6.1 CELLULAR CONTROL
1/73
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
74 Terms
6.1.1
GENE MUTATIONS
What is a mutation?
a random change to the genetic material, in a cell by base deletion, addition, substitution or by inversion or repeat of a triplet. Most likely to occur during DNA replication in the S phase of interphase.
How may a mutation occur?
caused by certain chemicals (mutagens) e.g. found in tobacco smoke and ionising radiation such as UV light, x-rays and gamma rays.
What are the three effects of mutations?
neutral, harmful, beneficial
Beneficial mutations
They are mutations that offer an individual a selective advantage. The final protein shape provides a better function.
Neutral mutations
- If the mutation occurs in the non-coding part of the DNA.
- Silent mutation (base triplet is changed, but has no effect on the amino acid due to there being more base triplets that code for the same DNA since the DNA code is degenerate).
The structure of the protein is unaffected—there is no change—and it has the same function. If it's an enzyme, then there's no change to the active site.
Harmful mutations
Produce characteristics that are harmful to the organism. Final shape of the protein can not fulfill its function. If its an enzyme, then the active site is deformed.
Examples of harmful mutations
70% of cystic fibrosis sufferers, the mutation is a deletion of a triplet of base pairs.
Proto-oncogenes can be changed into oncogenes by a point mutation, which
promotes uncontrolled cell division.
Huntington's disease is caused by a stutter, which repeats sections of CAG sequences.
What are the types of mutations?
1) Substitution - point mutation
2) Deletion
3) Addition
What's a substitution mutation?
When a nucleotide is replaced by one with a different base.
What is a deletion mutation?
An existing nucleotide is removed in the normal sequence, which shortens the sequence
What is a addition mutation?
an extra nucleotide is inserted, so an extra base is added to the sequence.
Which two mutations can cause a frameshift?
Addition and deletion.
What are the three types of point mutations?
silent, missense, nonsense
What reduces the effect of point mutations?
Due to the DNA code being degenerate, there are more than one base triplet that codes for the same amino acid, so point mutations do not always cause a change to the sequence of amino acids in a protein.
What is a silent mutation?
A point mutation involving a change to the base triplet, where that triplet still codes for the same amino acid. The primary structure of the protein, and therefore the secondary and tertiary structure is not altered.
What is a missense mutation?
Point mutation in which a single nucleotide is changed, resulting in a codon that codes for a different amino acid. Within a gene, such a point mutation may have a significant effect on the protein produced. The alteration to the primary structure leads to a change to the tertiary structure of the protein, altering its shape and preventing it from carrying out its usual function.
What is a nonsense mutation?
A point mutation may alter a base triplet, so that it becomes a termination (stop) triplet. This particularly disrupts point mutation results in a truncated protein that will not function. This abnormal protein will most likely be degraded within a cell. The genetic disease Duchenne muscular dystrophy is the result of a nonsense mutation.
6.1.2
REGULATION OF GENE EXPRESSION
Exon
the coding, or expressed region of DNA.
Intron
the non-coding region of DNA
Operon
a group of genes that function as a single transcription unit; first identified in prokaryotic cells.
Transcription factor
Protein or short non coding RNA that can combine with specific site on length of DNA and inhibit or activate transcription of gene
Regulation of gene expression at the transcriptional level
In eukaryotes
1) The transcriptional level
During mRNA production
What is a gene?
A gene is a section of DNA which codes for a particular protein.
All the cells of your body contain the same DNA but express different genes.
What are transcription factors?
Transcription factors are proteins which move in from the cytoplasm and bind to DNA.
They create a loop in the DNA molecule that activates the gene.
RNA polymerase will then attach to the DNA chain and transcription will start making mRNA.
What are the two types of transcription factors?
- Activators = the loop forms and transcription increases.
- Repressors = the loop doesn't form and transcription decreases.
What are structural genes?
Genes that code for polypeptides that function as enzymes, membrane carriers, hormones etc. are
structural genes
What are regulatory genes?
Genes that code for polypeptides and various forms of RNA that control the expression of structural genes are regulatory genes.
What is the promotor region?
part of a gene that controls the expression of a gene and lies 100 base pairs upstream from the start of the gene.
How is transcription initiated?
When proteins (transcription factors) bind to the promoter region of a gene to either inhibit or help RNA polymerase from binding to the promoter region which will either cause transcription to not begin or begin.
Diagram of the transcriptional control
More about transcription factors
- A specific combination of transcription factors is needed to activate a gene.
- Transcription factors are regulated by signals produced from other molecules.
- Inhibitor molecules prevent the transcription factors binding to the DNA and so prevents transcription and polypeptide synthesis.
What attaches to the promotor in order for transcription to begin?
RNA polymerase
What are genes that are transcribed all the time known as?
Constitutive
What are enzymes that are produced only when needed by
the presence of a chemical signal known as?
inducible
2) The post-transcriptional level
After mRNA production
Diagram of the post-transcriptional level
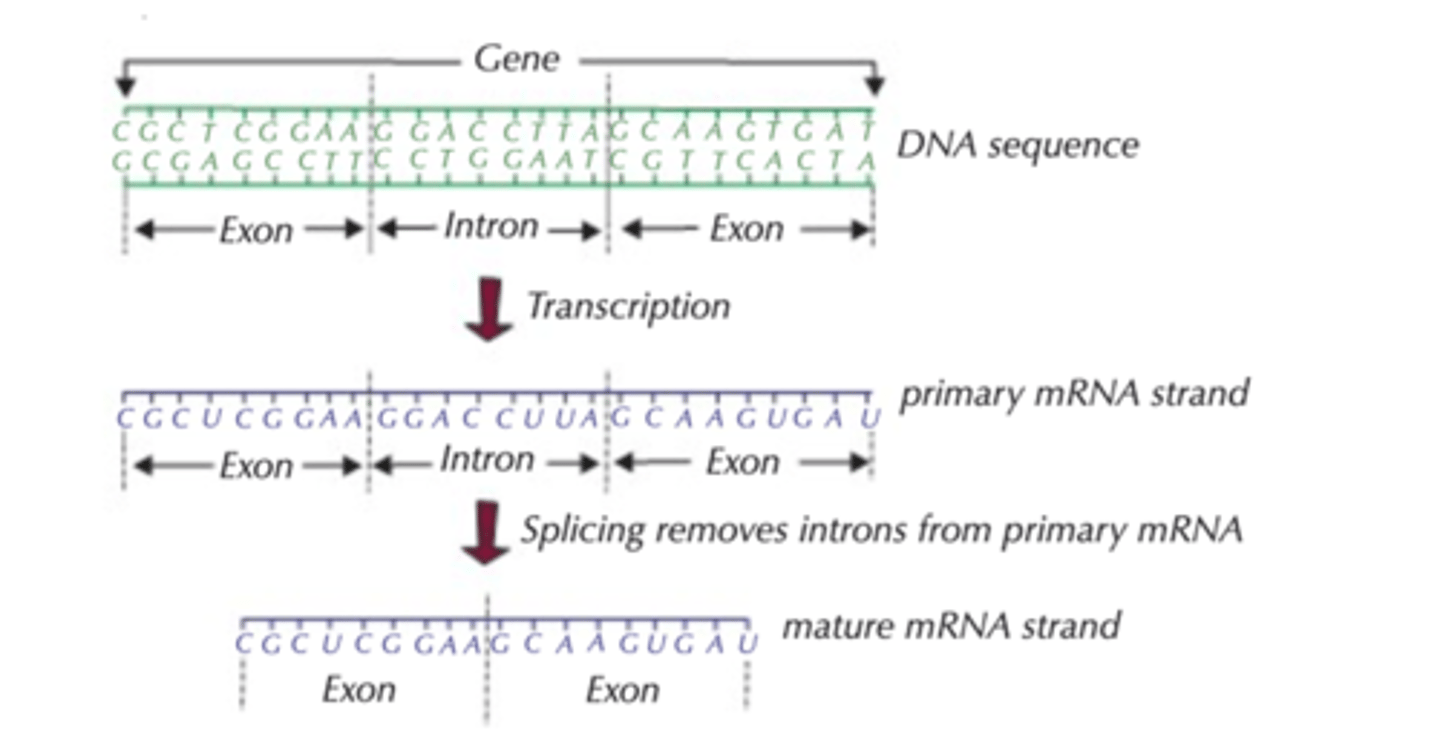
Post transcriptional gene regulation
Both introns and exons are initially transcribed resulting in a primary mRNA molecule.
The primary mRNA molecule becomes edited by the removal of introns and the remaining mRNA exons which correspond to the original DNA exons are joined together.
The enzyme endonuclease is involved in editing and splicing processes.
Some introns can code for proteins, while others are non-coding lengths of RNA involved in gene regulation. Genes can be spliced in different ways = such that they encode other proteins.
3) Post-translational level of gene regulation
After protein production
What does the post-translational level of gene regulation actually do?
The control of gene expression via activation of proteins
This occurs through phosphorylation- i.e. addition of phosphate group
to enzymes to activate them
How does cAMP activate enzymes and may also stimulate transcription?
1) A signalling molecule, such as the protein hormone glucagon, binds to a receptor on the plasma membrane of the target cell.
2) This activates a transmembrane protein which then activates a G protein.
3) The activated G protein activates adenyl cyclase enzymes.
4) Activated adenyl cyclase enzymes catalyse the formation of many molecules of cAMP from ATP.
5) cAMP activates PKA (protein kinase A).
6) Activated PKA catalyses the phosphorylation of various proteins, hydrolysing ATP in the process. This phosphorylation activates many enzymes in the cytoplasm, for example those that convert glycogen to glucose
7) PKA may phosphorylate another protein (CREB, cAMP response element binding).
8) This then enters the nucleus and acts as a transcription factor, to regulate transcription.
Regulation of gene expression at the transcriptional level
in prokaryotic cells
What do prokaryotic transcription factors do?
Bind to operons to control gene expression.
Operons are sections of DNA which contain a cluster of structural genes.
These are transcribed together alongside control elements and regulatory genes.
Structure of an operon and its regulatory gene
Structural genes code for useful proteins - enzymes.
The control region contains a promoter and an operator, RNA polymerase and TF bind these respectively.
A regulatory gene coding for an activator or repressor.
The lac operon
What is E.coli
a bacterium that respires glucose but can use lactose if glucose isn't available
What happens when E.coli is grown in a culture medium without lactose and then placed in a medium with lactose?
At first they cannot metabolise the lactose because they only have tiny amounts of the two enzymes needed to metabolise it.
A few minutes after lactose is added to the culture medium, E. coli bacteria increases the rate of synthesis if the enzymes by about 1000 times.
Lactose must trigger the production of the two enzymes, and is known as the
inducer.
What is the lac operon?
The lac operon is a section of DNA within the bacterium's DNA.
What are the structural genes of the lac operon?
Structural genes (lacZ and lacY) that code for the enzymes:
β-galactosidase - breaks down lactose to glucose and galactose.
Lactose permease - helps the cell to absorb lactose/increase
uptake of lactose.
What are the control sites of the lac operon?
Control sites:
Operator region (lac0) - binds to the repressor and can switch on and off the structural genes.
Promoter region (P) - binds to RNA polymerase and controls
What does E.coli do when glucose isn't available?
If glucose isn't available E.coli uses lactose for respiration.
The genes required for this are found in the lac operon.
The lacZ, lacY and lacA genes are required for this process.
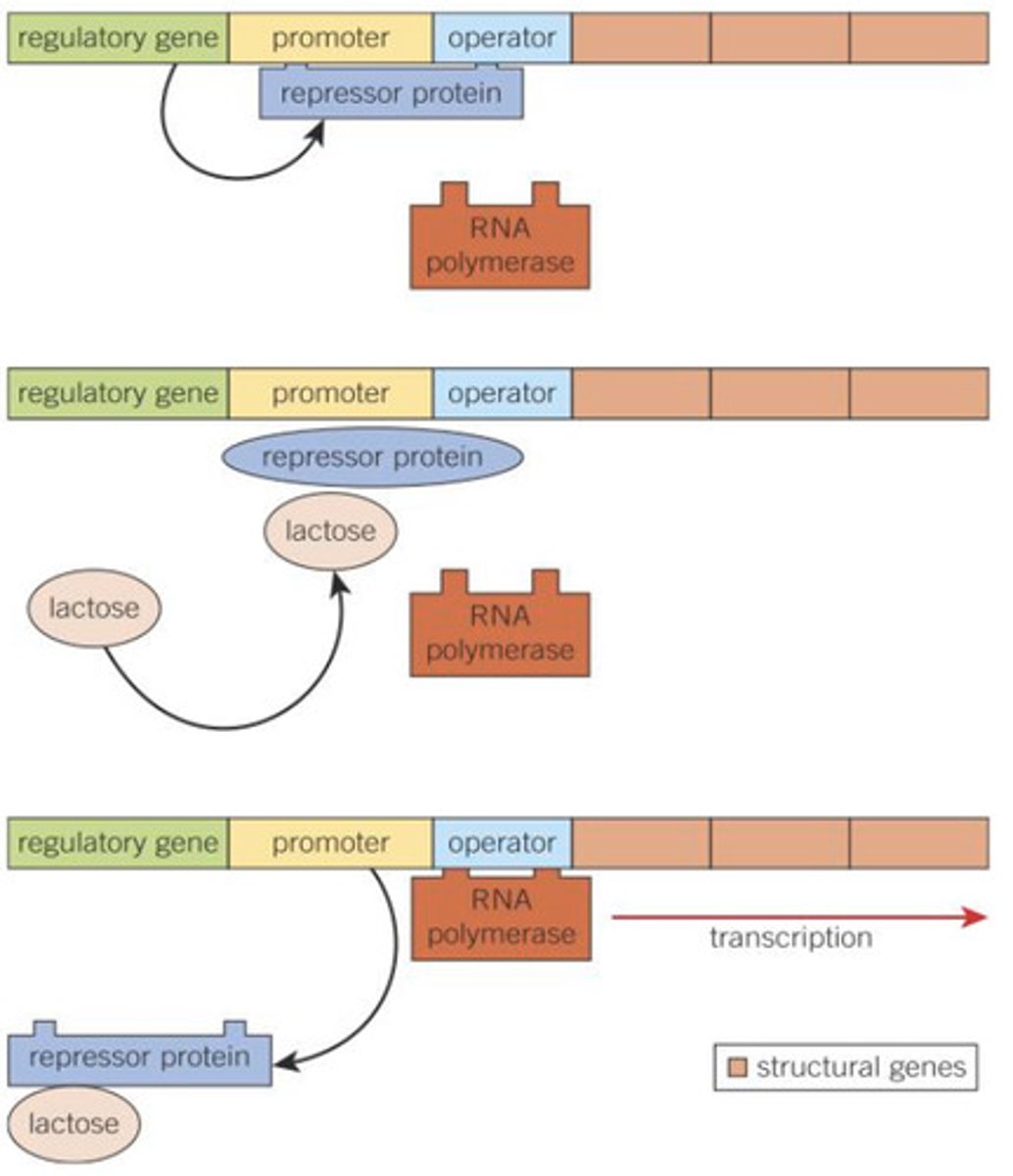
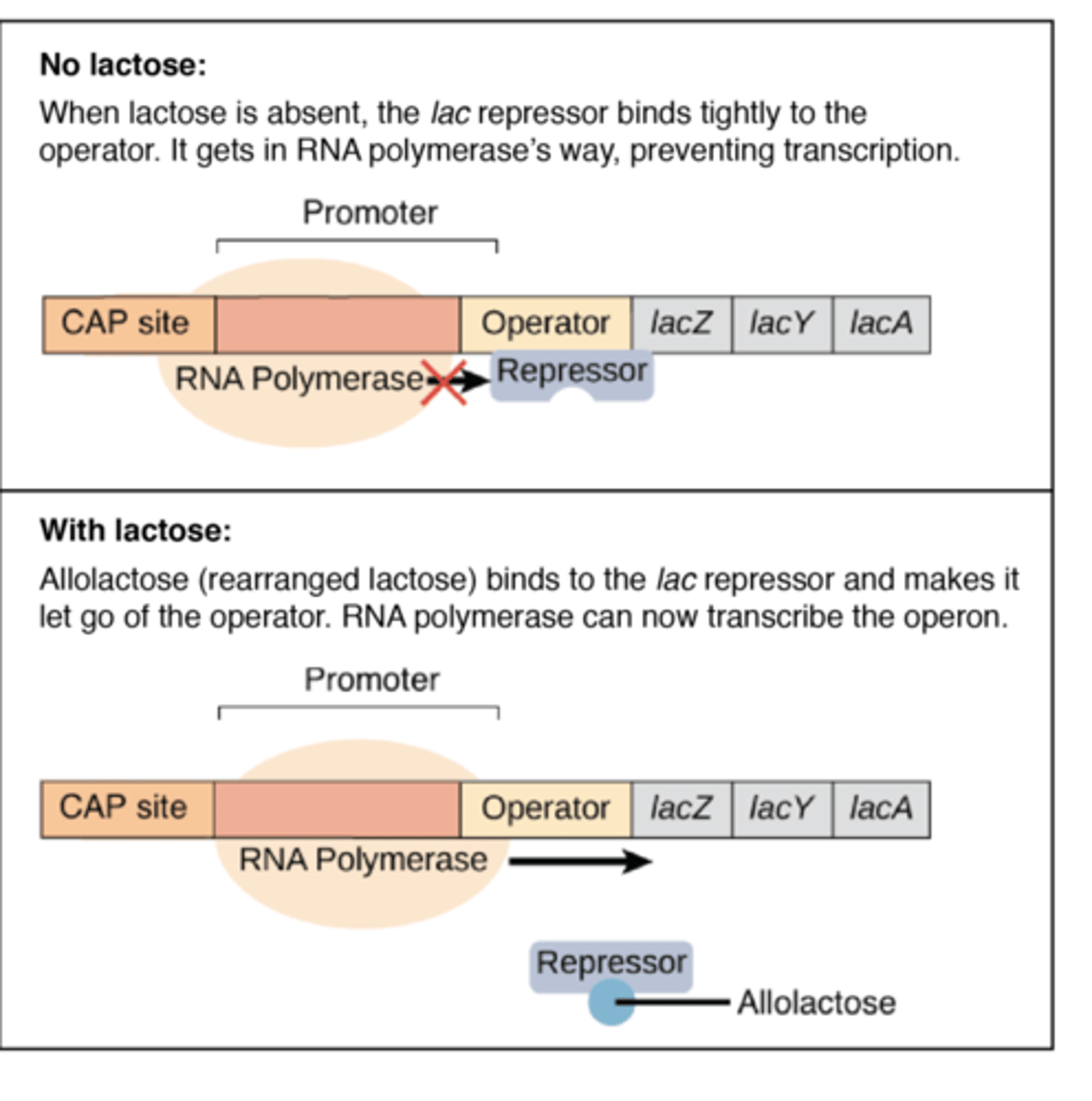
How does the lac operon work in the absence of lactose?
The enzymes β-galactosidase
and - lactose permease is not synthesized.
The regulator gene LacI is expressed (transcribed and translated) and the regulator/repressor protein is synthesized.
The repressor (regulator) protein binds to the operator region, blocking the binding site of the promoter region and so preventing RNA polymerase from binding.
Transcription cannot occur because RNA polymerase does not bind to the promoter region. Therefore, the structural genes are switched off.
How does the lac operon work when lactose is present?
The regulator gene is expressed (transcribed and translated) and the regulator protein is synthesized.
Lactose molecules bind to the repressor protein, causing the repressor protein to change shape so that it can no longer bind to the operator region.
RNA polymerase can bind with the promoter region, initiating the transcription of mRNA. The structural genes are switched on.
The enzymes β-galactosidase and - lactose permease are synthesized, converting lactose to glucose and galactose.
Diagram of an lac operon
6.1.3
GENETIC CONTROL OF BODY PLAN DEVELOPMENT
Apoptosis
programmed cell death
Conserved
Has remained in all descendent species throughout evolutionary history
Homeobox sequence
Sequence of 180 base pairs (excluding introns) found within genes that are involved in regulating patterns of anatomical development in animals, fungi and plants.
Hox genes
Subset of homeobox genes, found only in animals; involved in formation of anatomical features in correct locations of body plan.
What is the function of homeobox genes?
- They regulate the polarity of the whole organism, head to tail.
- Homeobox genes act as master genes controlling which genes will be switched on at each stage during early development.
Homeobox sequence
A homeobox sequence is 180 bases
including introns encoding/forming a
60 amino acid polypeptide called a
homeodomain sequence. The protein folds into a 3D shape and binds to DNA- i.e. they are transcription factors. TFs work in the nucleus and can regulate transcription of other genes like those of early development of animals, plants and fungi ensuring the sequence of gene expression is correctly ordered.
How are hox genes regulated?
- Hox genes are similar across different classes of animals
- Hox genes are regulated by other genes called gap genes.
- Gap genes are regulated by mRNA from the egg's cytoplasm.
Irregularities in Homeobox genes.
Homeobox genes that do not function
properly can result in the disruption in the
sequence of development.
Disruptions can result in conditions such as
antennapedia (legs grow where the
insect's antennae should be) in Drosophila
Mutation in binding regions
All DNA binding regions have the same
shape.
Any mutations in the binding regions
leaves the transcription factors unable
to regulate the transcription of genes
and the organisms produced will either
not be viable or will have reduced
survival.
Organisms with mutations are unlikely to
survive to reproduce and are therefore
selected against.
Mutations in transcription binding sites
= negative selection pressure.
Hox genes are highly conserved over
millions of years of evolution
Structure and function of hox genes?
Homeobox genes are organised in groups called Hox gene clusters (in animals).
Hox genes regulate head-to-tail development in embryos. Hox genes are arranged in clusters containing up to 10 genes.
In early embryonic development, Hox genes are active and expressed in order along the anterior-posterior axis.
The gene expression is in sequence and time matching up to the sequence and timing of body part development is known as collinearity.
Hox A - D gene clusters are found on different chromosomes of the mouse and human genomes.
Homeobox genes are organised in groups called Hox gene clusters.
13 homeobox genes in the mouse control the development of the embryo in sequence: head, thorax, and abdomen.
The same genes are duplicated & and found only in animals such as humans.
Hox genes control the body plan of an embryo along the axis (head-to-tail).
Role of Hox genes?
Some Hox genes activate genes that cause apoptosis (cell suicide).
In Drosophila a Hox gene activates the
Rpr (reaper) gene leads to cell death in the head lobes thereby separating the
maxilla and mandible.
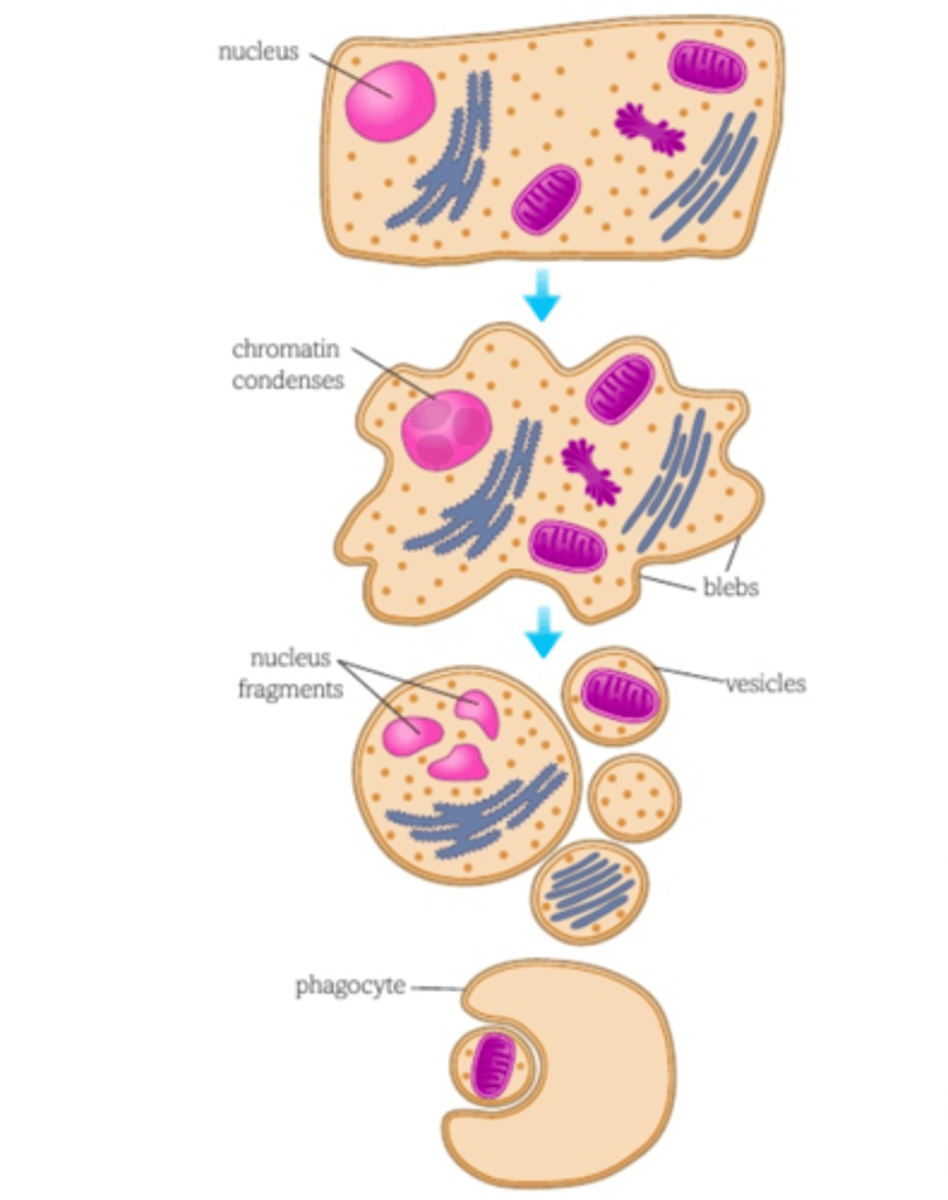
The sequence of events during apoptosis
1) Enzymes break down the cell cytoskeleton
2) Cytoplasm becomes dense with tightly packed organelles
3) Cell surface membrane changes and blebs form
4) Chromatin condenses, the nuclear envelope breaks and DNA breaks into fragments
5) Cell breaks into vesicles that are ingested by phagocytic cells - debris doesn't damage any other cells or tissues. The whole process happens quickly.
How is apoptosis controlled?
Cell signals help to control it, some of the signalling molecules may be released by cells when genes that are involved in regulating the cell cycle and apoptosis respond to internal cell stimuli and external stimuli such as stress. These signalling molecules include cytokines from cells in the immune system, hormones, growth factors and nitric oxide.
Examples of apoptosis
1) Removal of webbing between fingers and toes
2) Excess immune cells after an infection
3) Damaged cells
How does apoptosis help in the immune system?
Removes ineffective or harmful T-lymphocytes during the development of the immune system.
What happens if there is not enough apoptosis occurring?
Tumours could form
What happens if there is too much apoptosis occurring?
cell loss and degeneration