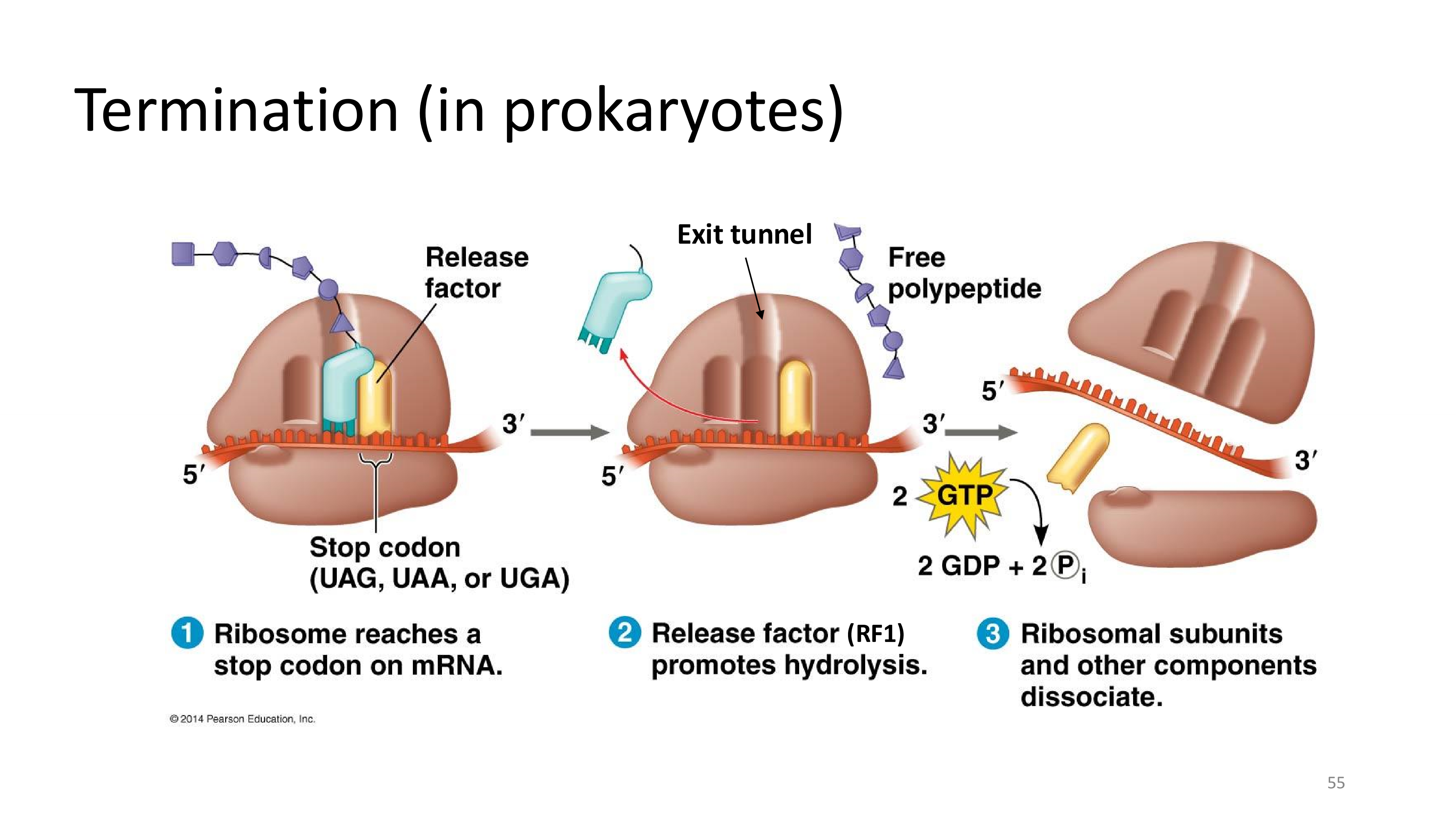
translation termination in prokaryotes
basic facts to remember about translation
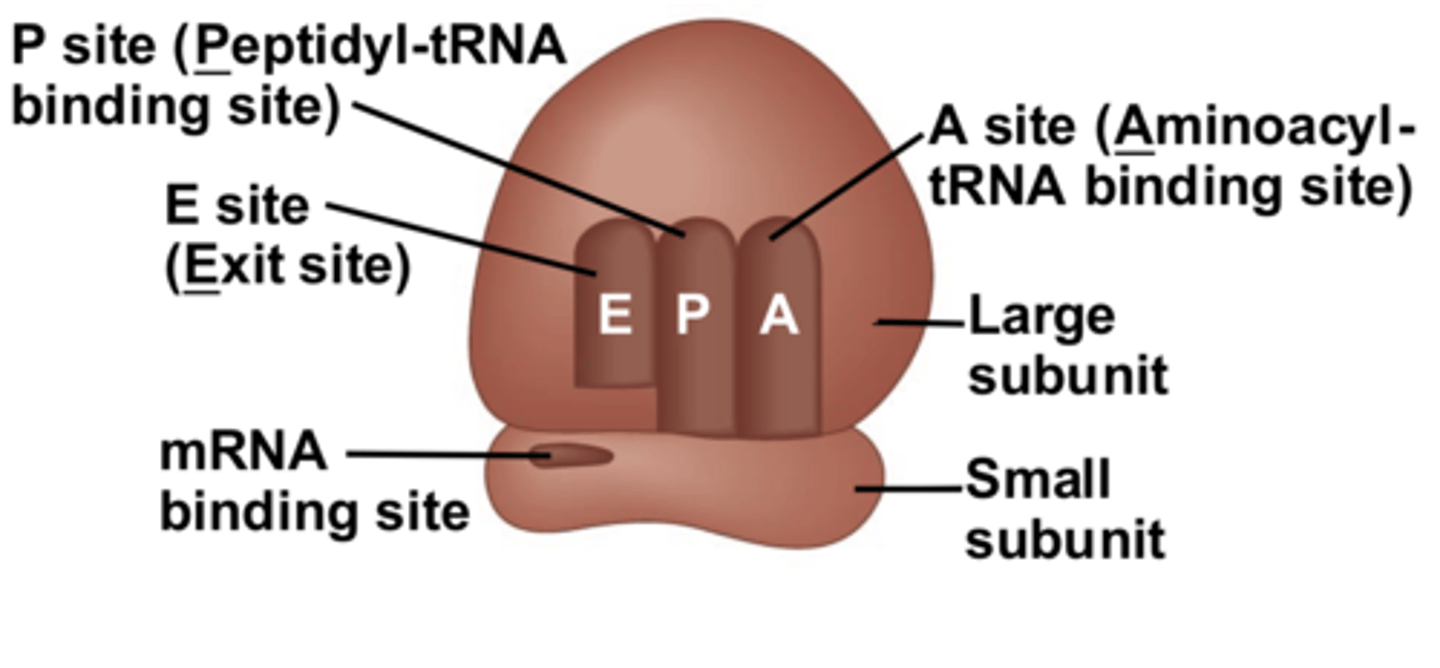
sites in the large subunit of the ribosome
- A site (Aminoacyl): where tRNA first attach to the ribosome
- P site (Peptidyl): where the newly arrived amino acid is removed from the tRNA and added to the peptide by a peptide bond
- E site (Exit): where the tRNA is ejected and where the polypeptide is guided and released
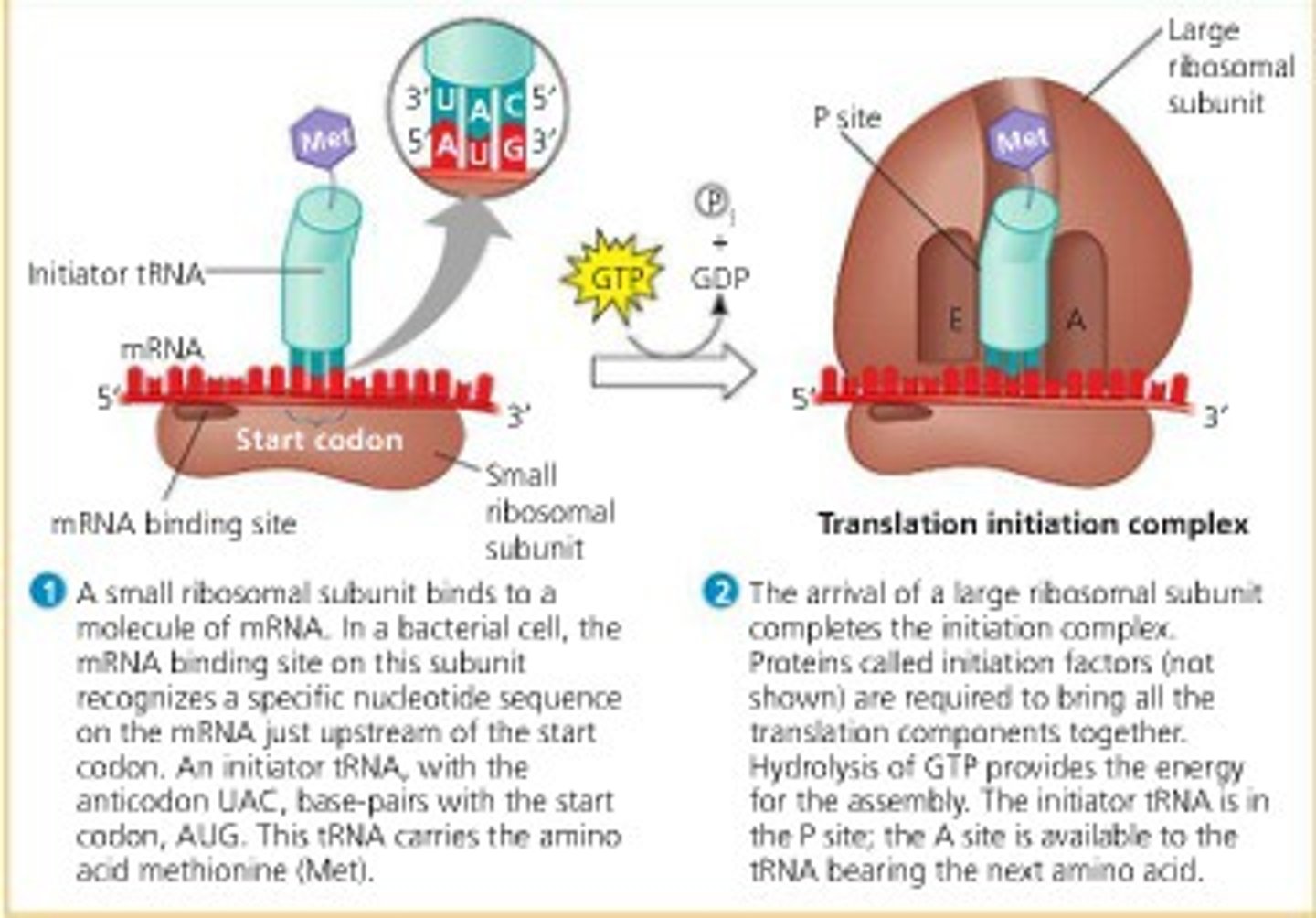
initiation in translation
mRNA binds to the small ribosomal subunit.
tRNA with methionine binds to the start codon (AUG).
Large ribosomal subunit attaches, completing the initiation complex.
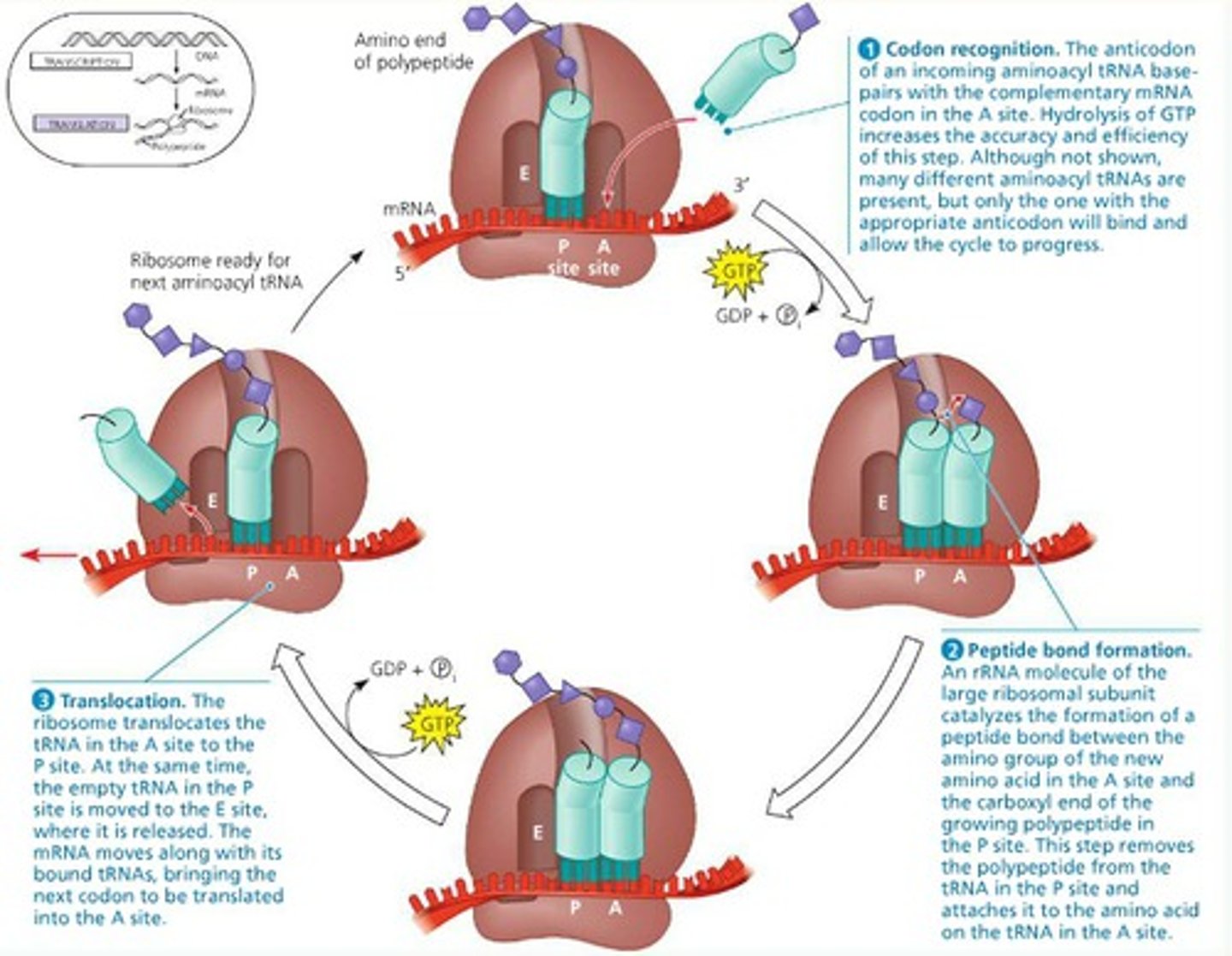
elongation in translation
tRNA brings amino acids to the A site.
A peptide bond forms between amino acids in the P and A sites.
Ribosome moves, shifting tRNA from A site to P site.
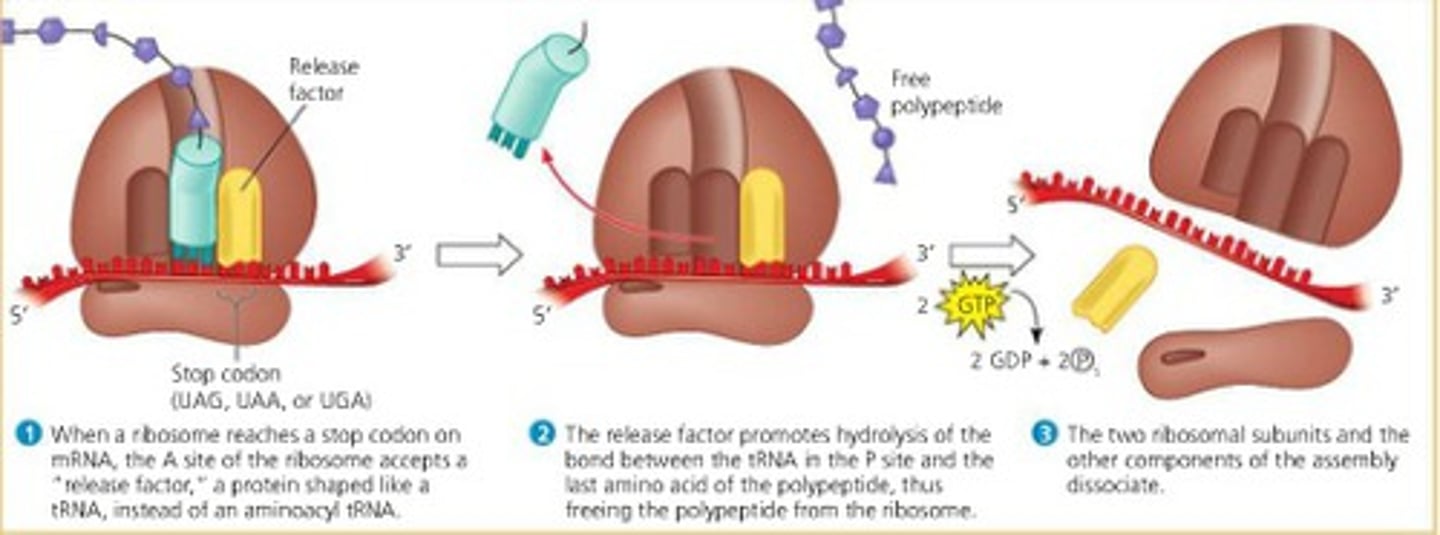
termination in translation
- occurs when the codon in A-site is a stop codon
- release factor places a water molecule on the polypeptide chain and thus releases the protein
why is the genetic code chart important?
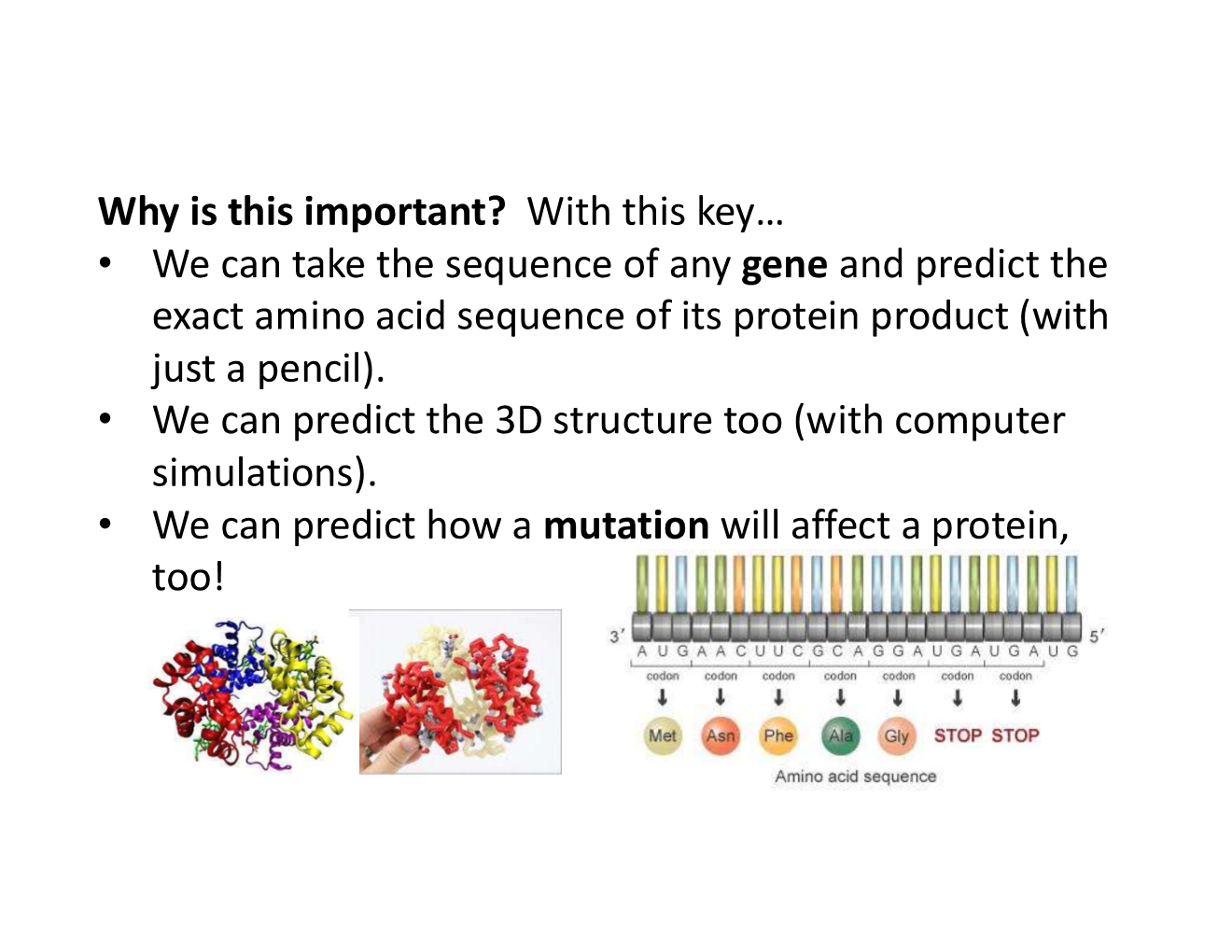
why was the genetic code chart challenging?

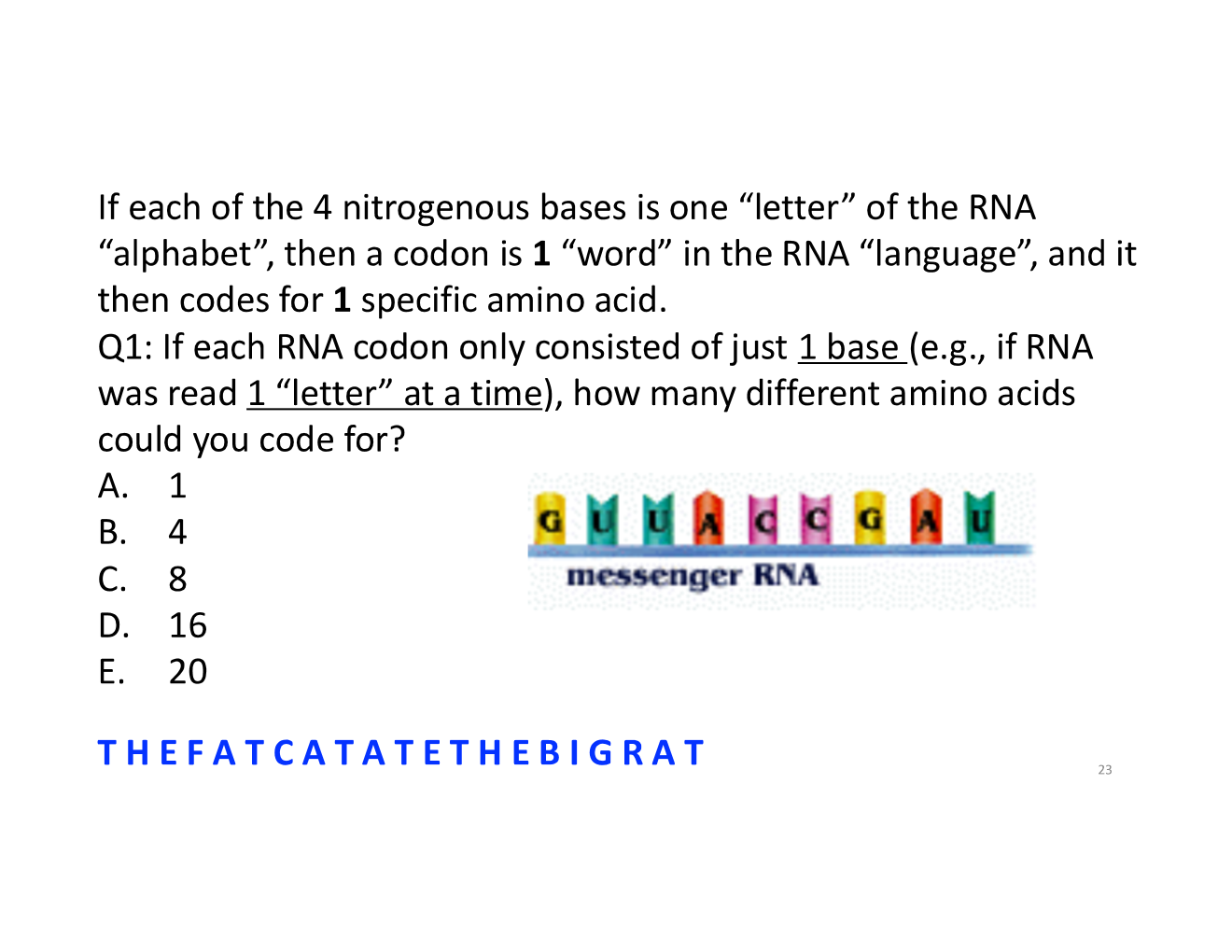
4; there are only 4 bases, so if it was read 1 letter at a time there are 4 possible amino acids to code for
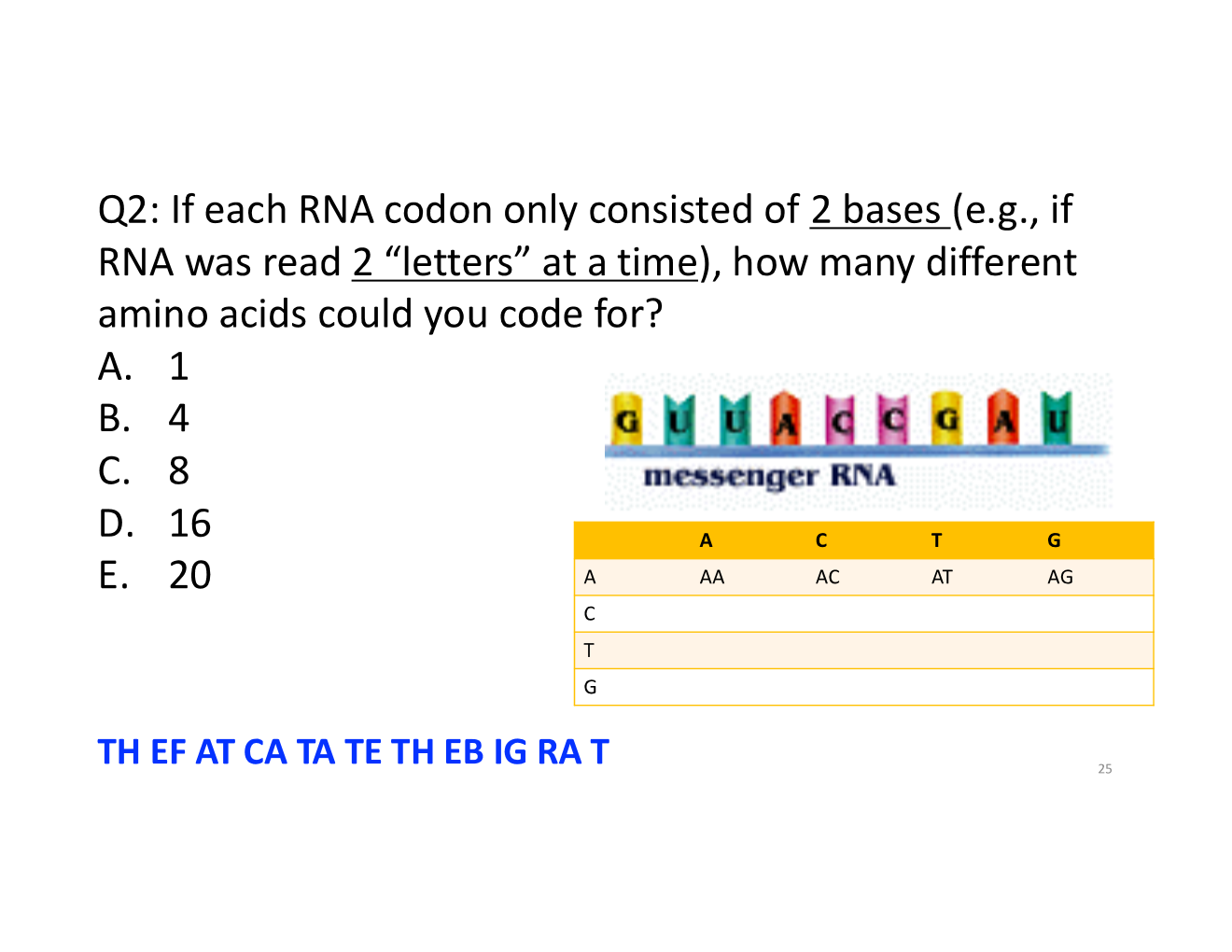
16

64
Brenner and Crick







How would genetic translation experiments differ in an organism with a 4-base codon reading frame?
Codons would consist of 4 bases, not 3.
There would be 256 possible codons.
Frameshifts would occur with insertions/deletions not in multiples of 4.
mRNA translation would rely on sequences divisible by 4.
Amino acid coding and mutation effects would differ from Earth-based systems.