Cell Biology Exam 1
1/58
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
59 Terms
How do amino acids come together to create proteins?
They link through the formation of a covalent peptide bond
What are the four categories of amino acids?
Acidic (negatively charged) polar
Basic (positively charged) polar
Uncharged polar
Uncharged nonpolar
How is the folding of proteins determined?
Steric restrictions confine the energy minima for the bond angles in polypeptides to a narrow range
The folding is determined by many different sets of weak noncovalent bonds that form between one part of the chain and another (backbone and side chain atoms)
hydrogen bonds, ionic bonds (electrostatic), van der Waals attractions, and hydrophobic forces
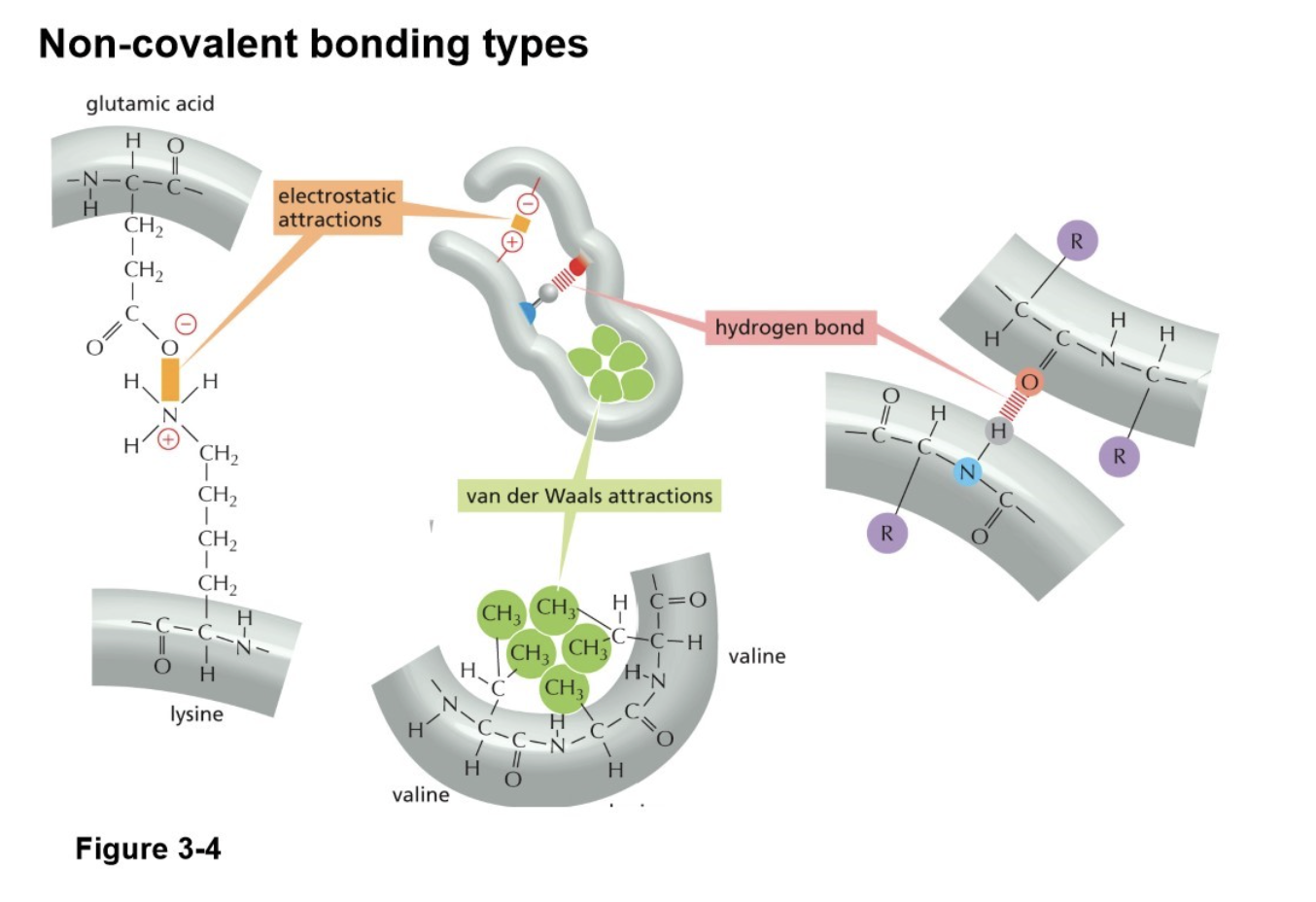
What is hydrophobic force?
Hydrophobic molecules, including nonpolar side chains of amino acids, tend to be forced together in an aqueous environment
Therefore, an important factor governing folding is the distribution of its polar and nonpolar amino acids, as nonpolars will cluster toward the inside
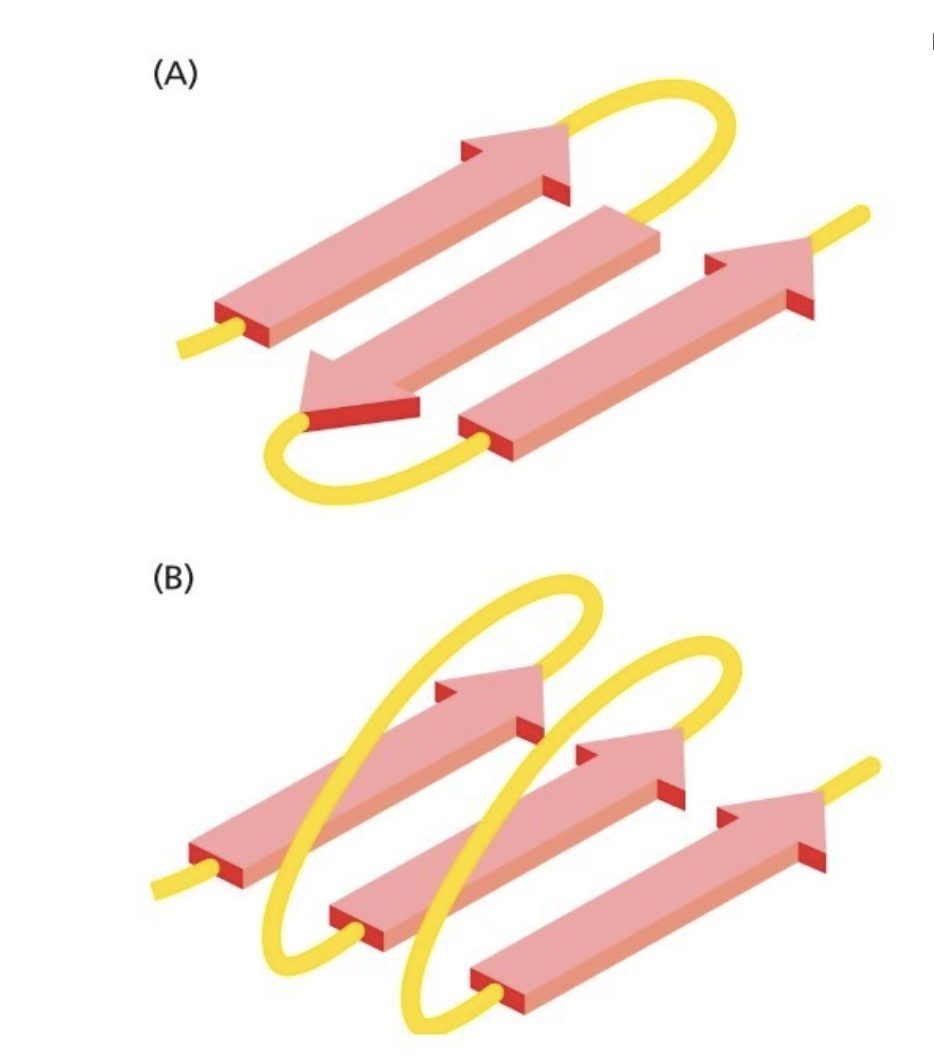
Parallel and antiparallel beta sheets
A is antiparallel, B is parallel
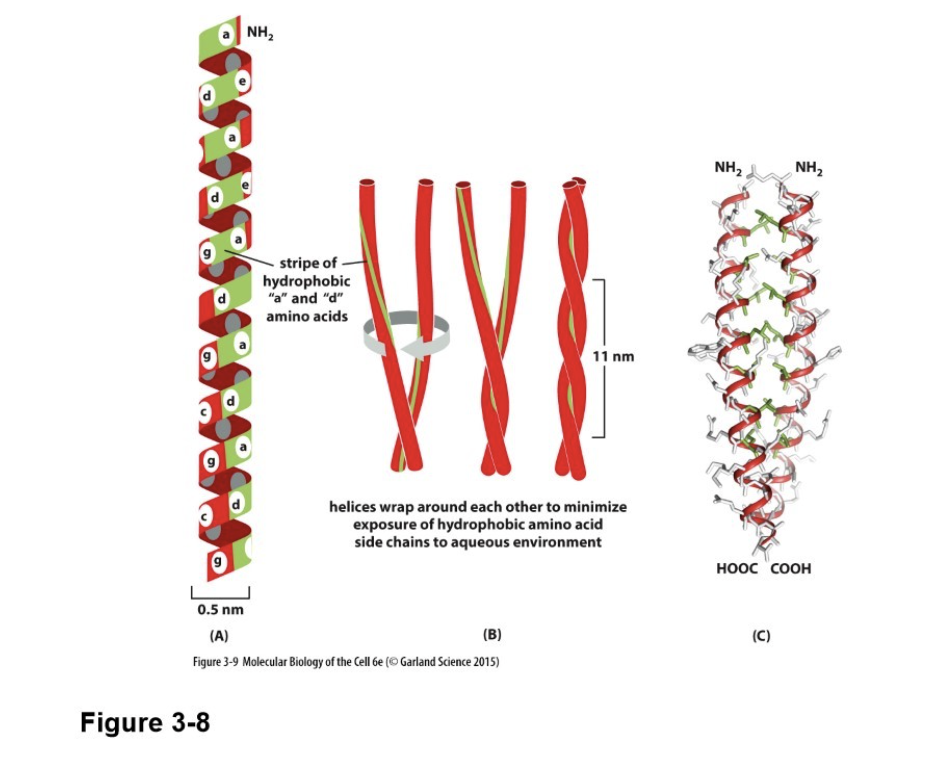
What is a coiled-coil, and what force causes it?
Alpha helices can wrap around each other to form a particularly stable structure known as a coiled-coil
Caused by hydrophobic forces to move nonpolar side chains towards inside
What is the primary structure of proteins?
The amino acid sequence
What is the secondary structure of proteins?
Alpha helices
Beta sheets
What is the tertiary structure of proteins?
Its final, 3D shape
What is the quaternary structure of proteins?
Individual proteins (subunits) binding to each other to create a larger protein complex, such as a dimer
Why are the secondary structures of proteins common?
Because they can form due to hydrogen bonds between backbone atoms
Why is the alpha helix “regularly” spaced?
Because the C=O of one amino acid bonds with the N-H of the amino acid exactly 4 amino acids away
What is special about an alpha helix with entirely hydrophobic side chains?
They can cross membranes if they have at least 20 amino acids
Membranes are super hydrophobic as well
Protein domains vs subunits
Domains: sub-structure of a protein that folds more or less independently (within a chain)
Typically encoded by a single exon
Subunits: an individual protein within a protein complex
Subunits exist within protein complexes, and domains exist within subunits/proteins
Protein complex levels
Dimer: two subunits
Trimer: three subunits
Tetramer: four subunits
Homodimer vs Heterodimer
Homodimer: forms from two identical subunits, symmetrical
Heterodimer: forms from different subunits
What is self-assembly?
The spontaneous organization of individual proteins into larger, functional structures driven by specific molecular interactions (like hydrogen bonds, hydrophobic effects, charge) and physical forces (like crowding) within a cell
What is exon shuffling/domain shuffling?
Eukaryotic proteins are often the result of exon shuffling, in which a sequence encoding a particular domain has been inserted into many different genes, creating proteins with a modular structure, which means that a protein has different regions that carry out specific types of function
Why the same domain shows up in many different proteins
What is a protein module?
A common protein domain
What are protein families?
Proteins can be classified into many families, each family member having an amino acid sequence and 3D conformation that resembles other members
Each is encoded by genes derived from the same ancestral gene (homologous)
Evolutionary process: arises primarily through gene duplication, where an extra gene copy accumulates mutations and diverges to form new functions
Must be at least 30% identical
What are serine proteases?
A large, essential family of enzymes that cut (cleave) peptide bonds in proteins
What is a cell culture?
A homogeneous population of cells growing in a lab (the cells are all the same)
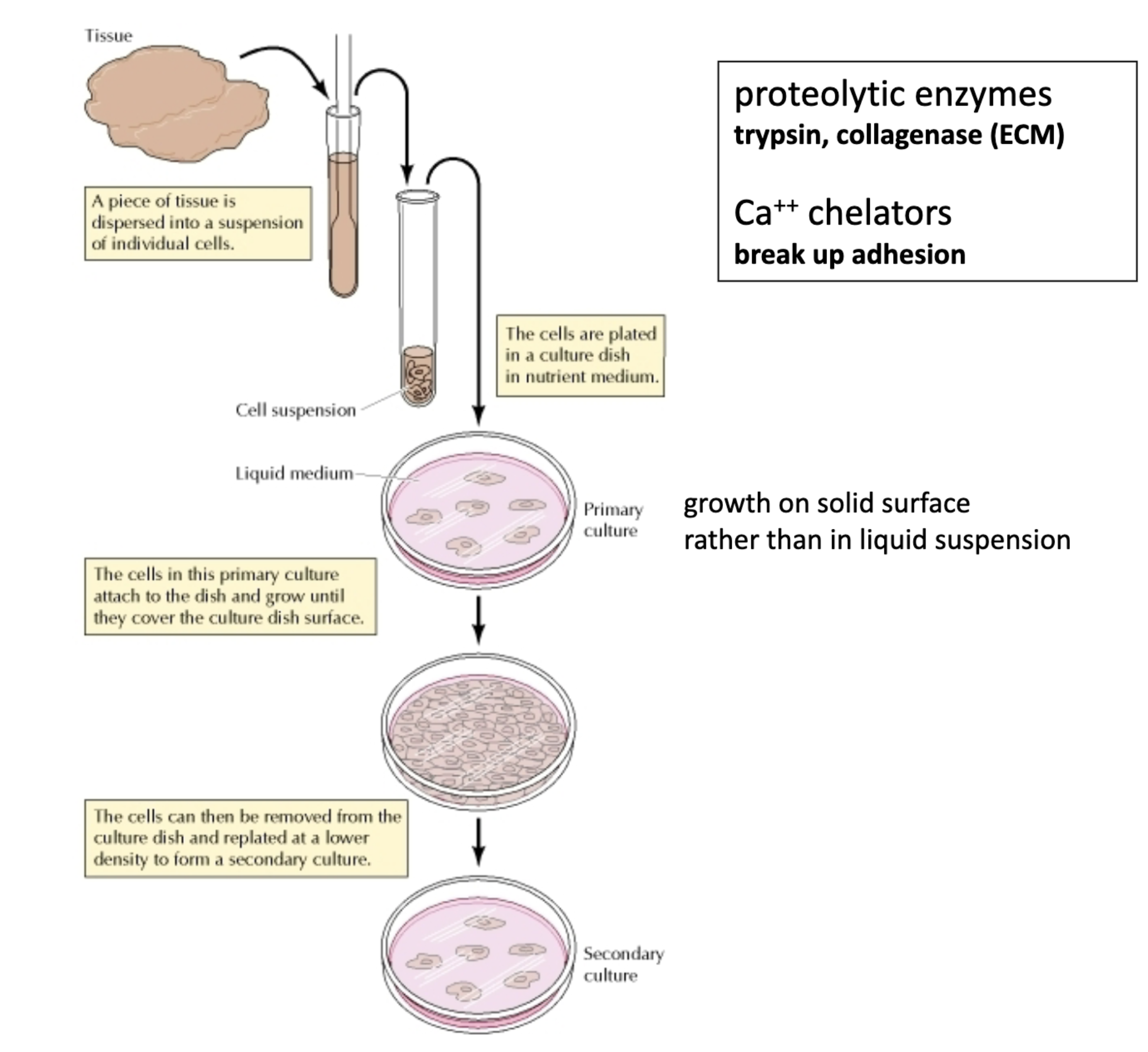
How is a cell culture made from a tissue?
Isolate and separate the cells
Disrupt ECM --> cell suspension
Proteolytic enzymes – degrade ECM proteins (ex: collagen (collagenase))
EDTA – binds to calcium ions (Ca2+), which are required for cell-cell and cell-ECM junctions; EDTA breaks these contacts
Make homogenous (ie. One cell type)
Centrifugation (isolated by size)
Grow in a defined medium (isolated by growth properties)
Fluorescence-activated cell sorting (FACS) (isolated by light scattering and fluorescent characteristics)
Expand cells in the lab
Plastic petri dish (growth medium, bottom coated with ECM, helps give cells something to grow on/stick to
Standard medium: growth factors include salts, glucose, amino acids, vitamins, and bovine serum
Defined medium: contains all the above growth factors, but no serum, and includes a specific growth factor
Make cell line (a cell clone, derived from one cell)
Can also make cells immortal and freeze in liquid nitrogen (permanent stock)
Done by providing cells with the gene that encodes the catalytic subunit of telomerase
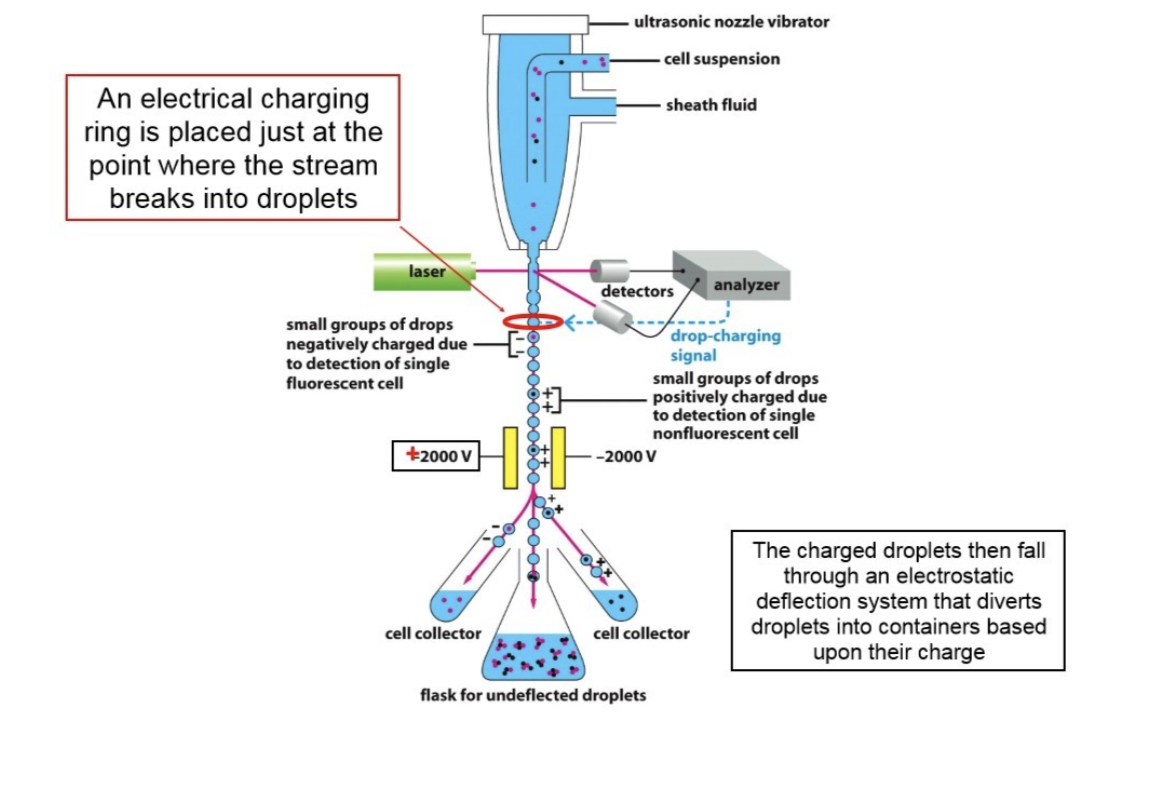
How does fluorescence-activated cell sorting (FACS) work?
Uses an antibody bound to the cell surface
Identifies and sorts heterogeneous cell mixtures into distinct, highly pure populations based on specific light scattering and fluorescent characteristics
Standard medium vs Defined medium
Standard medium: growth factors include salts, glucose, amino acids, vitamins, and bovine serum
Defined medium: contains all the above growth factors, but no serum, and includes a specific growth factor
What is a cell line? How can they be made immortal?
Cell clones derived from a single cell that are all genetically homogeneous
Can also make cells immortal and freeze them in liquid nitrogen (permanent stock)
Done by providing cells with the gene that encodes the catalytic subunit of telomerase
Immortal cell lines usually derived from tumors (tumor cell lines)
What is fractionation of cells and in what ways can it be done?
Cell lysis, disrupting the cell membrane to get cell extract (cell components)
Homogenization: shearing force mechanically disrupts the membrane
Sonication
Pestle in cell suspension (high-frequency sound waves)
Osmotic lysis (hypotonic solution)
What is fractionation of cell extract and in what ways can it be done?
Done on cell extract (rather than cell as a whole) to iolate organelles and protein complexes
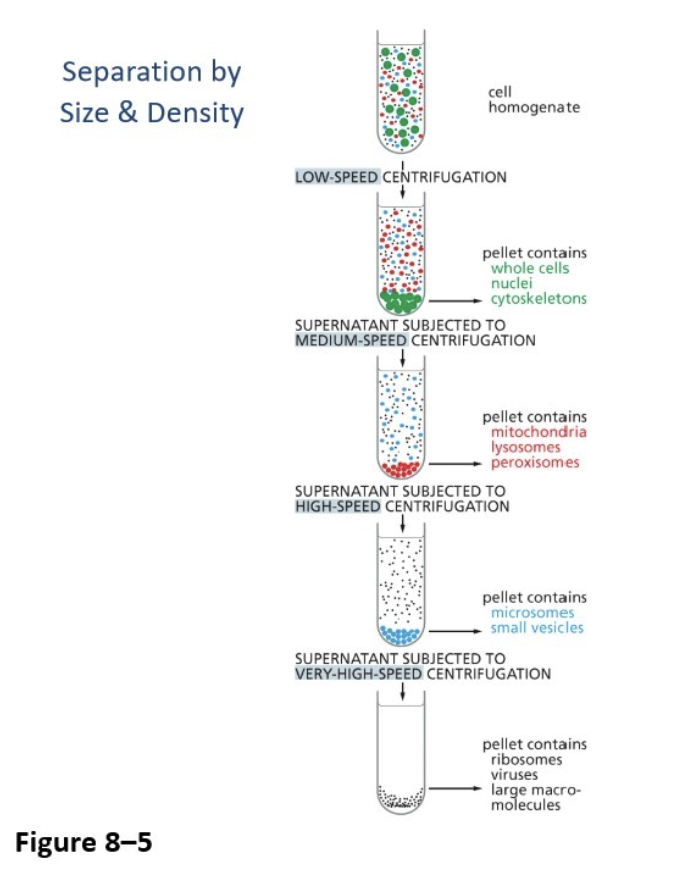
Ultracentrifugation (differential centrifugation)
Separates cell components by size and density based on the spin speed
Pellet is created at the bottom, and then you centrifuge the supernatant again (liquid above pellet)
Velocity centrifugation (sedimentation)
Provides a finer degree of separation (e.g., protein complexes), separating further by size and shape
Utilizes a solution that contains a gradient of sucrose or salt (for example, the top may be 5% while the bottom is 25%)
The larger and rounder components go to the bottom (no pellet)
Poke a hole at the bottom and let it drip through
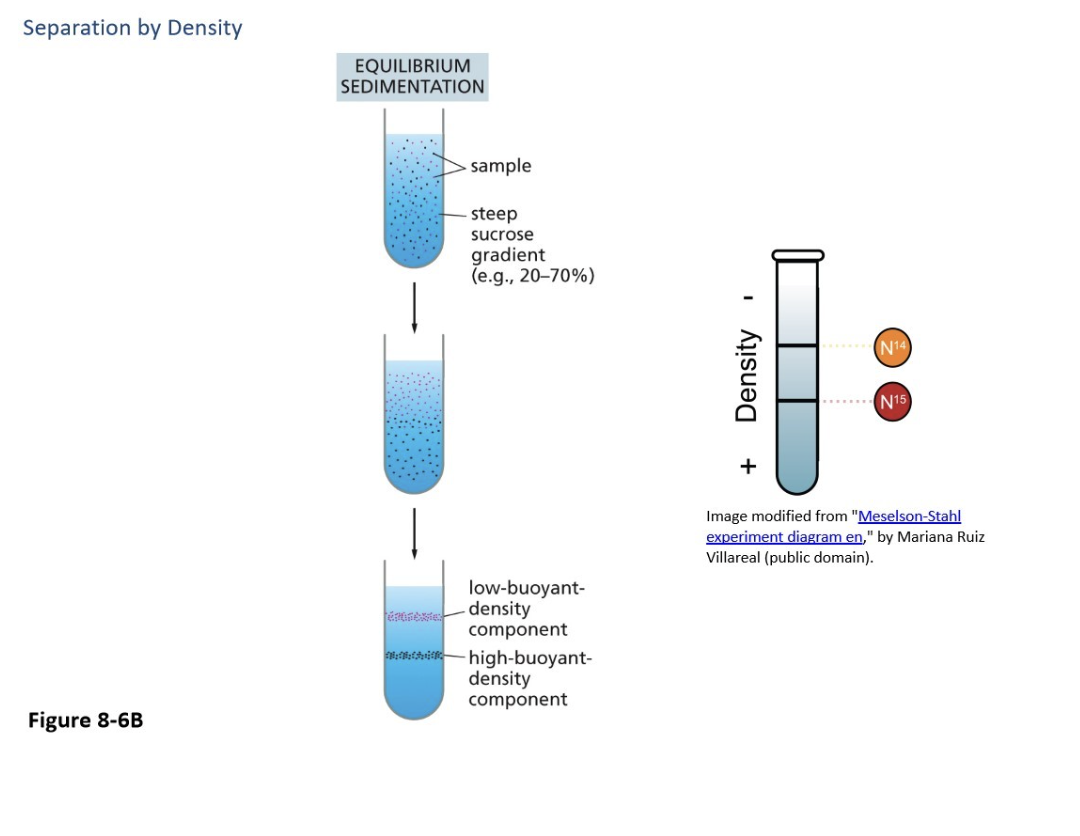
Equilibrium sedimentation
Provides the most fine degree of separation, separating by DENSITY rather than size and shape, can separate the same protein complexes
Also uses a sucrose or salt gradient, but is much higher (but more like 25% at top and 75% at bottom)
The denser ones move to the bottom
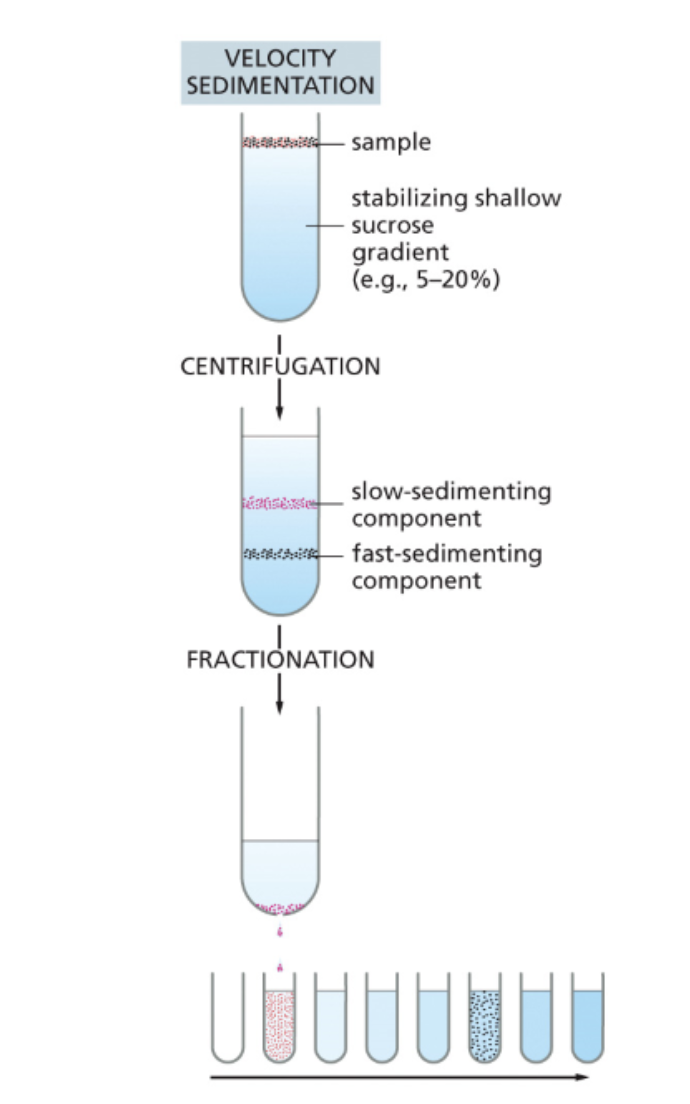
Velocity centrifugation
Provides a finer degree of separation (e.g., protein complexes). separating further by size and shape
Utilizes a solution that contains a gradient of sucrose or salt (for example, the top may be 5% while the bottom is 25%)
The larger and rounder components go to the bottom (no pellet)
Poke a hole at the bottom and let it drip through
Equilibrium sedimentation
Provides the most fine degree of separation, separating by DENSITY rather than size and shape, can separate the same protein complexes
Also uses a sucrose or salt gradient, but it is much higher (but more like 25% at the top and 75% at the bottom)
The denser ones move to the bottom
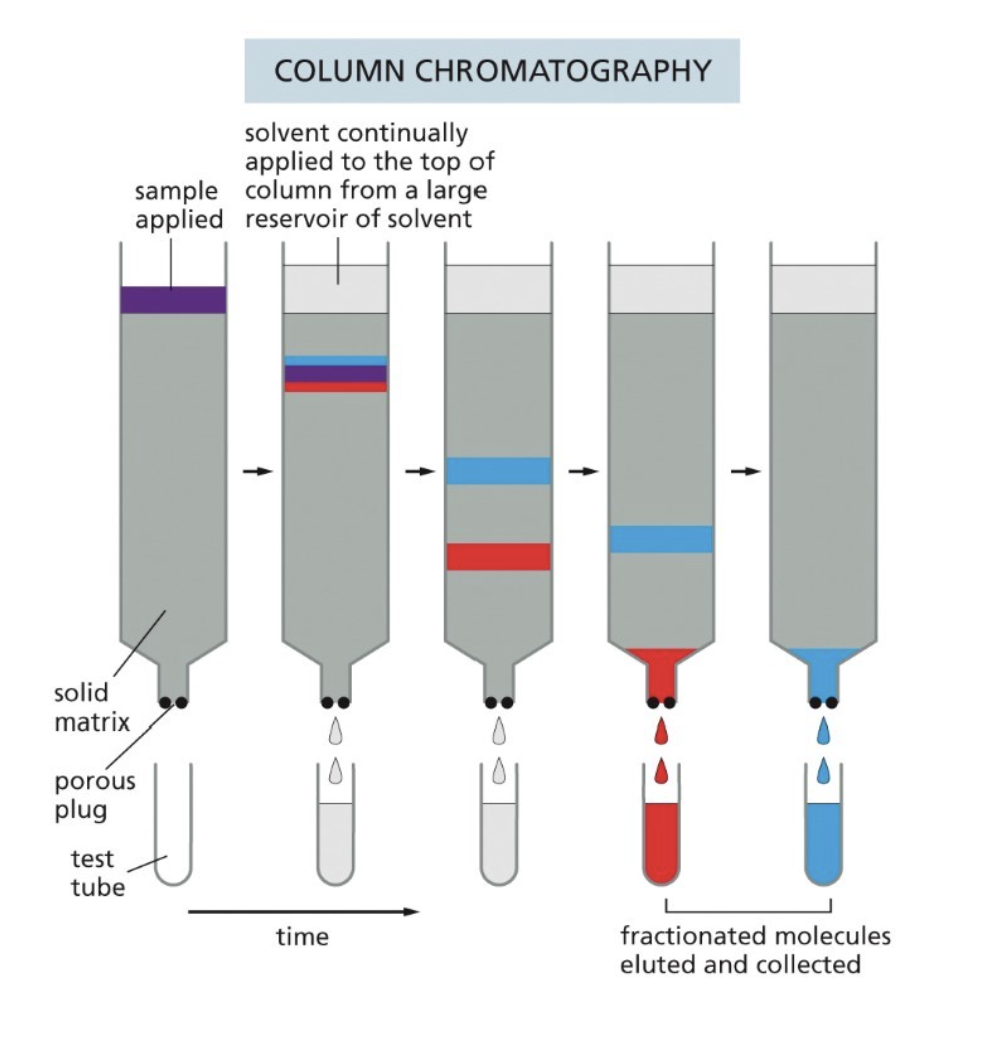
What is fractionation of proteins and in what ways can it be done?
Separates complex protein mixtures into simpler, distinct fractions based on physical and chemical properties like size, charge, solubility, and affinity
Column chromatography
Rate of passage through the column depends on interaction with the beads
The surface of the beads are designed to interact with proteins in some particular ways (such as with charges), three ways they can interact:
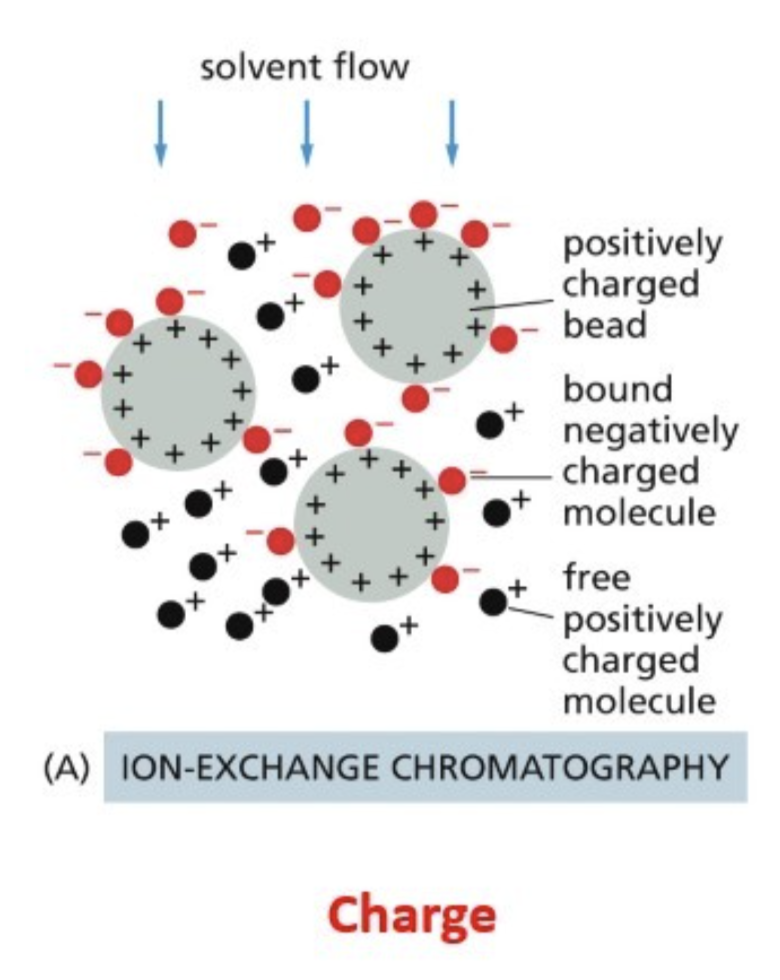
Ion exchange — beads interact with the charge of a protein
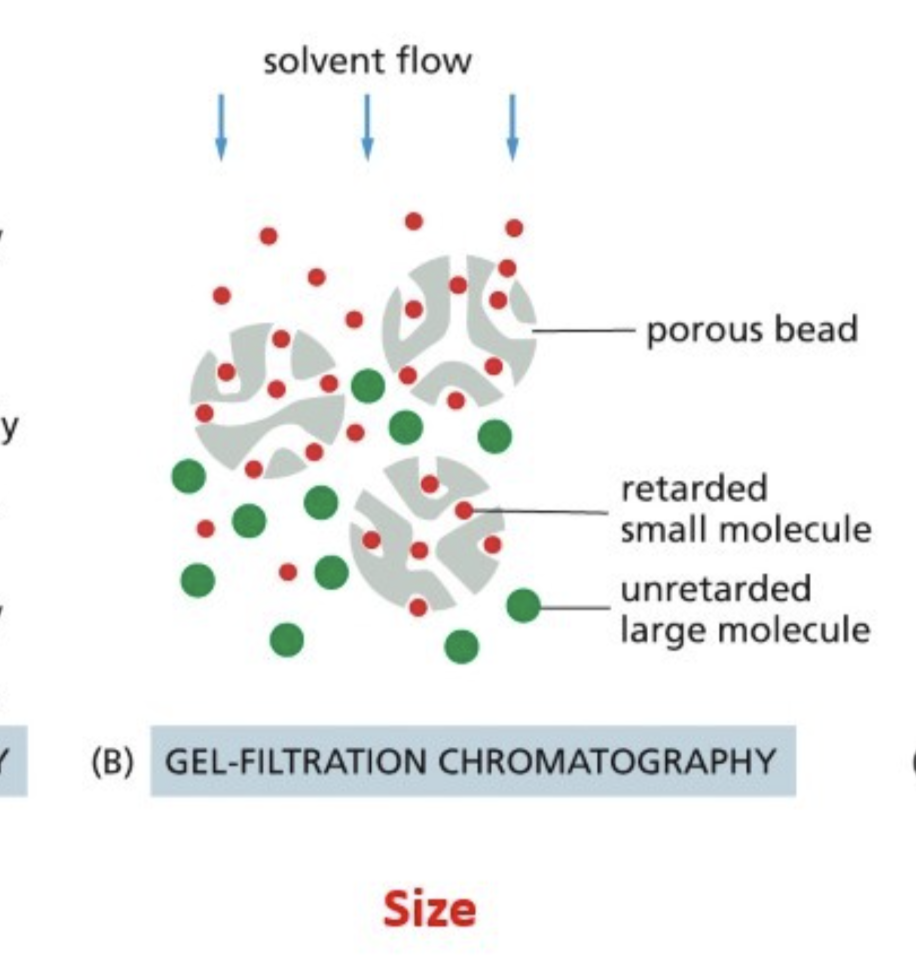
Gel filtration — beads have pores of a certain size, which trap proteins of a certain size
Affinity — beads have a conjugate that binds to the protein of interest, specific affinity to a conjugate
Such as an antibody (Ab) against the protein of interest
Antibody that binds to “affinity tag”
DNA - gene promoter sequence your protein binds to (for transcription factor)
Ion exchange chromatography
Beads interact with the charge of a protein
Gel-filtration chromatography
Beads have pores of a certain size, which trap proteins of a certain size
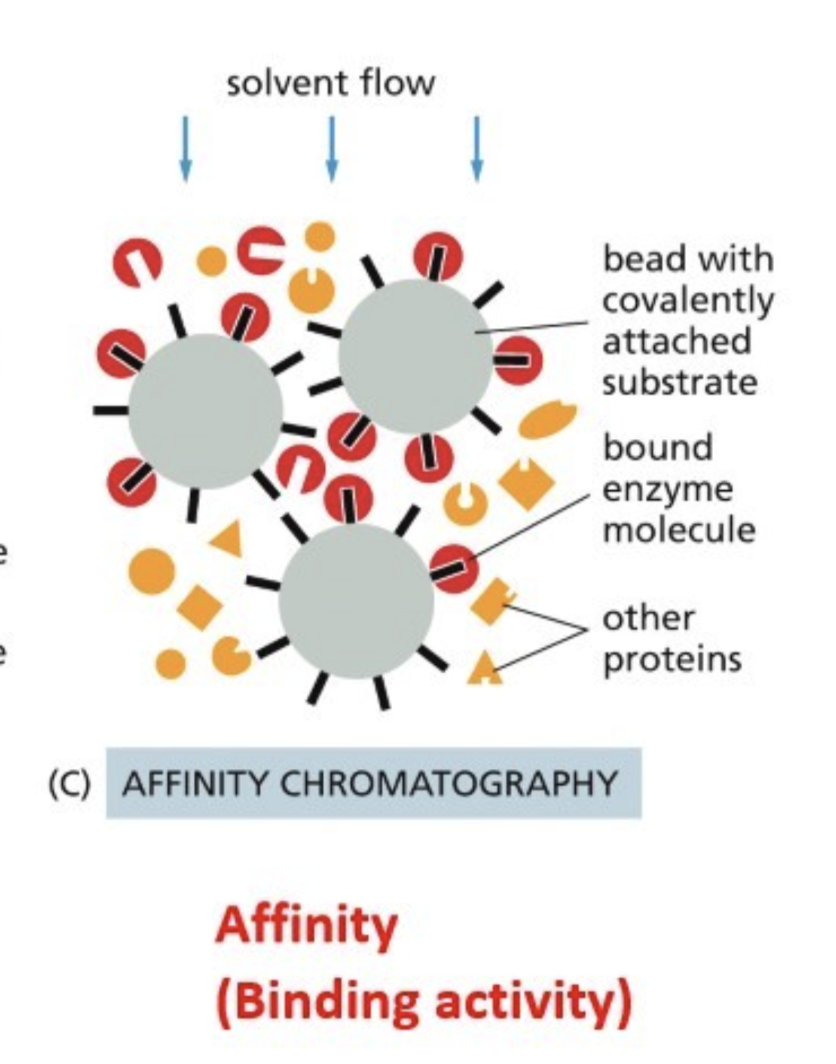
Affinity chromatography
Beads have a conjugate that binds to the protein of interest, specific affinity to a conjugate
Such as an antibody (Ab) against the protein of interest
Antibody that binds to “affinity tag”
DNA - gene promoter sequence your protein binds to (for transcription factor)
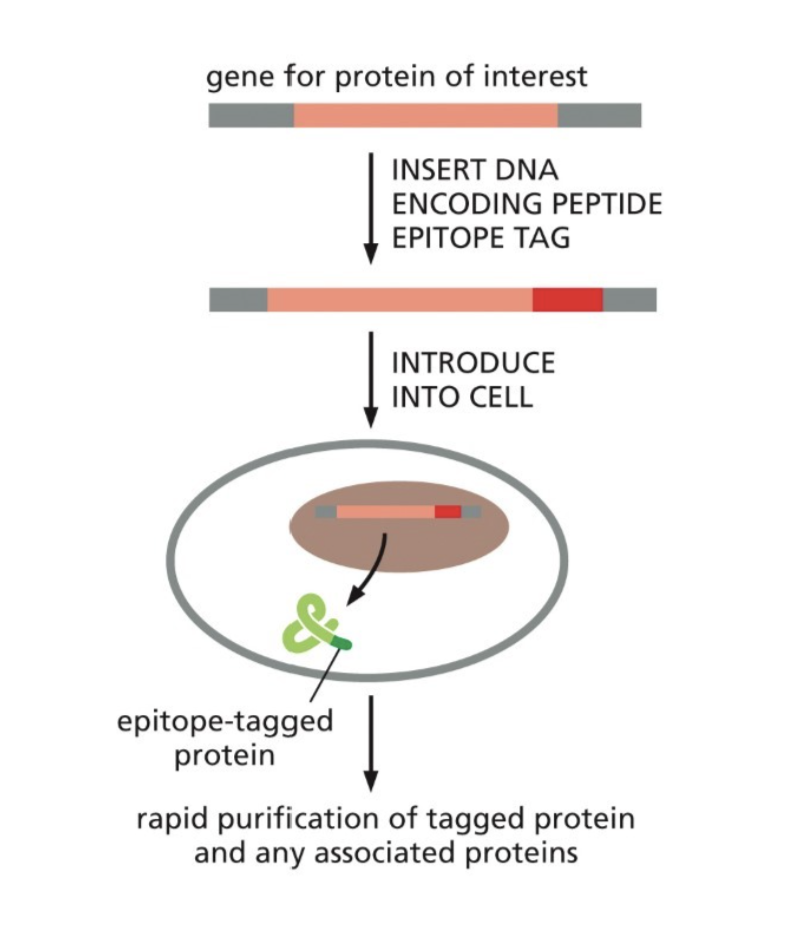
What is an affinity tag?
Used in affinity chromatography when there is no antibody for the protein
Allows for the use of an antibody that binds to the epitope tag
What is immunoblotting (western blotting) used for?
Identifies specific proteins by exposing all the proteins on the gel to a specific antibody that has been labeled with a radioactive isotope or fluorescent dye
What is two-dimensional gel electrophoresis used for?
Separates complex protein mixtures based on two independent properties: isoelectric point and molecular weight
What is mass spectrometry?
Used to identify unknown compounds, quantify known materials, and elucidate the chemical structure of molecules by measuring the mass-to-charge ratio of ionized particles
Resolving power
The ability of an optical instrument (microscope, telescope) or imaging system to distinguish and separate the images of two closely spaced objects
Human eyes — 0.2 mm
Light microscope — 0.2 um
Electron microscope — 2 nm (uses electron beam instead of lightwaves)
(Don’t need to know specific numbers)
Amplitude vs Wavelength
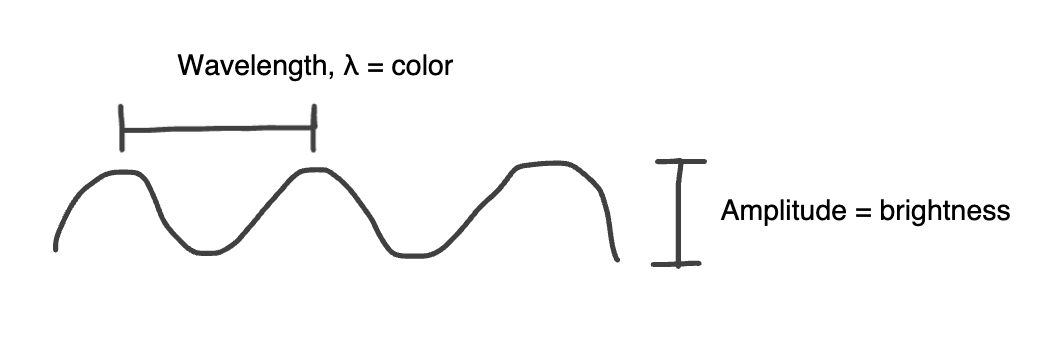
Bright field microscopy (light microscope method)
Because cells are transparent:
Fix cells: chemically treat cells, making holes in the membrane to make them more permeable to dyes and antibodies (Ab)
Stain cells: dyes and antibodies
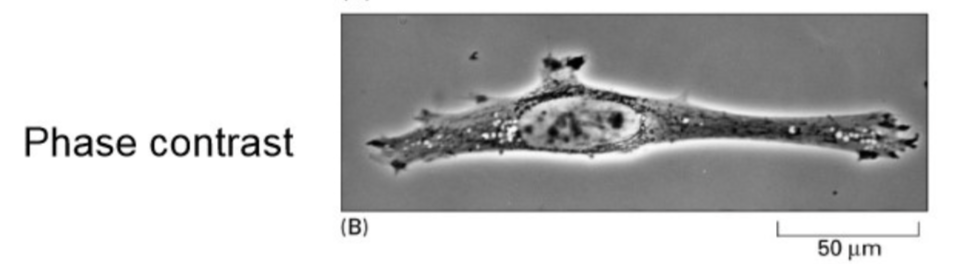
Phase contrast microscopy (light microscope method)
Converts phase differences in light into brightness (amplitude) differences
Light enters the specimen in phase, but waves travel at different rates through the sample depending on density, exits specimen in different phases
The microscope converts these differences in wavelengths into brightness differences
Increases contrast
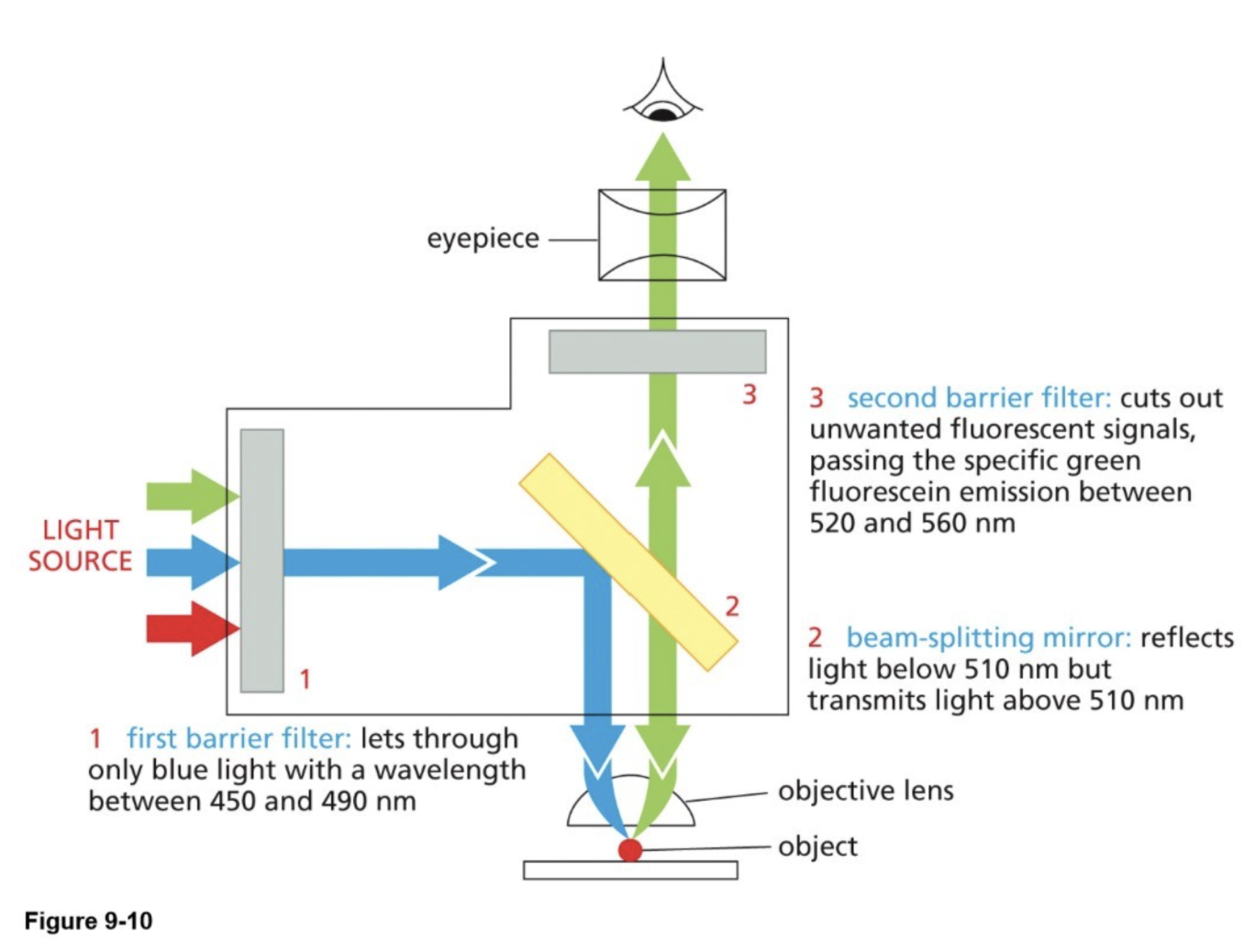
Fluorescence microscopy (light microscope method) & parts of a fluorescent microscope
A fluorescent molecule absorbs light of a specific wavelength (λ) and then emits light of a longer wavelength (λ)
Fluorescent molecules (GFP) absorb light at 460 nm and emit light at 520 nm
Parts of the microscope:
Barrier filters: transmit (allow through) specific wavelengths
Excitation wavelength — one that we excite the fluorescent molecule with
Emitted wavelength — one that the fluorescent molecule emits
Beam splitting mirror (dichroic): transmits light above a specific wavelength and reflects light below
Outcome: bright image (1 color) against dark background
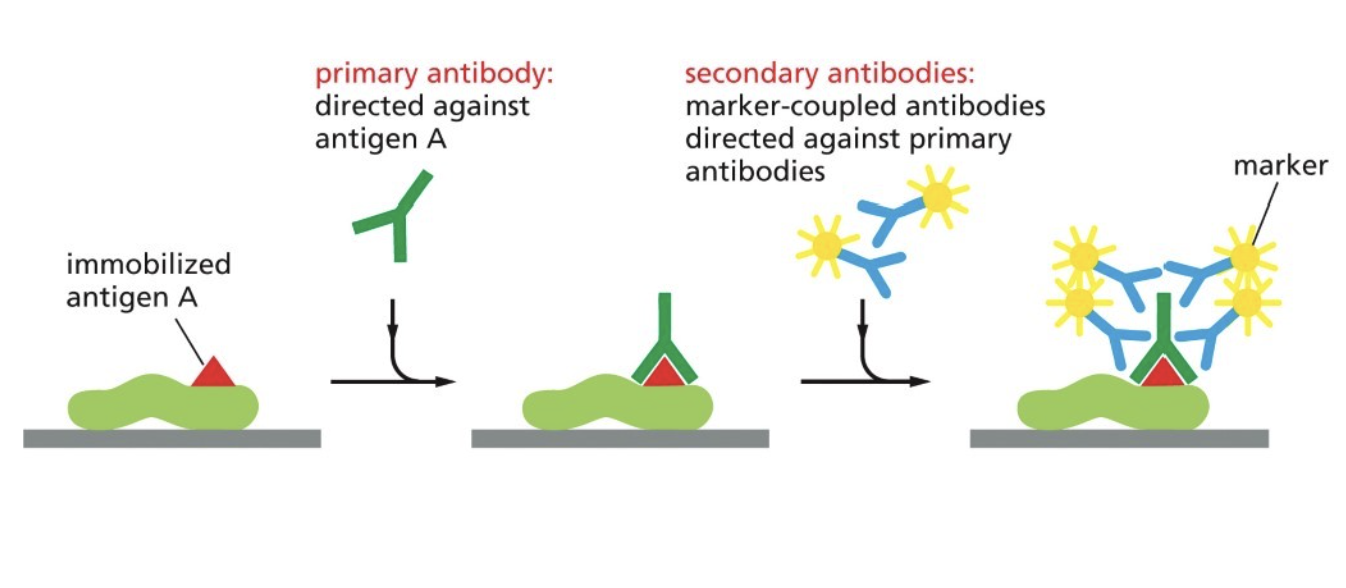
What is indirect immuno-fluorescence (immuno-staining)
What you’re actually seeing is indirectly binding to the protein (why it's considered indirect), but allows you to alter level of fluorescence
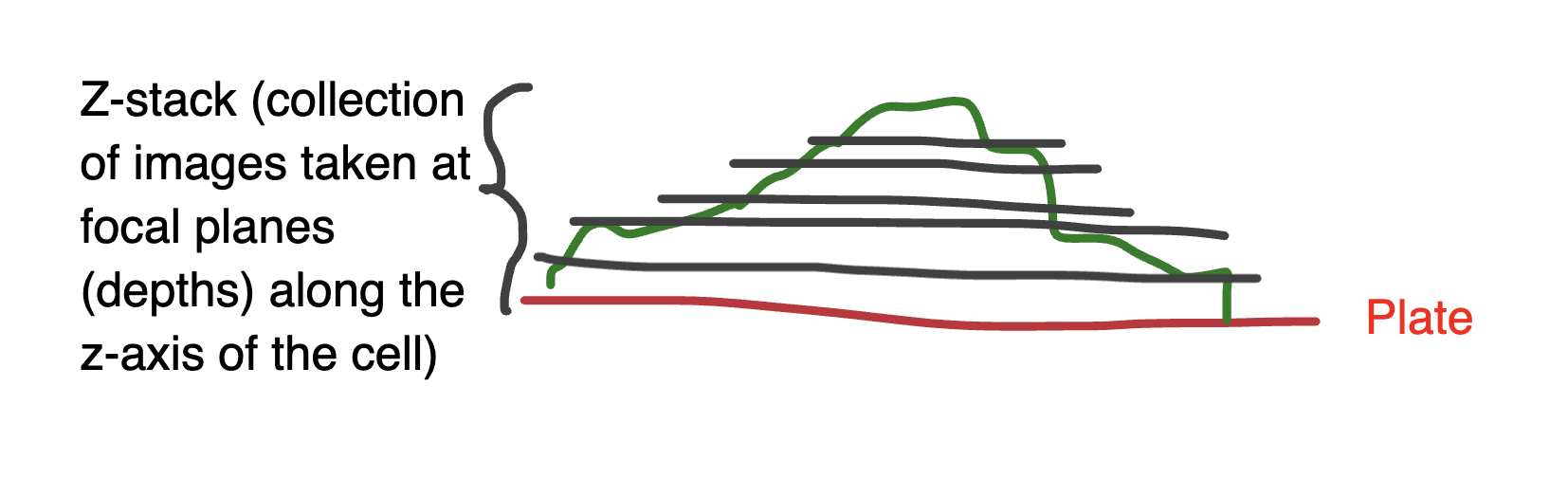
Deconvolution microscopy (a computational method)
Computational, post-processing technique that improves the resolution, contrast, and signal-to-noise ratio of fluorescence images by removing out-of-focus light and blurring
Uses the standard fluorescence microscope
Collects a z-stack of images (a collection of images taken at focal planes (depths) along the z-axis of the cell)
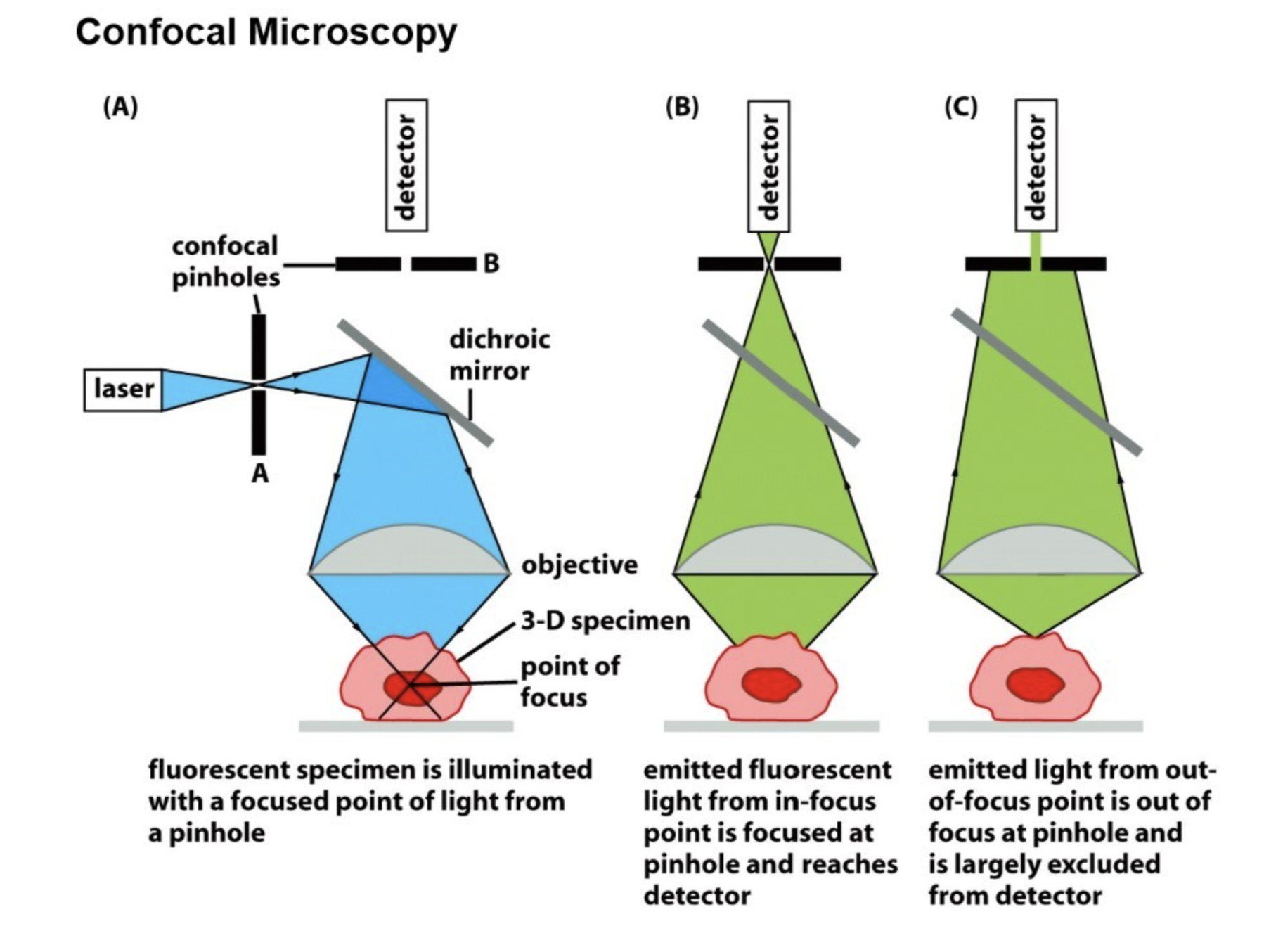
Confocal microscopy
Also removes out-of-focus light, but unlike deconvolution microscopy, does this through optic techniques rather than computational ones
Only allows light through in a very specific path
Uses a laser (allows for a specific wavelength), e.g., using 460nm for GFP
2 confocal pinholes (exclude out-of-focus light)
Dichroic mirror
Uses z-stack method: produces “optical sections” in a z-stack
Detector
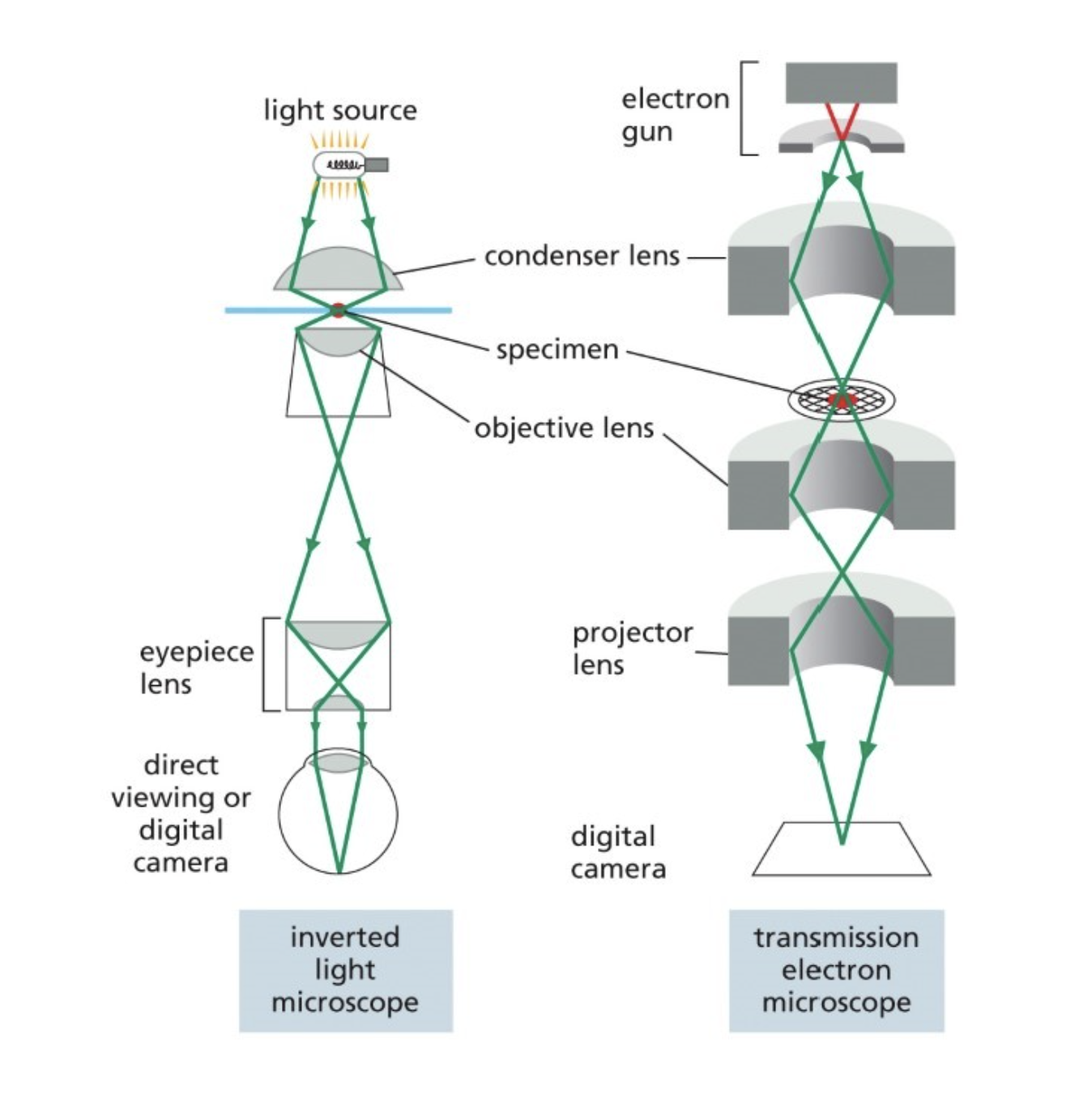
Electron microscopy
Uses electron beam
Magnetic coils (focus the electrons)
Scanning EM gathers an image of the surface of the sample
What are the functions of cell membranes?
Define cells (plasma membrane/PM) and compartments/organelles
Are barriers, help create ion gradients (which are needed for things like ATP synthesis, nerve impulses)
Membrane protein functions (roughly 30% of proteins are in membranes)
What are the properties of membranes?
Made of lipids (phospholipids, glycolipids, cholesterol), proteins, and carbs
They are dynamic (fluid bilayer, 5mm thick)
Impermeable to most H20 soluble molecules
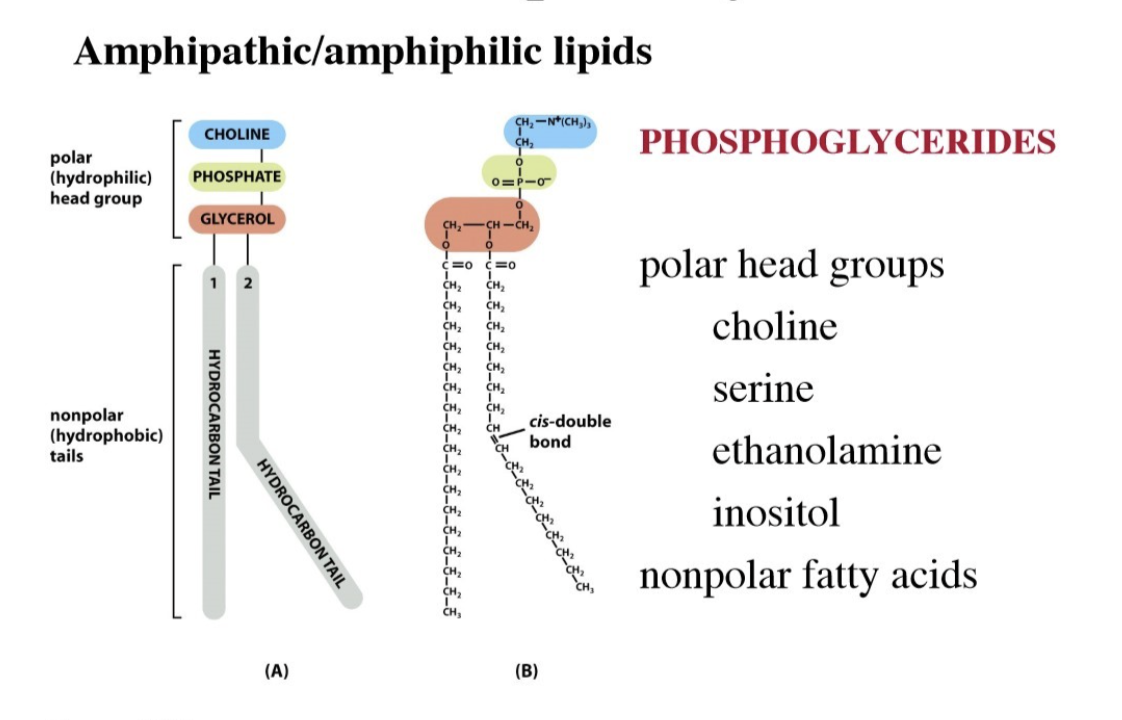
Lipids are amphipathic/amphiphilic, one side (head) is polar (hydrophilic) and one side (tail) is nonpolar (hydrophobic)
What are the types of lipids covered in this course?
Phospholipids
Phosphoglycerides
phosphatidyl-ethanolamine
phosphatidyl-serine
phosphatidyl-choline
phosphatidyl-inositol
Sphingolipids
sphingomyelin
Glycolipids
Cholesterol
What are phosphoglycerides?
The primary structural lipids found in eukaryotic cell membranes, consisting of a glycerol backbone esterified to two fatty acids, a phosphate group, and an alcohol
Form the lipid bilayers
The double bonds make the tails kinked (unsaturated fatty acid)
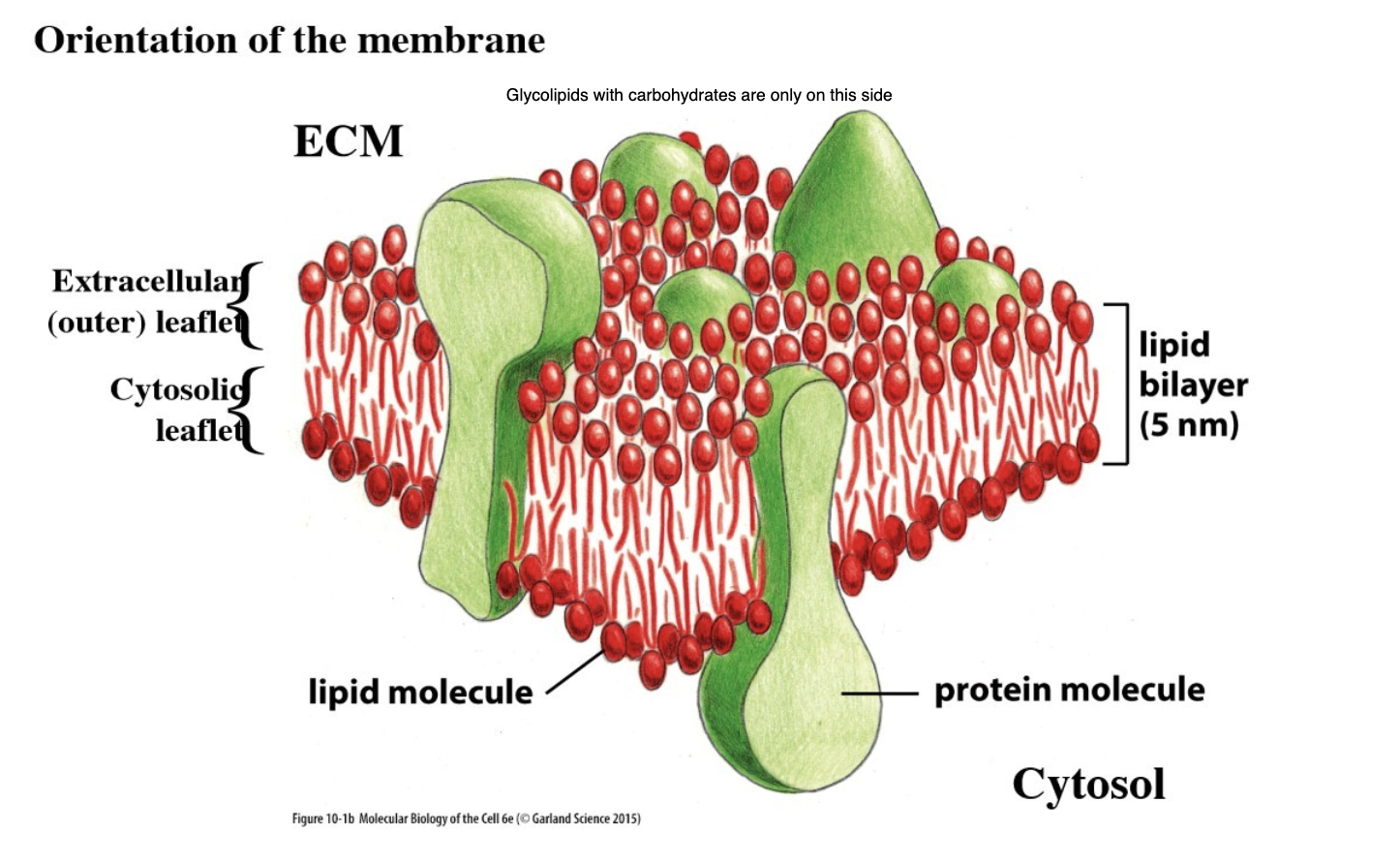
What are glycolipids?
Have carbohydrate (sugar) on the polar head (added in golgi lumen)
These carbohydrates are only present on the heads in the outer leaflet of the plasma membrane (not the cytosolic leaflet)
Use different sugars, and different sugars attached to each other in chains
Tend to be in “lipid rafts”
Function in cell-cell junctions
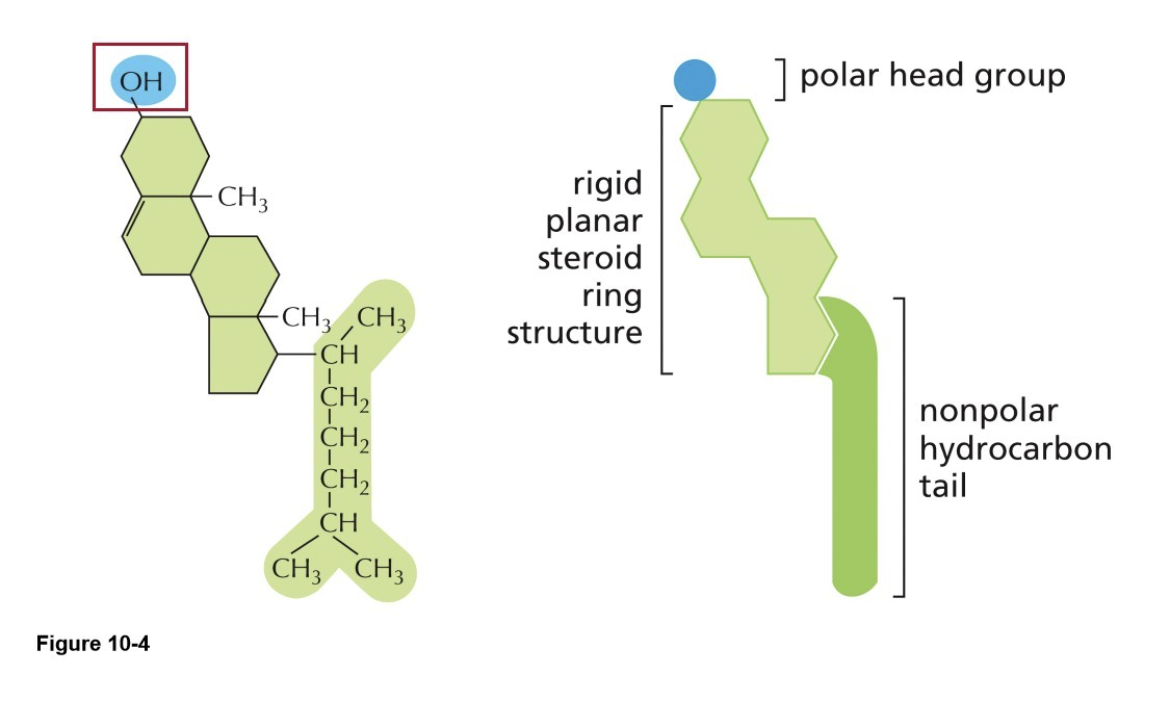
What is cholesterol?
Rigid ring structure
Is bilayer formation spontaneous? Why?
Spontaneous in solution due to two properties of the lipids
Shape of the lipid
Amphiphilic
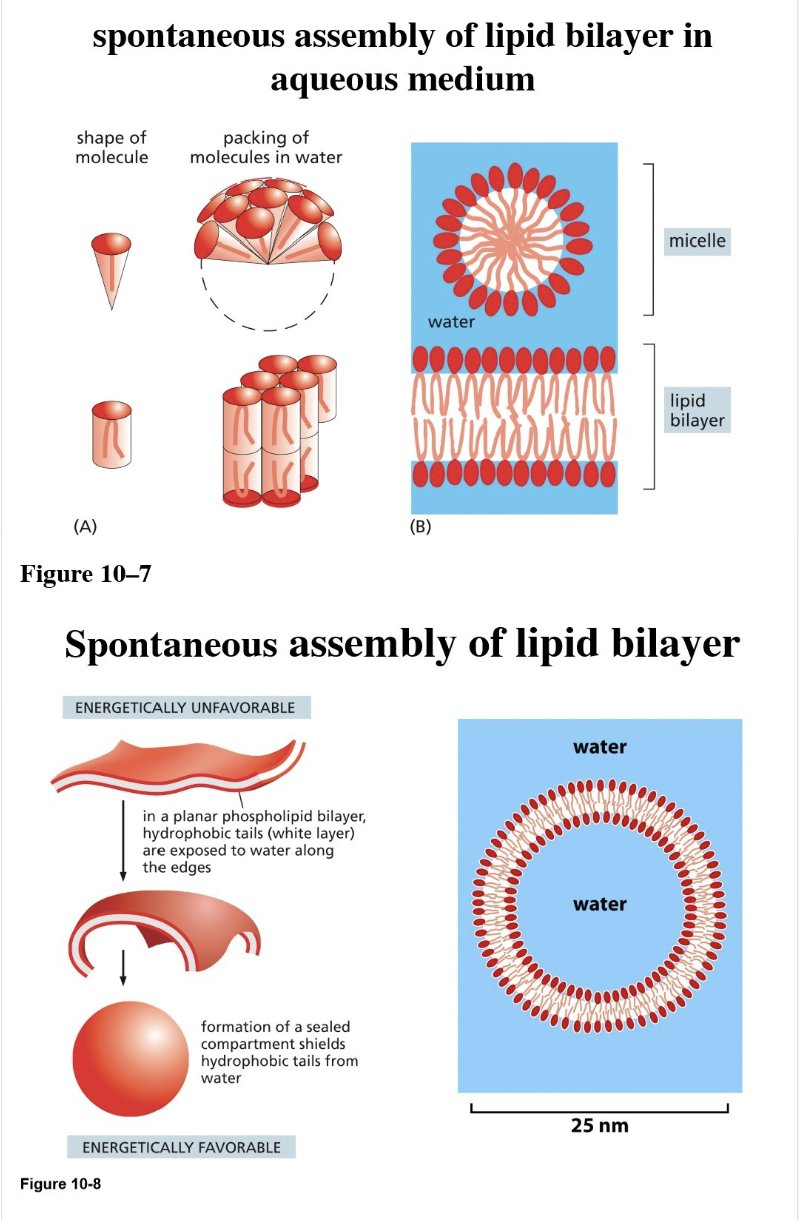
What is a liposome?
Spherical vesicles made from synthetic lipid bilayers in a lab, used to study membrane fluidity
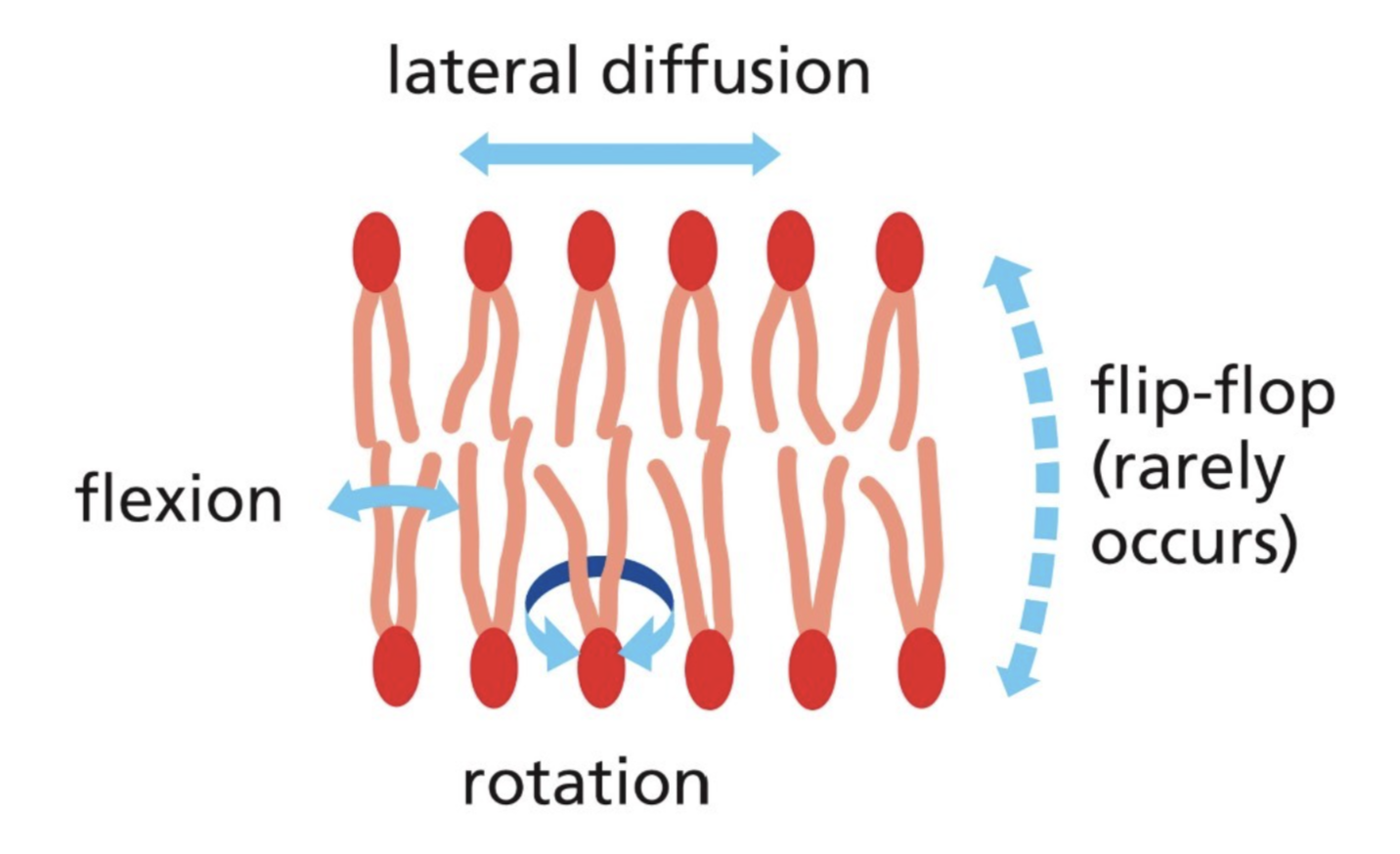
Characteristics of bilayer fluidity (how can lipids move within the bilayer?)
Through experiments with liposomes, found that there is:
Rapid lateral diffusion of lipids (lipids switch places with neighboring lipids in the same leaflet)
Rotation of lipids (flexion) is common
Flip-flop (moving from one leaflet to another) is very rare, energetically unfavorable
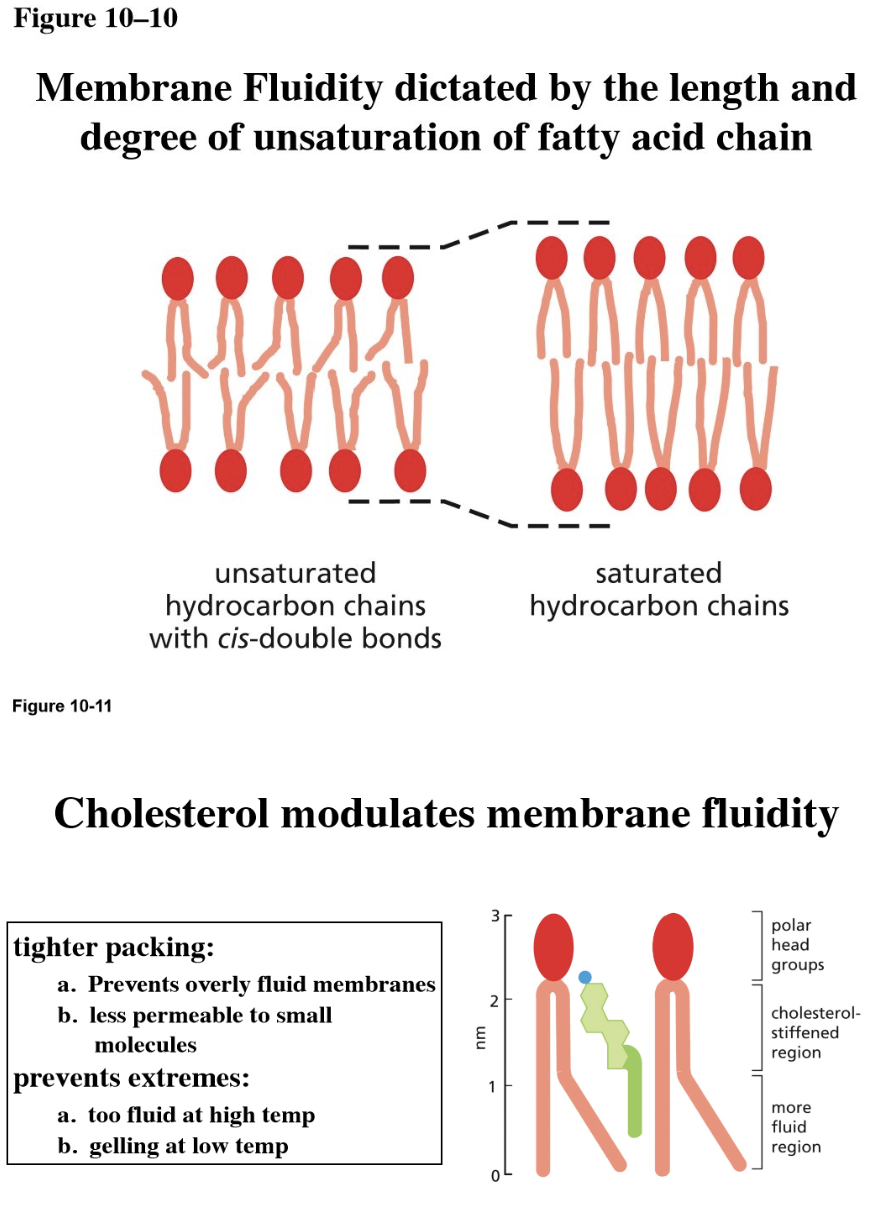
In what cases would there be increased membrane fluidity? And how?
Increased fluidity (useful in colder climates/cold adaptation): increased fluidity increases temperature to counteract a cold environment
Lipids with shorter tails (can move around more easily)
Lipids with kinked tails/unsaturated fatty acid chain
More cholesterol
What is lateral asymmetry?
Lateral asymmetry: non-uniform distribution of lipids and proteins within the plane of a single leaflet
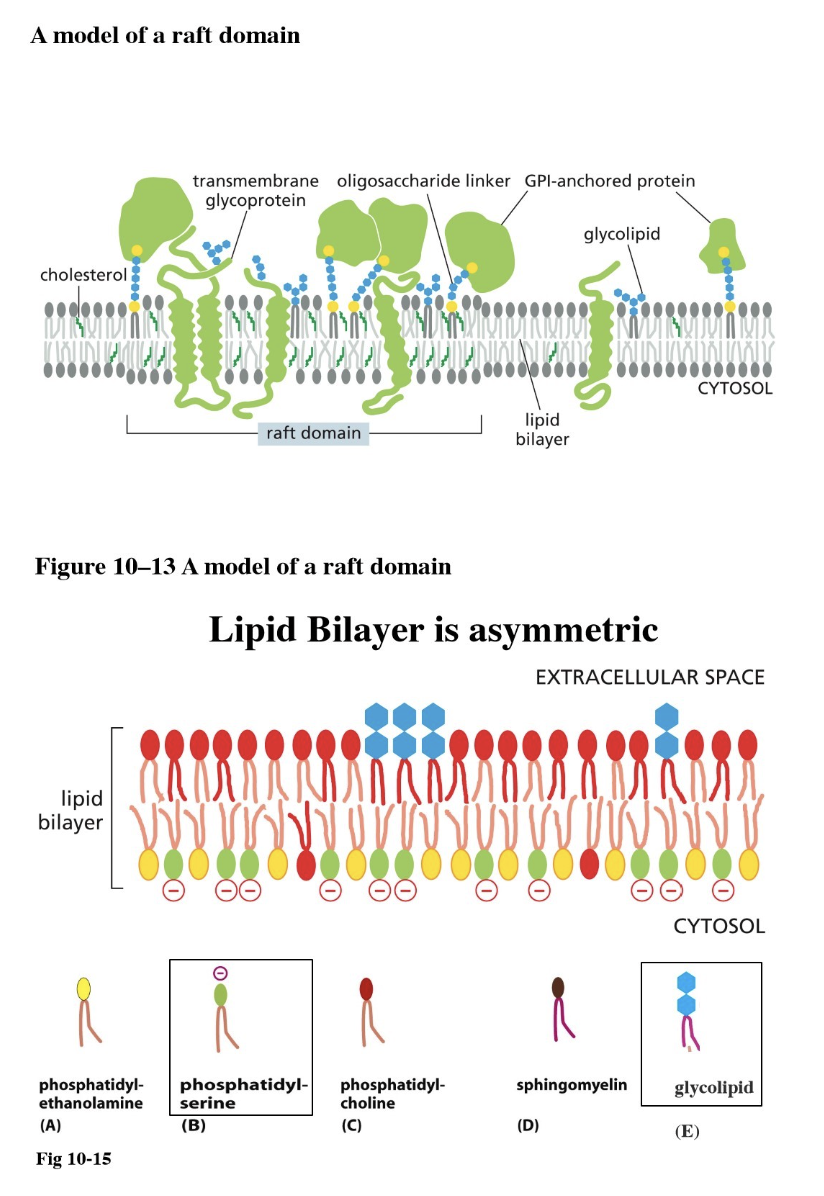
Lipid raft: a patch of membrane with a specific function (such as for cell signaling)
They are immobile — attached to ECM proteins or the cytoskeleton
Components:
Lipids have longer tails → thicker membrane
Transmembrane proteins have long hydrophobic alpha-helix
More cholesterol, more glycolipids, and more glycoproteins
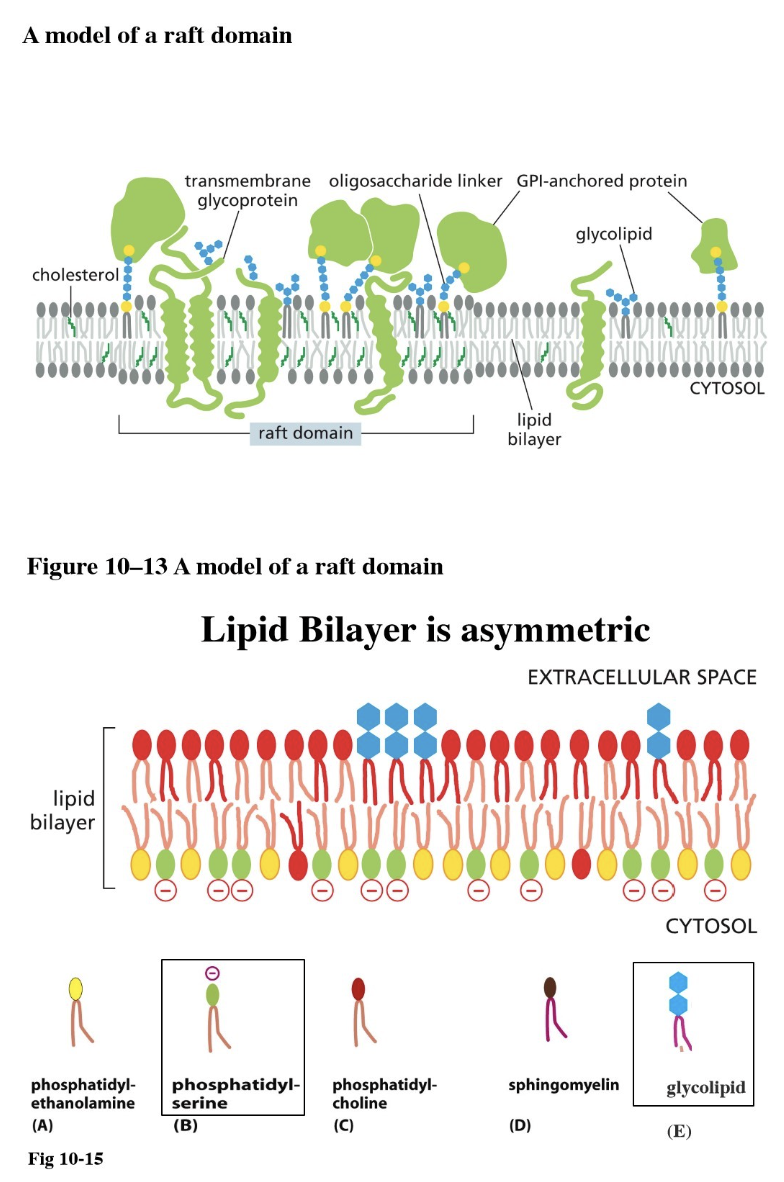
What is a lipid raft?
A patch of membrane with a specific function (such as for cell signaling)
They are immobile — attached to ECM proteins or the cytoskeleton, contributes to membrane asymmetry
Components:
Lipids have longer tails → thicker membrane
Transmembrane proteins have long hydrophobic alpha-helix
More cholesterol, more glycolipids, and more glycoproteins