Molecular Biology II
1/42
Earn XP
Description and Tags
Transcription and Translation (aka Gene Expression)
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
43 Terms
Learning Objectives
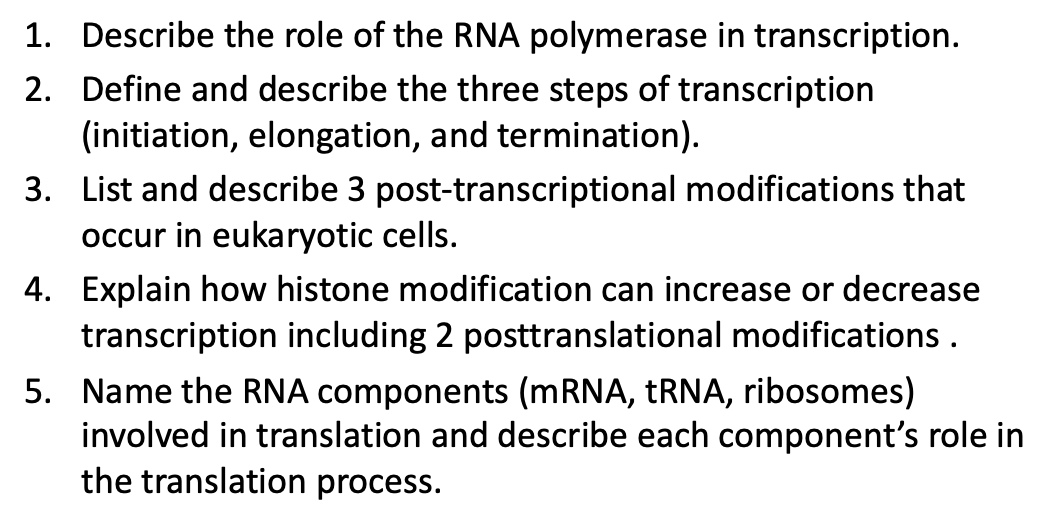
Gene Expression
The combined process of transcription of a gene into mRNA, the processing of that mRNA, and its translation into protein
Required for:
Adaptation
Tissue specific differentiation
Development
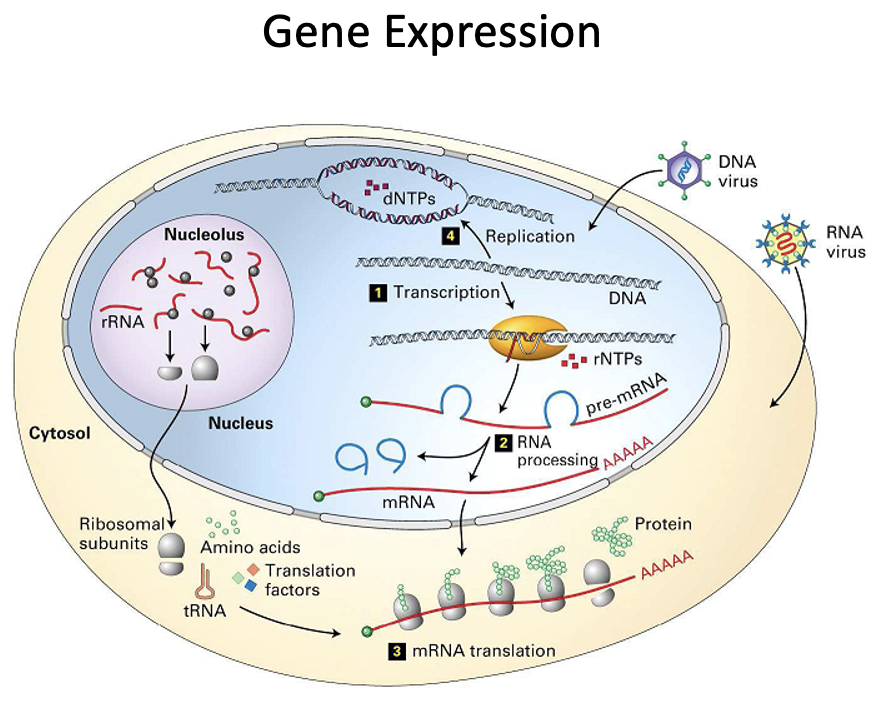
RNA Transcription
Transcribe DNA into RNA (slightly different language)
dsDNA → ssRNA
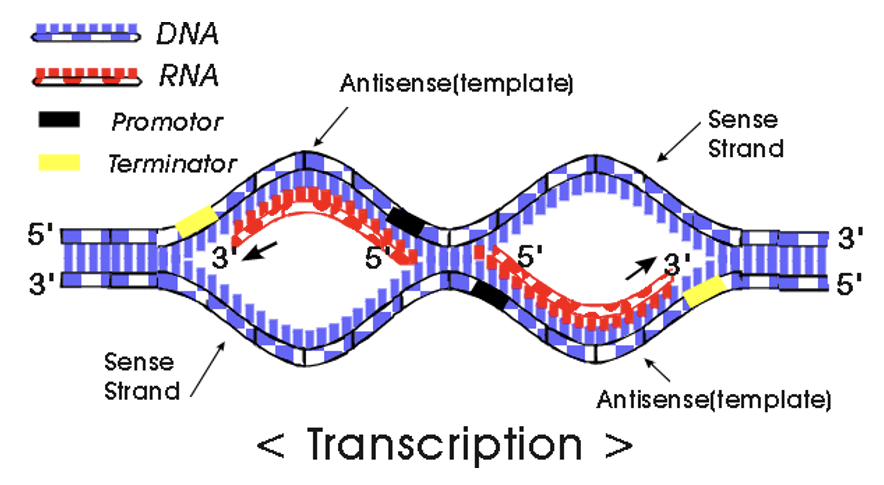
RNA polymerase
enzyme that catalyzes the reaction of rNTP polymerization
always synthesized in the 5’ → 3’ direction
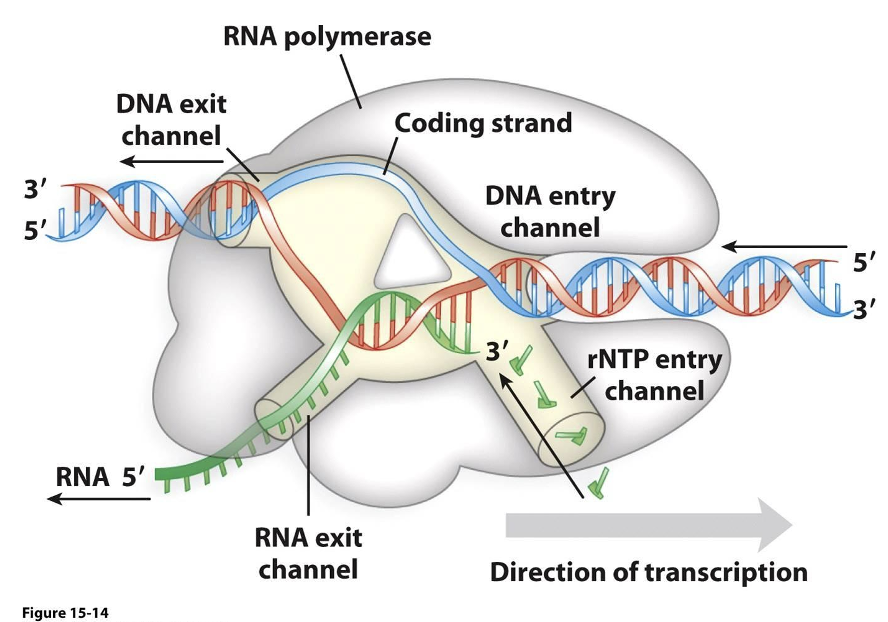
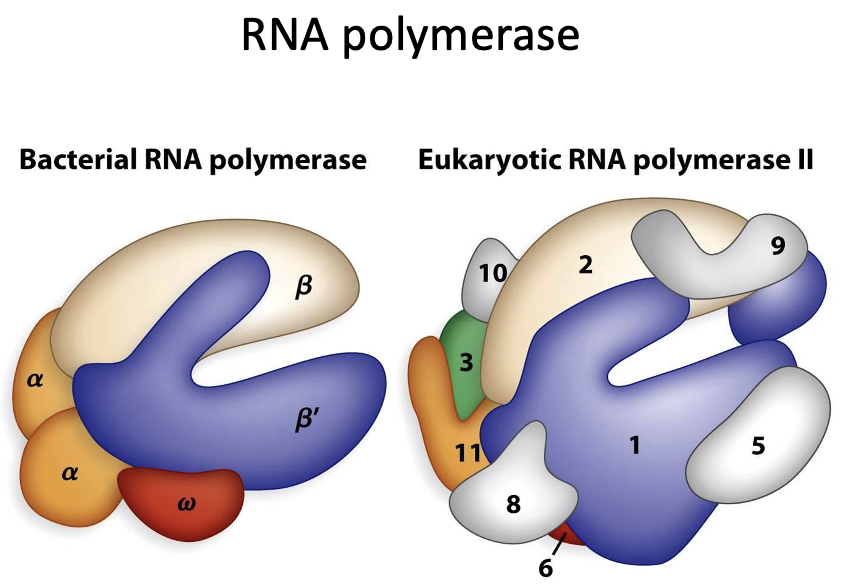
RNA polymerase in prokaryotes
•The sigma region of the bacterial RNA polymerase recognizes specific regions of DNA called promoters
•At approximately 10 and 35 base pairs from the start site there are similar DNA sequences termed the -10 and -35 sequence
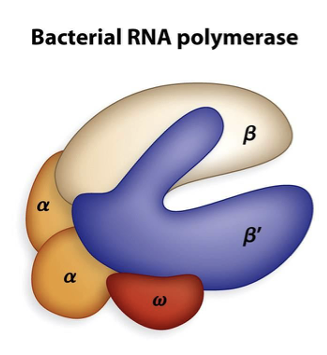
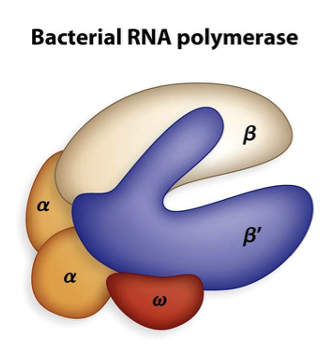
Which region of bacterial RNA polymerase recognizes specific regions of DNA called promoters?
the sigma region
At approximately 10 and 35 base pairs from the start site there are similar DNA sequences termed the -10 and -35 sequence
In other words, …
Bacterial RNA polymerase recognizes specific DNA sequences known as promoters, particularly the -10 (Pribnow box) and -35 consensus sequences, which are upstream of the transcription start site.
The recognition is primarily mediated by the sigma (σ) subunit of the RNA polymerase, which binds to these sequences and facilitates the opening of the DNA double helix to begin transcription
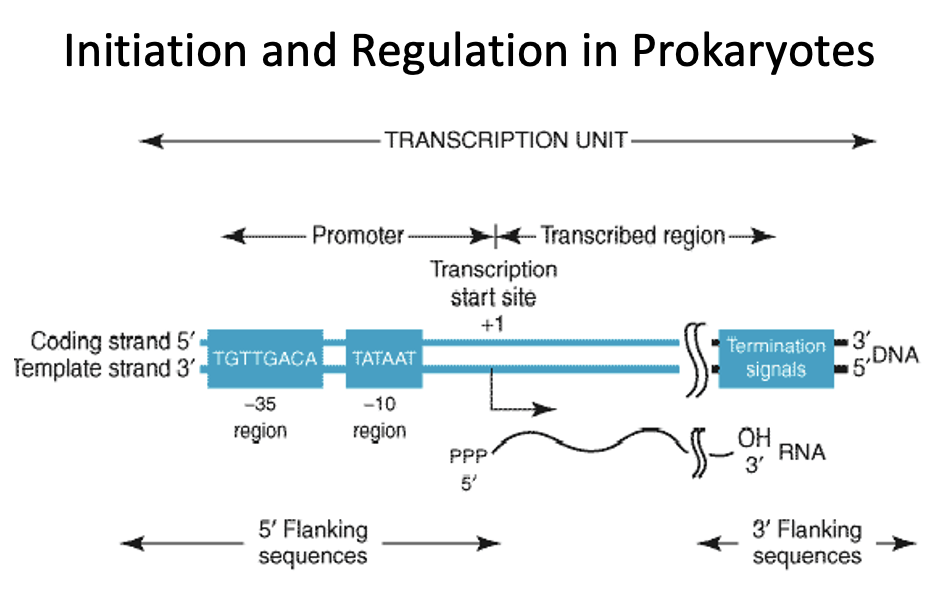
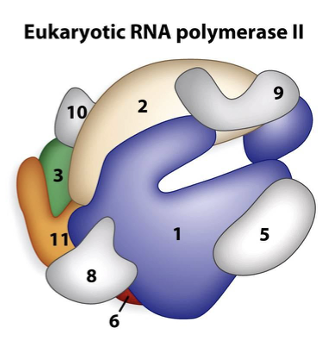
RNA polymerase in Eukaryotes
The majority of eukaryotes initiate transcription using the TATA box region at approximately -30bp
TATA box consensus sequence is recognized by the RNA polymerase and directs it where to initiate transcription
Pol I : large ribosomal RNA
Pol II: messenger RNA (mRNA)
Pol III: small rRNA, tRNA and other small RNA
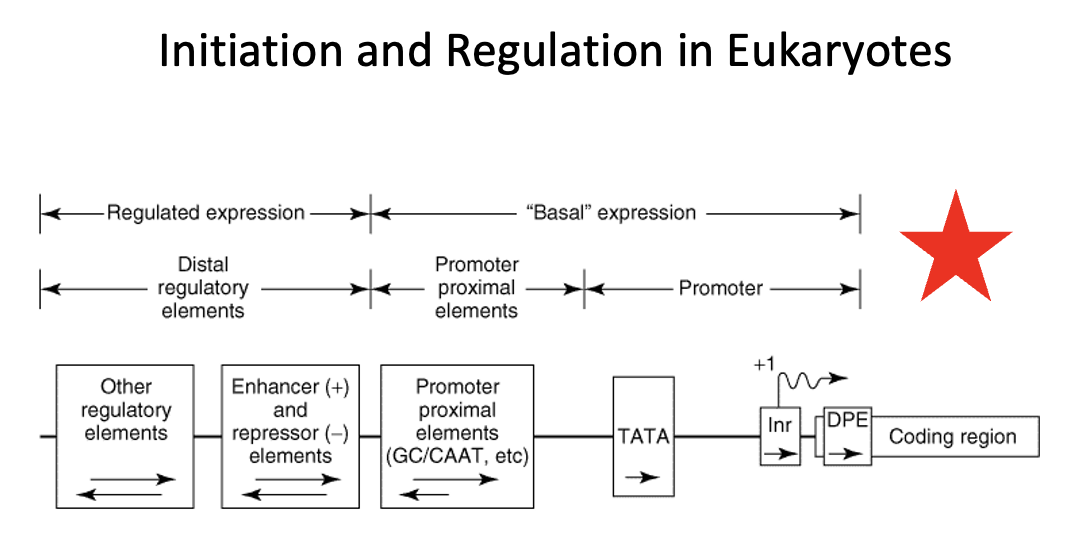
TATA box
a DNA sequence, most commonly TATAAA, located in the promoter region of many genes, about 25-35 base pairs upstream (-30bp) of the transcription start site.
It serves as a recognition site for the TATA-binding protein (TBP), a key transcription factor that recruits RNA polymerase and initiates transcription.
The TATA box's sequence is rich in adenine and thymine (A-T), which allows for easier unwinding of the DNA strands for transcription.
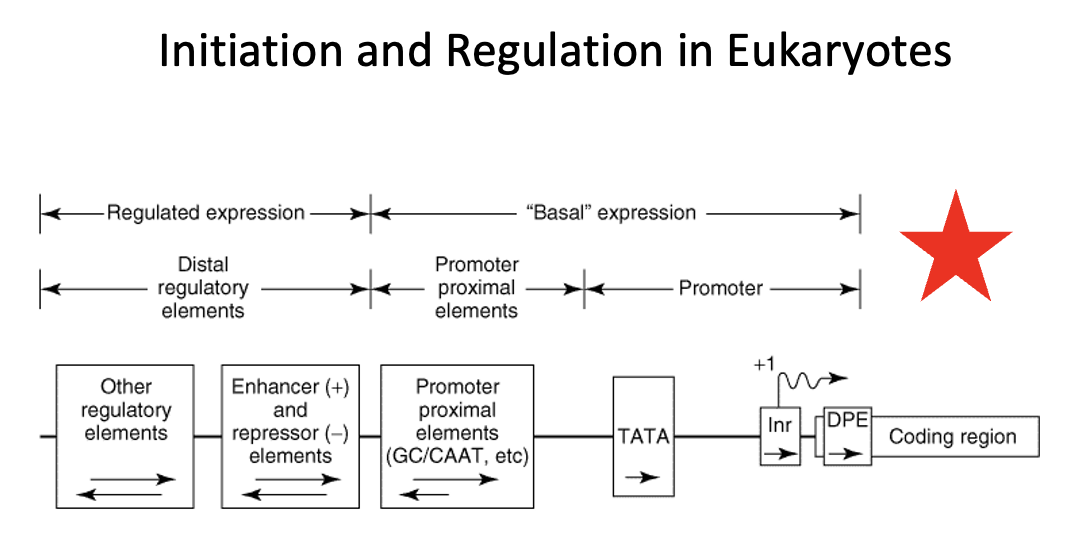
Coding region in Eukaryotes
region that contains the DNA sequence that is transcribed into mRNA, which is ultimately translated into protein
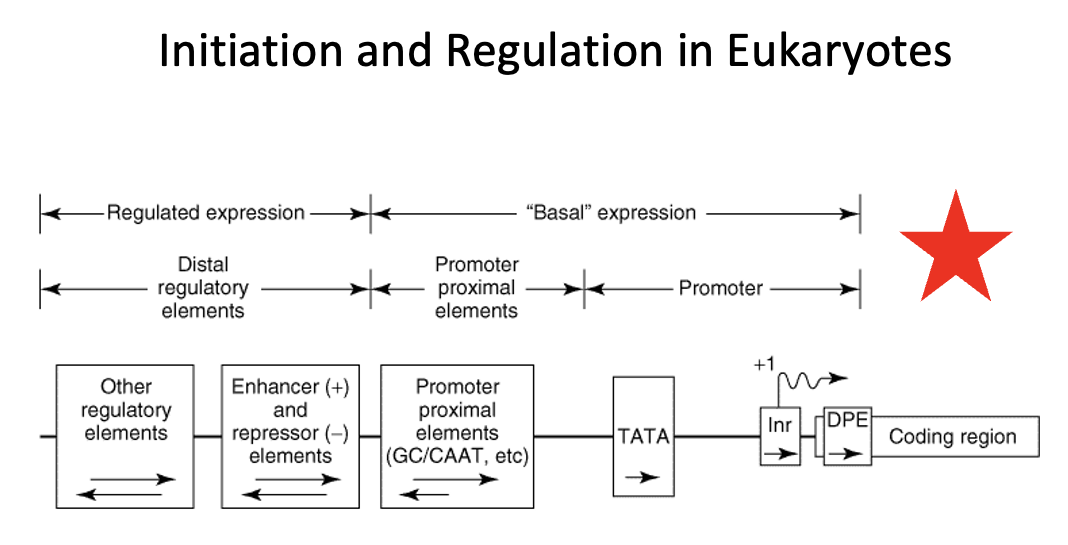
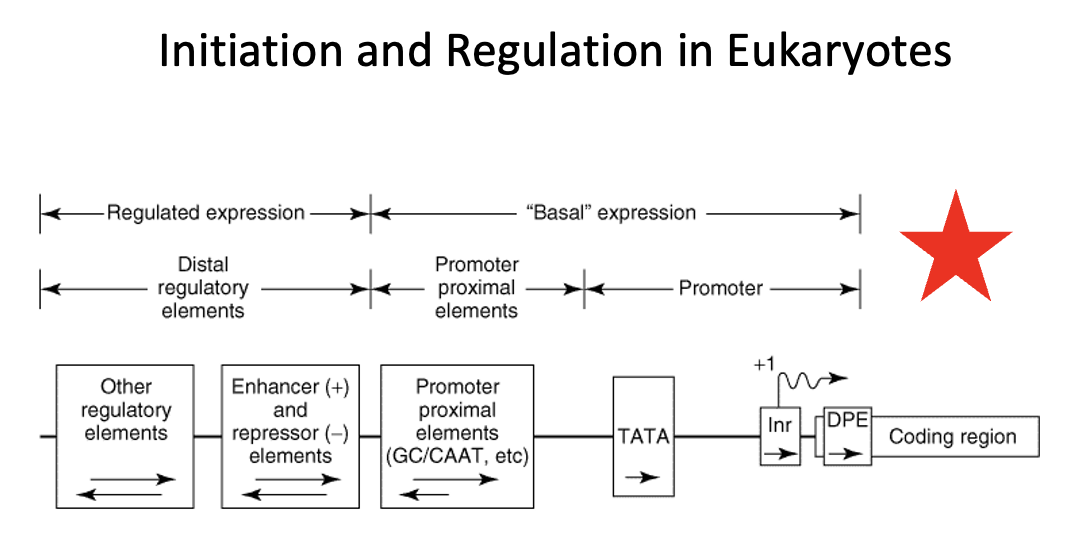
Regulatory region in Eukaryotes
region that consists of two classes of elements
one class is responsible for ensuring basal expression
→ these elements generally have two components (the proximal component = TATA box or Inr or DBP elements direct RNA polymerase to the correct site. In TATA-less promoters, an initiator (Inr) element that spans the initiation site may direct the polymerase to the appropriate site. The upstream elements, specifies the frequency of initiations, for example the CAAT box.
the other class is responsible for ensuring regulated expression
→ the distal regulatory elements consist of enhancers and repressors and other regulatory regions. Enhancers and repressors regulate transcription expression based on various signals including hormones, heat shock, heavy metals, and chemicals
The proximal promoter elements are required for:
a. basal expression of a gene
b. up regulation of a gene
c. down regulation of a gene
d. allowing transcription factors and RNA polymerase access to the DNA template
e. none of the above
a. basal expression of a gene
basal expression refers to the baseline level of gene transcription that occurs even without strong activating signals.
It is established by the core promoter and the binding of general transcription factors (GTFs) to recruit RNA polymerase to initiate transcription at a low but consistent level. This basal rate serves as a foundation upon which other regulatory mechanisms, such as enhancers and enhancers and repressors, can build to increase or decrease gene activity as needed
Pol I
one of the 3 RNA polymerases in eukaryotes
large ribosomal RNA
Pol II
one of the 3 RNA polymerases in eukaryotes
messenger RNA (mRNA)
often uses the TATA box consensus sequence to initiate transcription
Pol III
one of the 3 RNA polymerases in eukaryotes
small rRNA, tRNA, and other small RNA
RNA polymerase
the primary enzyme responsible for transcription (always in the 5’-3’ direction)
Proximal promoter elements
Basal expression in eukaryotes is controlled by _____ ______ _____
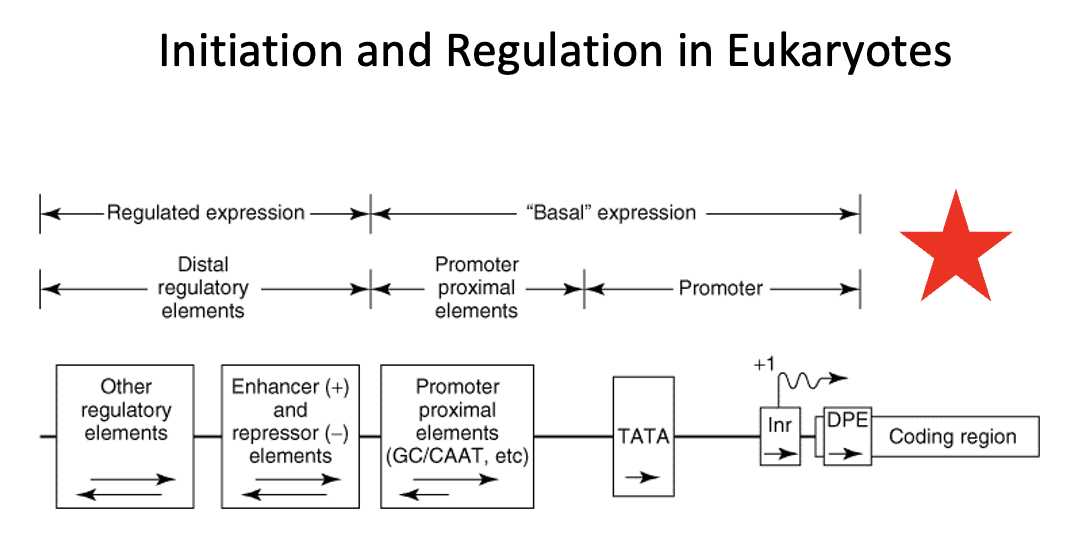
Distal regulatory elements
Regulated expression in eukaryotes is controlled by ____ _____ ______
What are the three steps of transcription?
Initiation = promoter, transcription factors and start point
Elongation = DNA is unwound, the template strand is read in the 3’-5’ direction and RNA is synthesized in the 5’-3’ direction by RNA polymerase
Termination = stop signal causes RNA polymerase to dissociate and the RNA transcript is complete
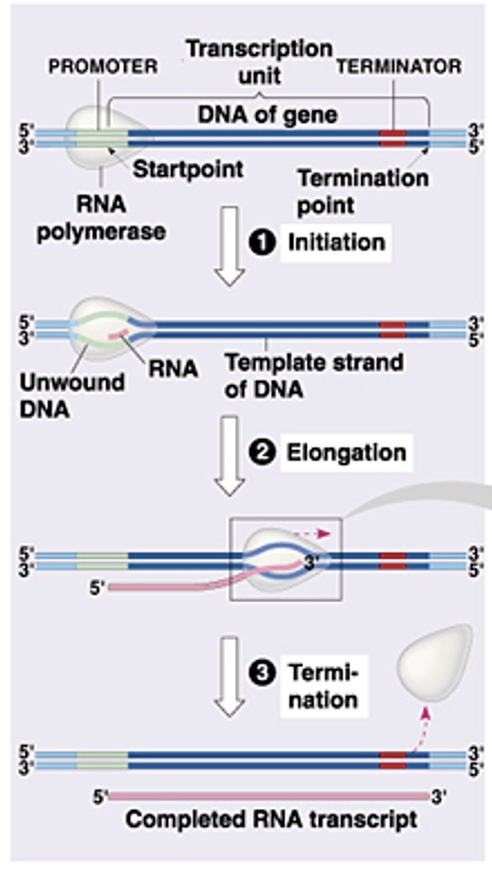
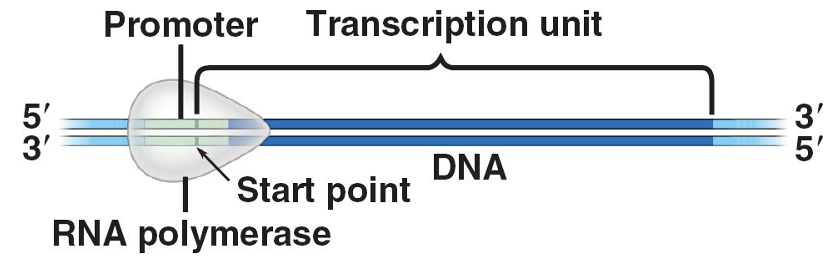
Initiation
•RNA polymerase recognizes and binds a specific promoter site
•RNA polymerase separates the DNA strands to expose the template and create the transcription bubble
•Initiation is complete when the first two ribonucleotides of an RNA chain are linked by a phosphodiester bond
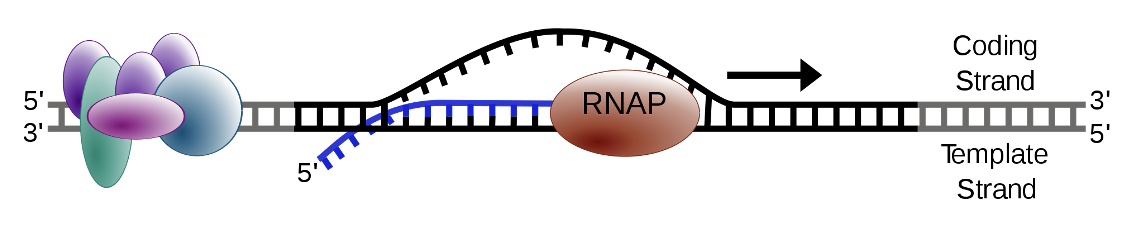
Elongation
occurs along the DNA molecule and therefore DNA unwinding must occur
Transcription bubble is approximately 20 base pairs per polymerase molecule
RNA polymerase has “unwindase” activity that opens the DNA helix
Topoisomerases precede and follow the progressing polymerase to prevent super helical complexes
RNA is elongated in the 5’-3’ direction, therefore DNA is read in the 3’-5’ direction
Termination
Certain factors cause the RNA polymerase to dissociate
Rho dependent, rho independent (prokaryotes)
poly A signal (eukaryotes)
Pre mRNA is released (but must still be processed)
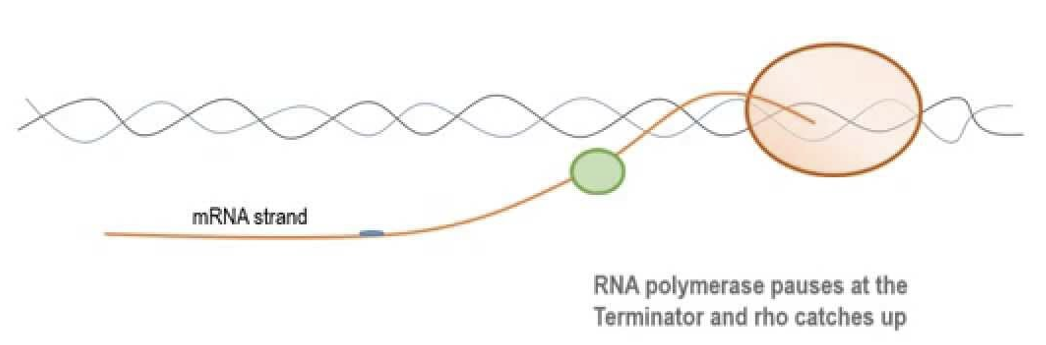
Rho-Dependent termination
occurs in prokaryotes
a sequence in the DNA molecule is recognized by a termination protein (the rho factor)
Rho is an ATP-dependent RNA stimulated helicase that disrupts the nascent RNA-DNA complex and causes RNApolymerase to dissociate from the template DNA.
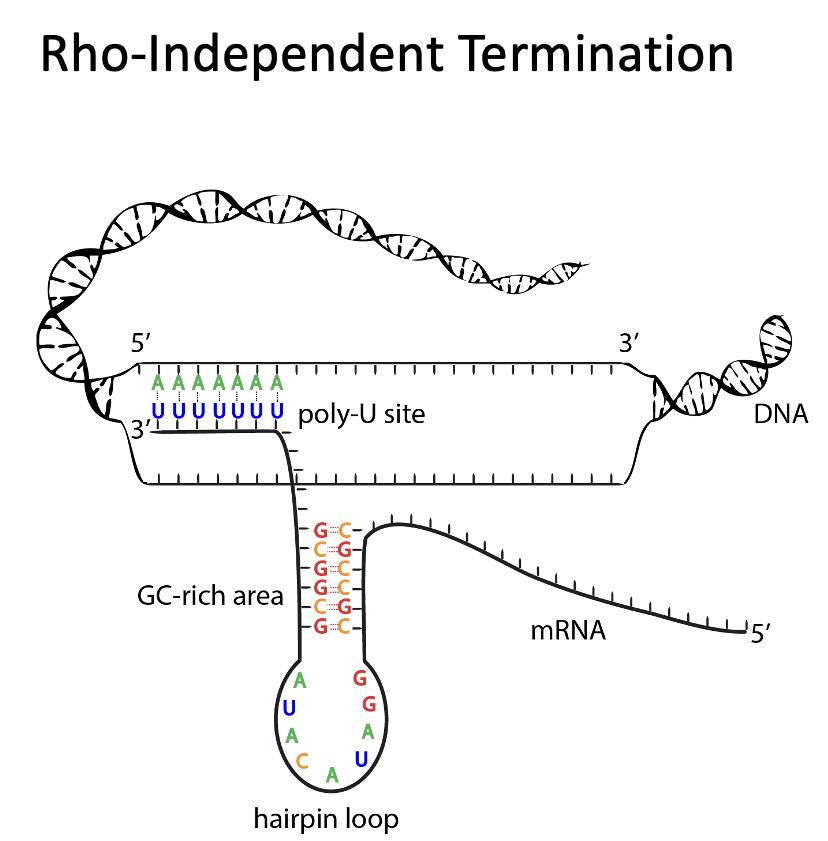
Rho-Independent Termination
occurs in prokaryotes
process that requires the presence of intrachain self-complementary sequences in the newly formed primary transcript so that it can acquire a stable hair pin turn that slows down the progress of the RNA polymerase and causes it to pause temporarily
near the stem of the hairpin, a sequence occurs that is rich in G and C (this stabilizes the secondary structure of the hair pin)
following the hair pin is a series of Us which provides a section of weak U-A bonding that facilitates the dissociation of the primary transcript from the DNA
Downstream Terminator sequence
occurs in eukaryotes
poly-A sequence
termination in eukaryotes is less well understood
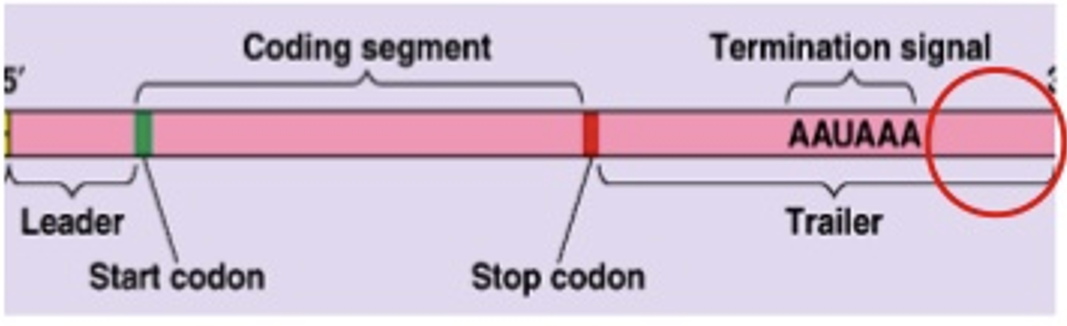
Post-transcriptional modification
In eukaryotes, mRNA must be modified before it is ready for transport to the cytoplasm for translation into proteins
The 5’ end of the RNA becomes capped (a methylated guanine residue is added to the end of the RNA transcript)
The 3’ end is modified by the addition of about 200 adenines by the poly-A polymerase
The RNA must undergo splicing to remove the introns
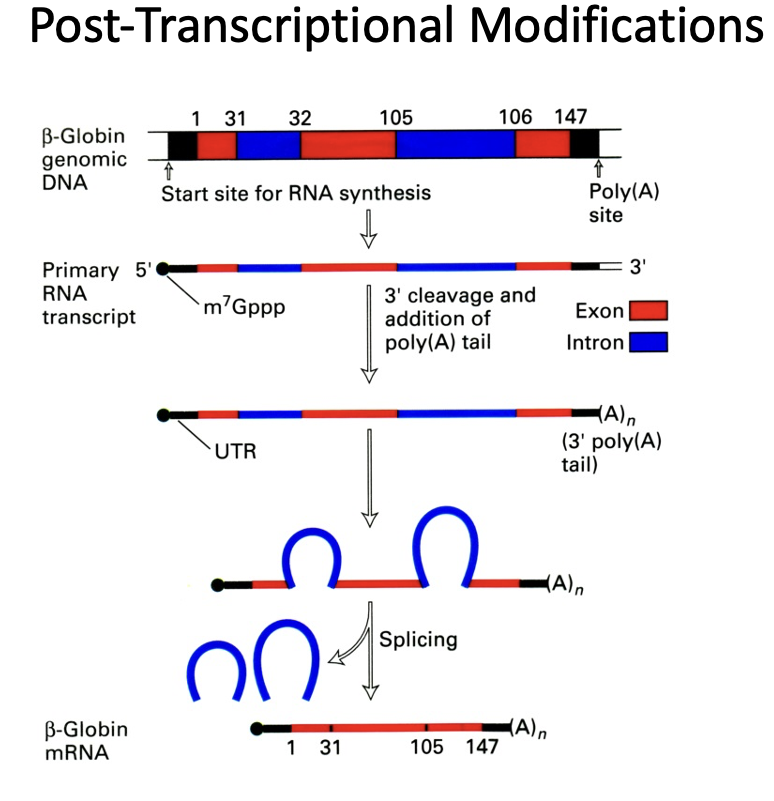
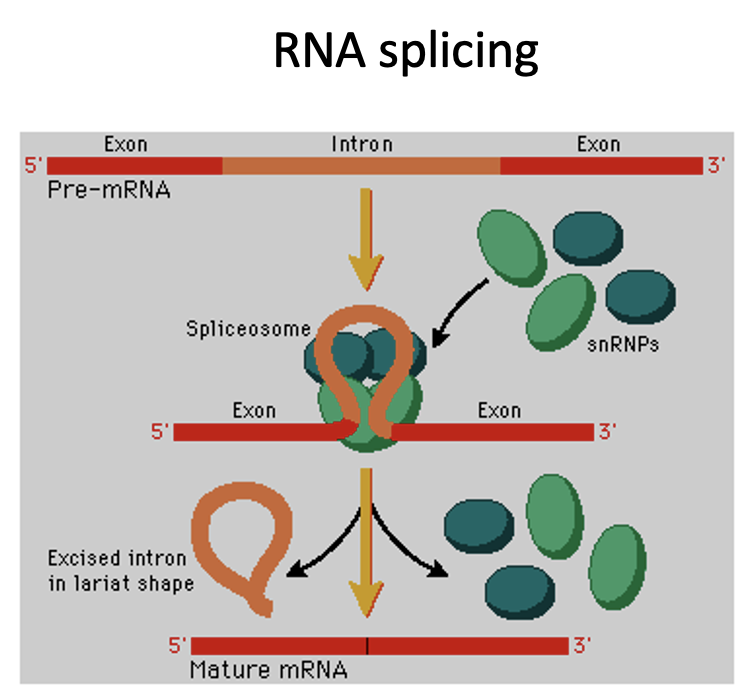
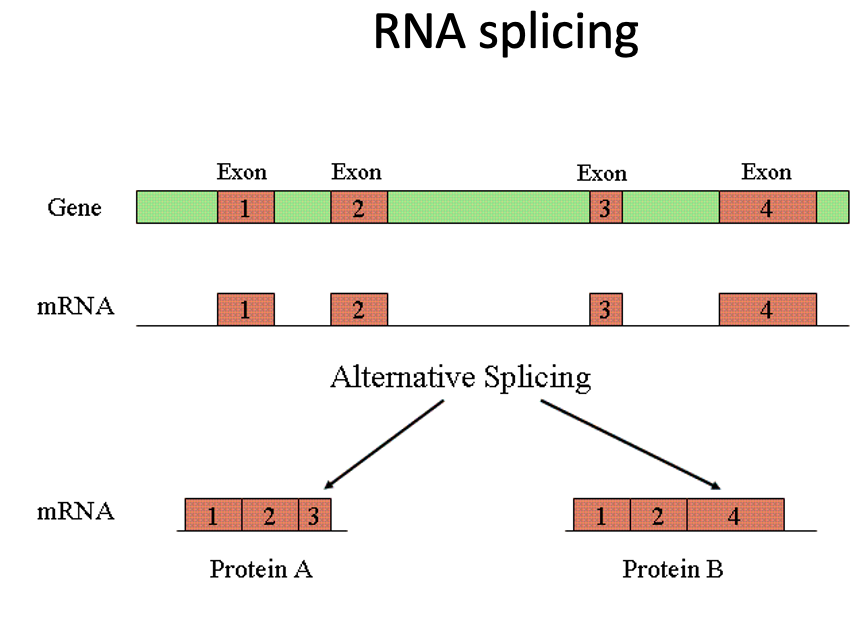
RNA splicing
one of the three post-transcriptional modifications that occur in eukaryotic cells
RNA is spliced to remove introns
alternative splicing can provide different protein products as well as a mechanism for differential regulation of the same protein
Methylated guanine cap
one of the three post-transcriptional modifications that occur in eukaryotic cells
it is added to the 5’ end of the mRNA
ribosomal subunits recognize the 5’ cap and use it to initiate translation
Poly A tail
one of the three post-transcriptional modifications that occur in eukaryotic cells
we take advantage of this in the lab and can use an oligo dT primer when examining expression levels of mRNA for use in PCR
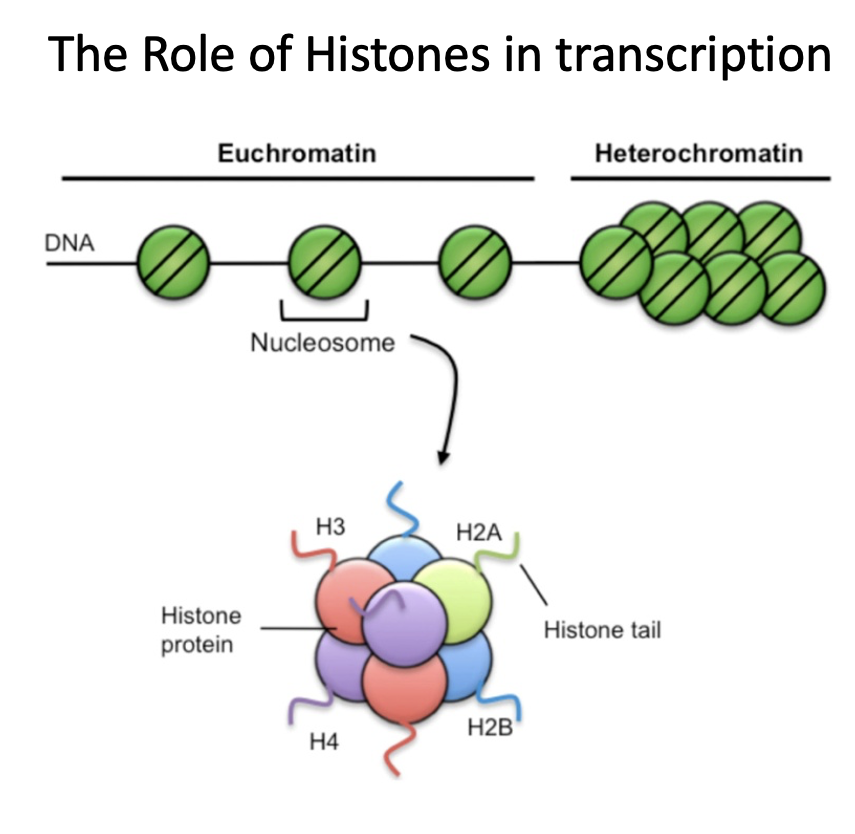
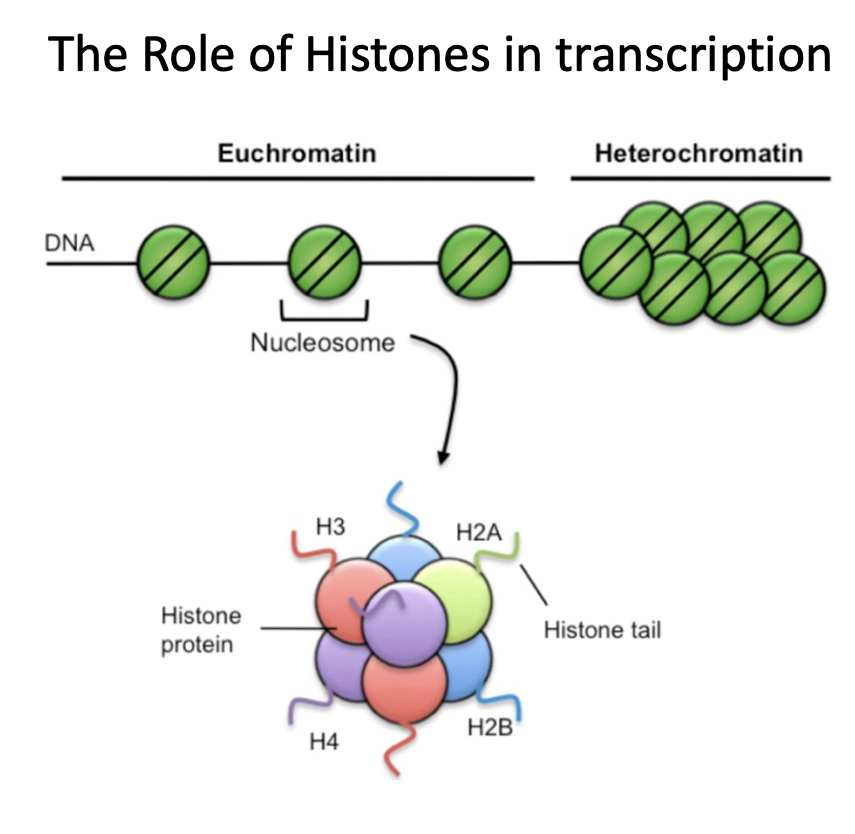
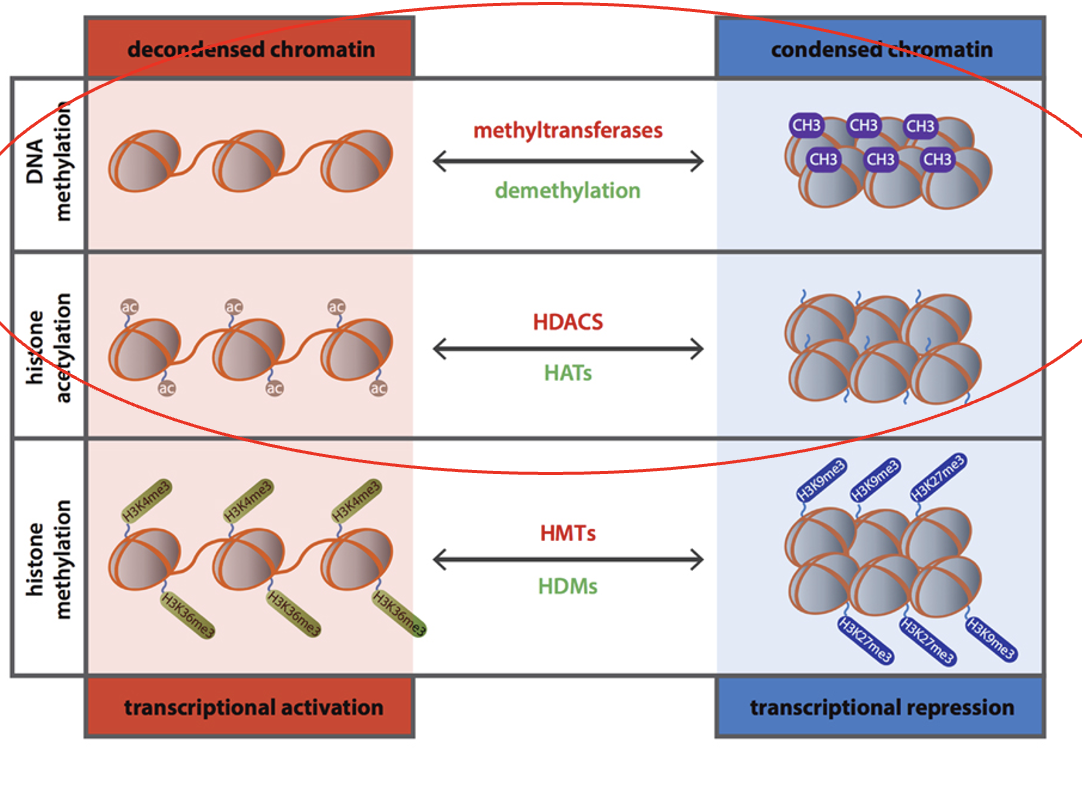
How does histone modification increase or decrease transcription?
Two post-translational modifications
acetylation = increases access to DNA, increases transcription
histone or DNA methylation = decreases access to DNA, decreases transcription
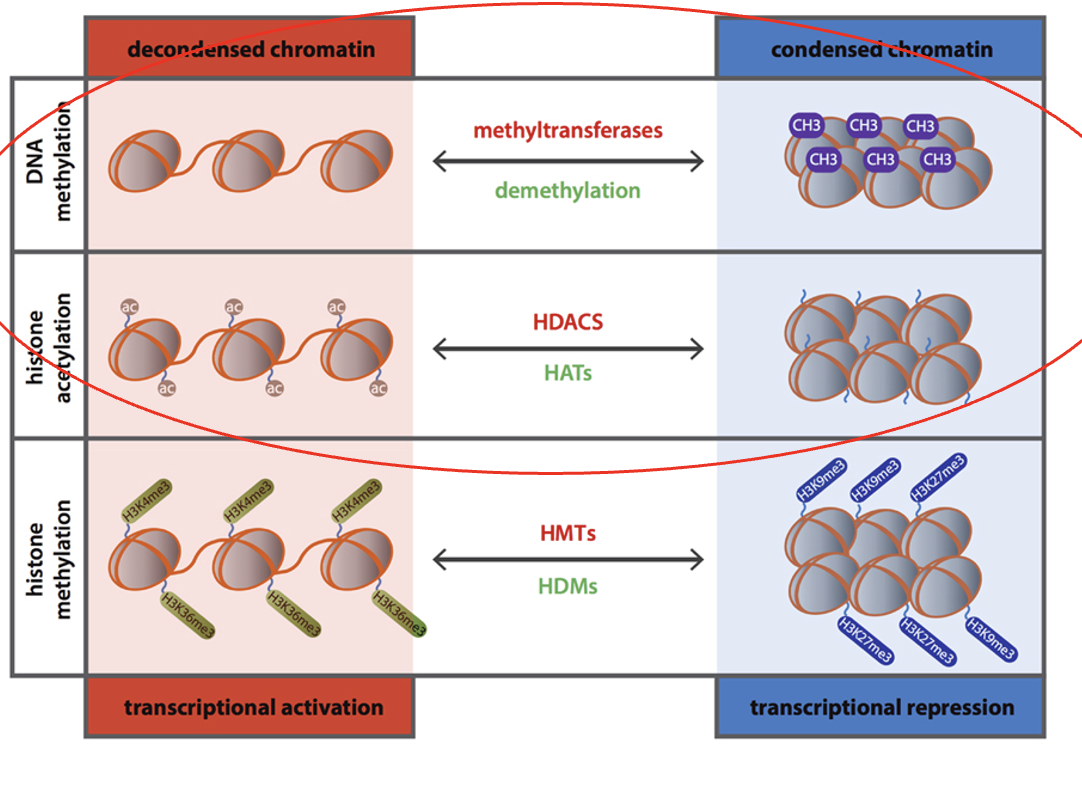
Acetylation
a type of histone modification
increases access to DNA, increases transcription
Histone Acetyl Transferase (HAT)
Histone Deacetylase (HDAC)
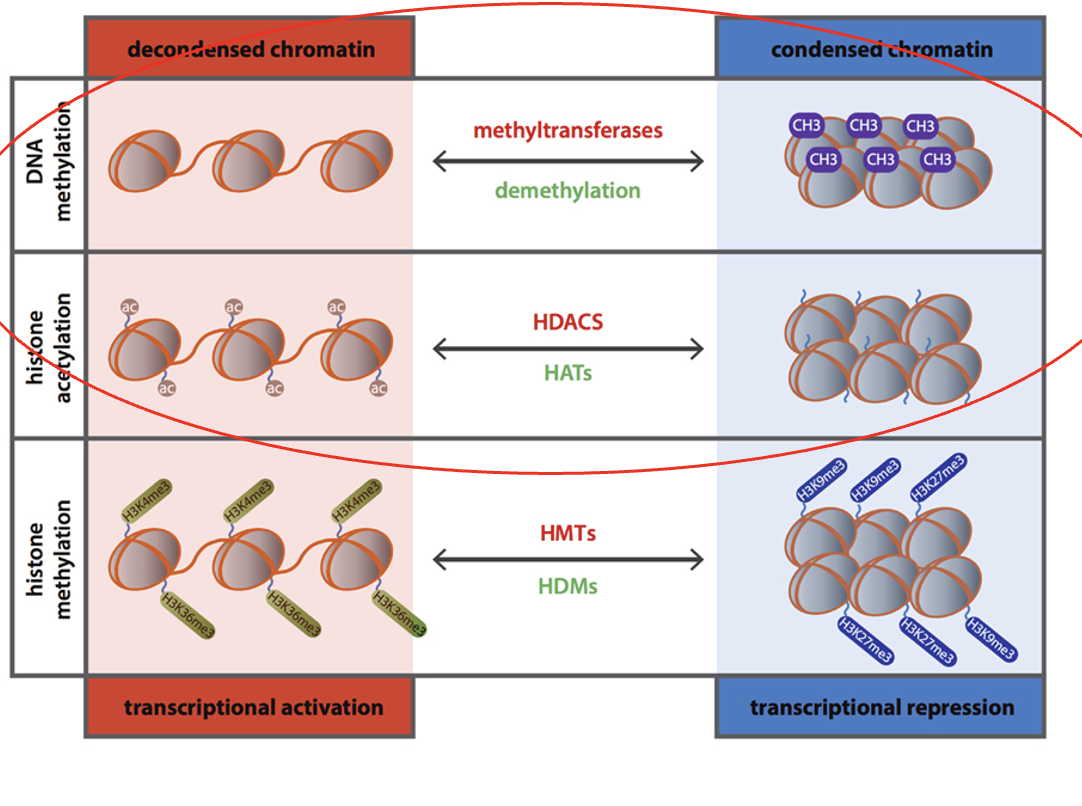
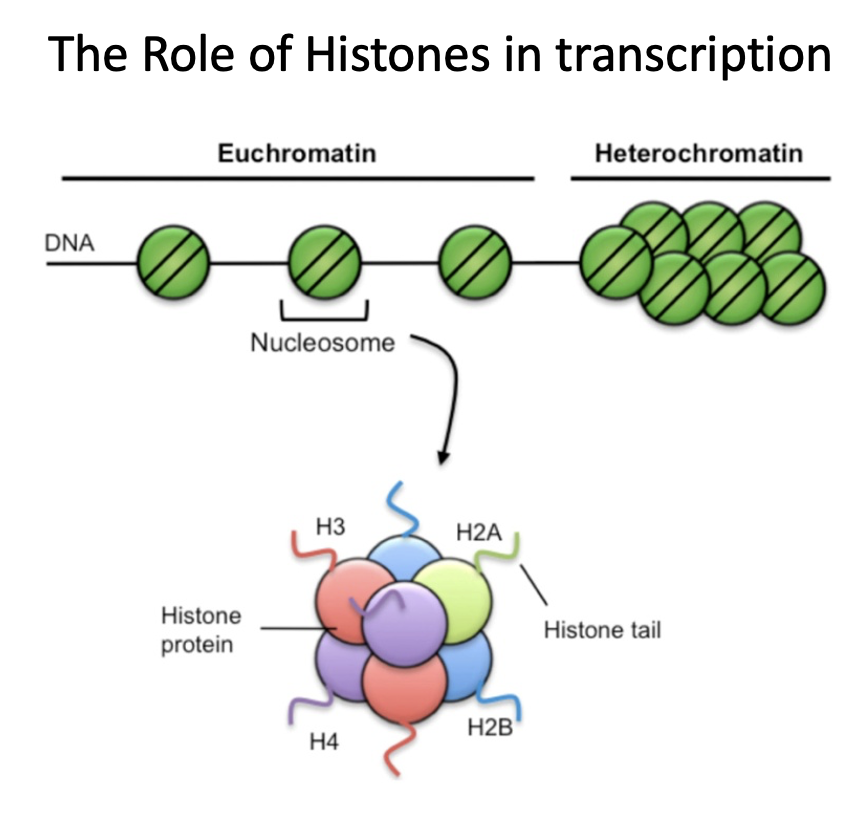
Histone or DNA methylation
a type of histone modification
decreases access to DNA, decreases transcription
DNA methyl transferase (DNMT)
Demethylase
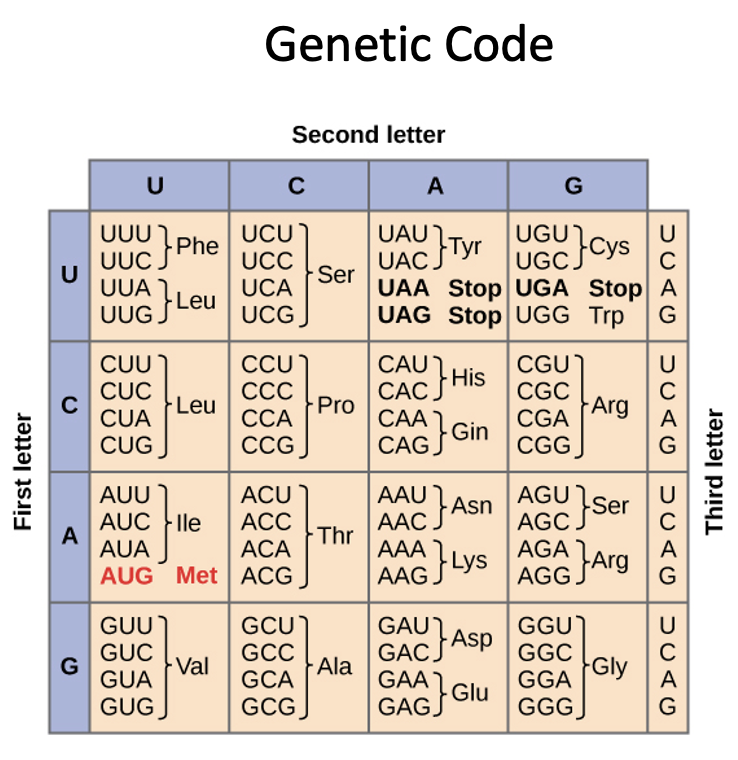
Start codon
AUG = Met
What are the three key components of translation?
tRNA
ribosomes
mRNA
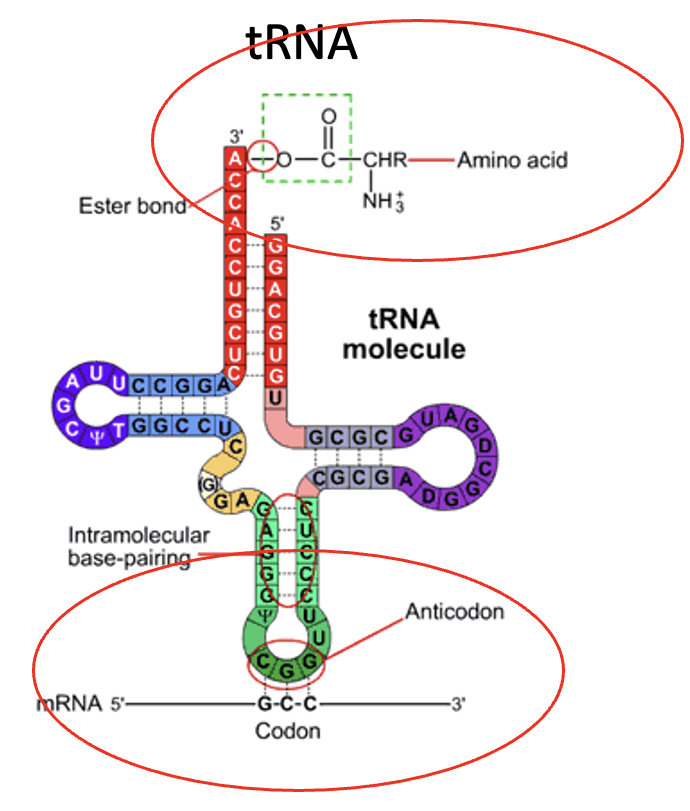
tRNA
responsible for transferring the amino acids to the ribosome
there are two important sites to remember on the molecule, the amino acid attachment (amino acyl) site and the anti-codon site
tRNA carries the amino acid and recognizes the codon
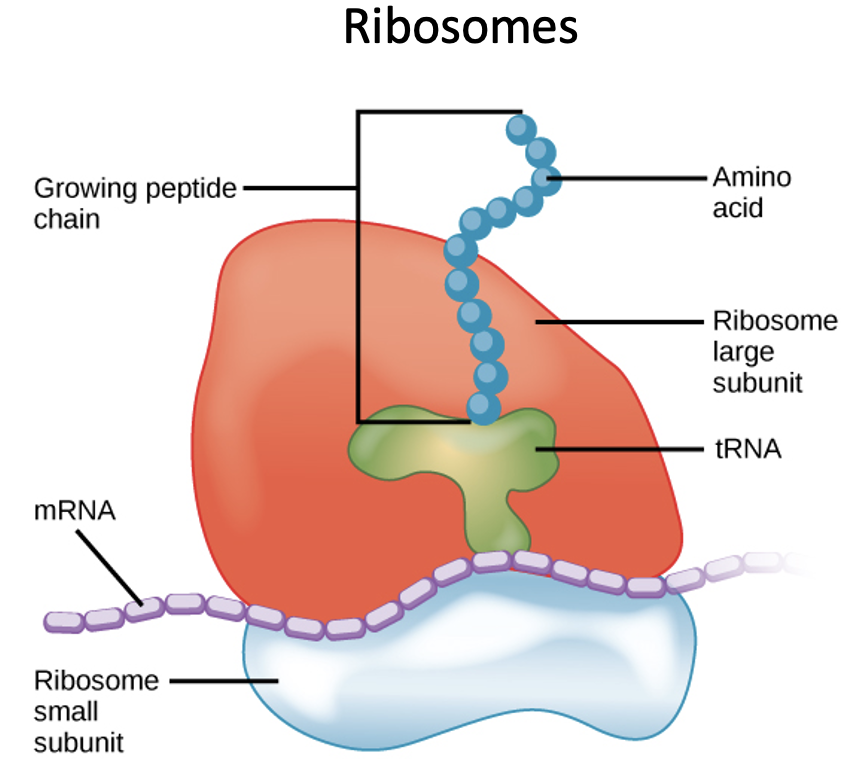
Ribosome
Site of protein synthesis
Has two binding sites for tRNA molecule: A and P sites, each of which extends over both subunits.
During translation, the A site binds an incoming Aminoacyl tRNA as directed by the codon currently occupying the site. The codon specifies the next amino acid to be added to the growing peptide chain.
The P site codon is occupied by the Peptidyl-tRNA. This tRNA carries the chain of amino acids that has already been synthesized.
An E site is also there that is occupied by the empty tRNA that is about to exit the ribosome
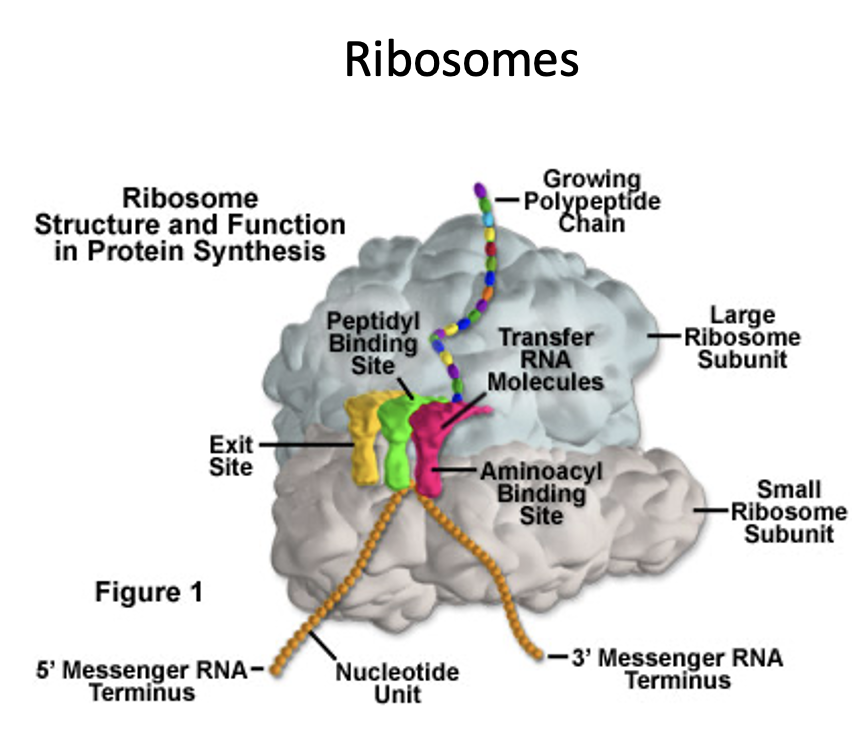
What are the steps of translation?
Requires two ribosomal subunits, mRNA, tRNA, and (in prokaryotes) 3 initiation factors.
First, the small ribosomal subunit binds to an initiation factor which then binds to the mRNA at the Shine-Dalgarno sequence (a specific sequence that the ribosomal subunit recognizes in prokaryotes).
Next IF-2 binds to MET-tRNA and promotes binding to the start codon.
The ribosome scans the mRNA until it encounters the start codon. This is called the initiation complex.
Next the large subunit attaches, the initiation factors are released and the complex is ready for protein synthesis.
Peptidyl transferase catalyzes the formation of a peptide bond.
The whole ribosome shifts over one codon and the A site is now free to accept the tRNA.
The peptide chain elongation process terminates when the stop codon is encountered on the mRNA. In prokaryotes, 15 aa can be added/sec.
Anti-codon site
a part of tRNA
3 bases that bind to the RNA and pair with the codon of the mRNA. Indicates which amino acid will be added to the peptide chain
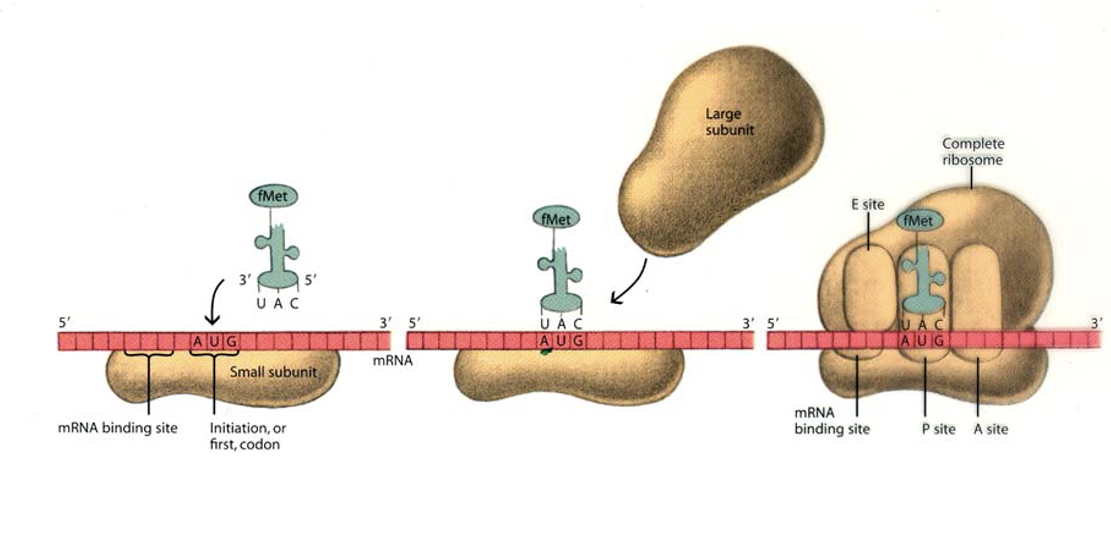
Initiation phase of translation
mRNA comes from transcription which is used to build a protein in the process of translation
A ribosome and initiator tRNA bind to the start codon on the mRNA
First, the small ribosomal subunit binds to an initiation factor which then binds to the mRNA at the Shine-Dalgarno sequence (a specific sequence that the ribosomal subunit recognizes in prokaryotes.)
First, the small ribosomal subunit binds to an initiation factor which then binds to the mRNA at the Shine-Dalgarno sequence (a specific sequence that the ribosomal subunit recognizes in prokaryotes.
Next the large subunit attaches, the initiation factors are released and the complex is ready for protein synthesis.
Next the large subunit attaches, the initiation factors are released and the complex is ready for protein synthesis.
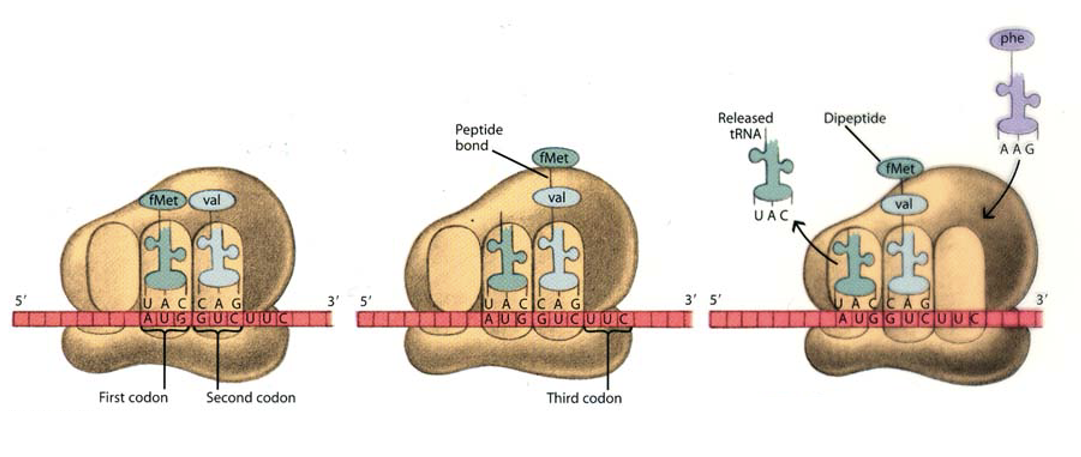
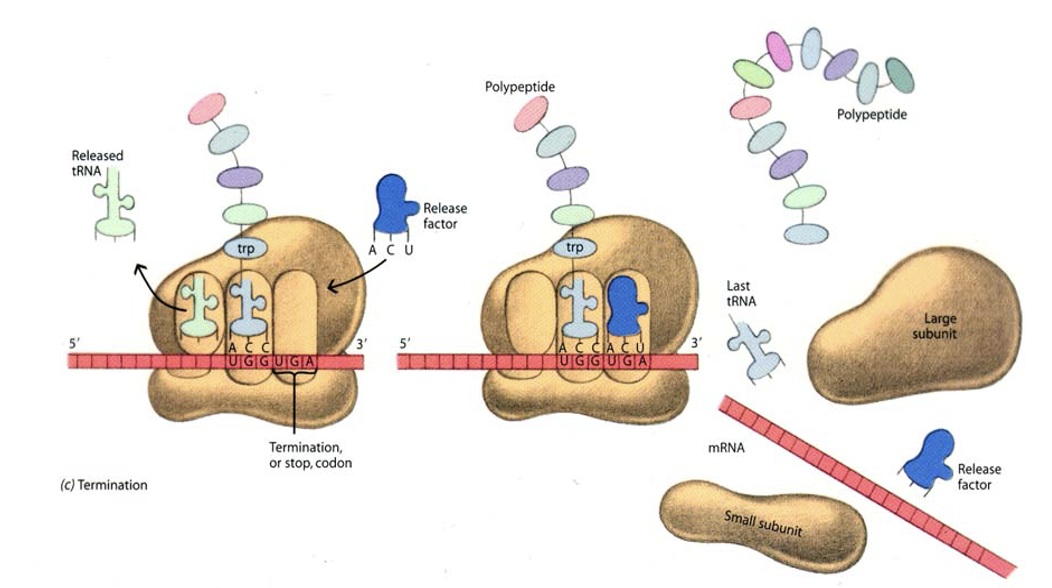
Elongation / Termination phase of translation
During translation, the A site binds an incoming Aminoacyl tRNA as directed by the codon currently occupying the site. The codon specifies the next amino acid to be added to the growing peptide chain.
The P site codon is occupied by the Peptidyl-tRNA. This tRNA carries the chain of amino acids that has already been synthesized.
An E site is also there that is occupied by the empty tRNA that is about to exit the ribosome
****The peptide chain elongation process terminates when the stop codon is encountered on the mRNA. In prokaryotes, 15 aa can be added/sec.
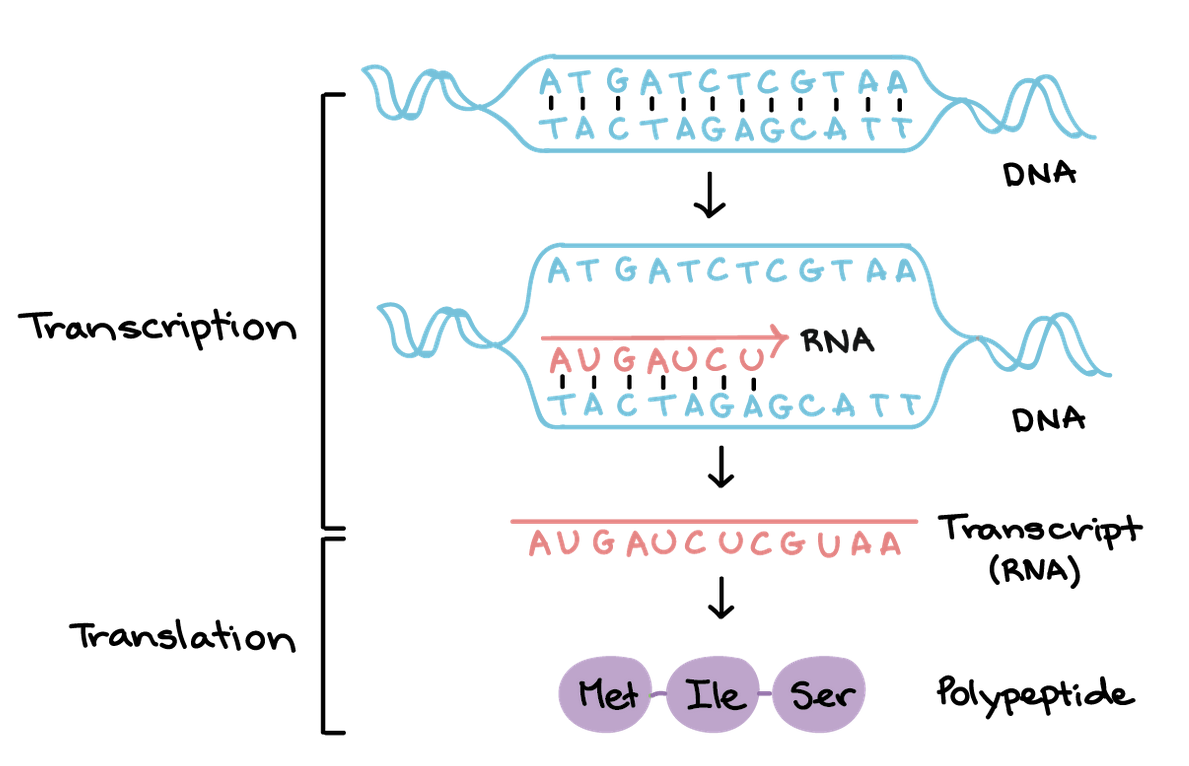
Transcription vs Translation Summary
Transcription involves initiation, where RNA polymerase binds to DNA to start copying a gene's code into messenger RNA (mRNA), followed by elongation, where RNA polymerase synthesizes the mRNA strand, and termination, where it stops and the mRNA is released.
Translation then uses this mRNA to build a protein, also in three phases: initiation, with a ribosome and initiator tRNA binding to the start codon on the mRNA; elongation, where amino acids are linked to form a growing polypeptide chain; and termination, where a stop codon signals the release of the finished protein
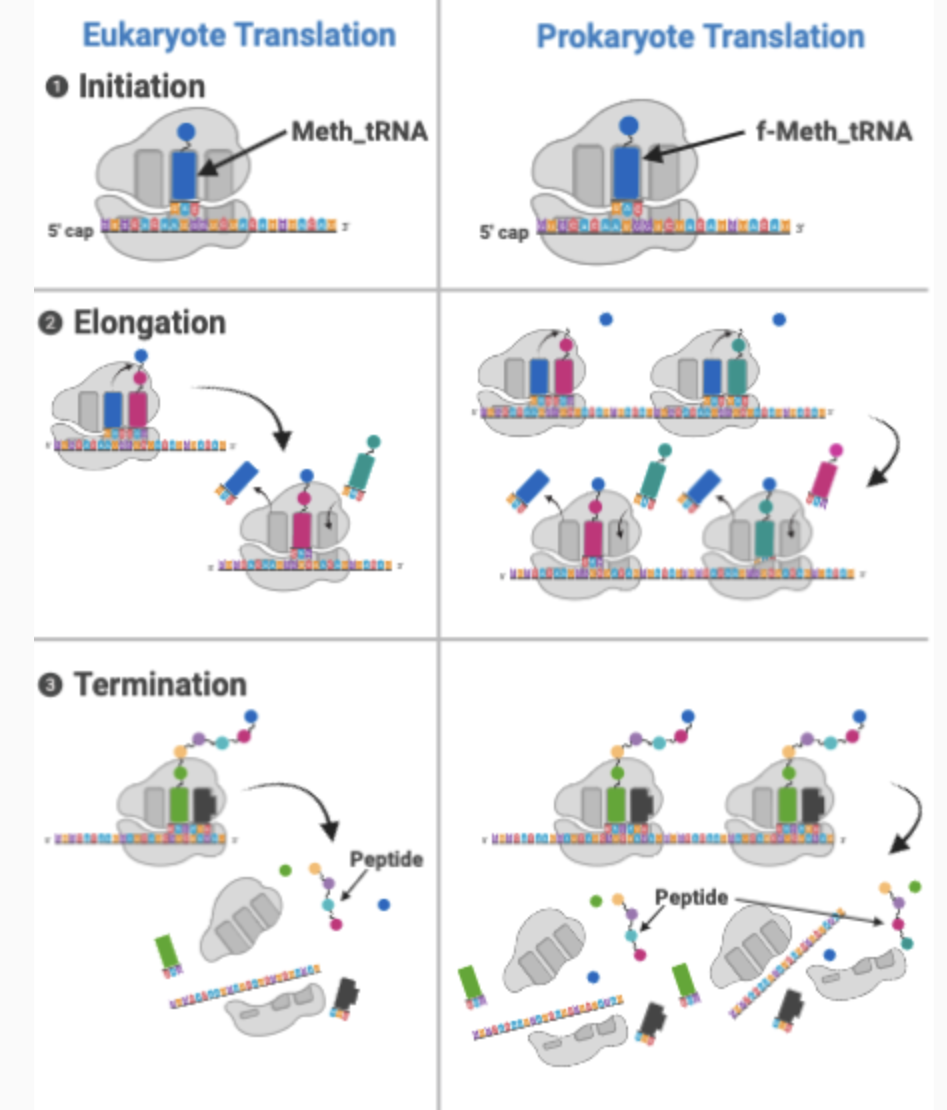
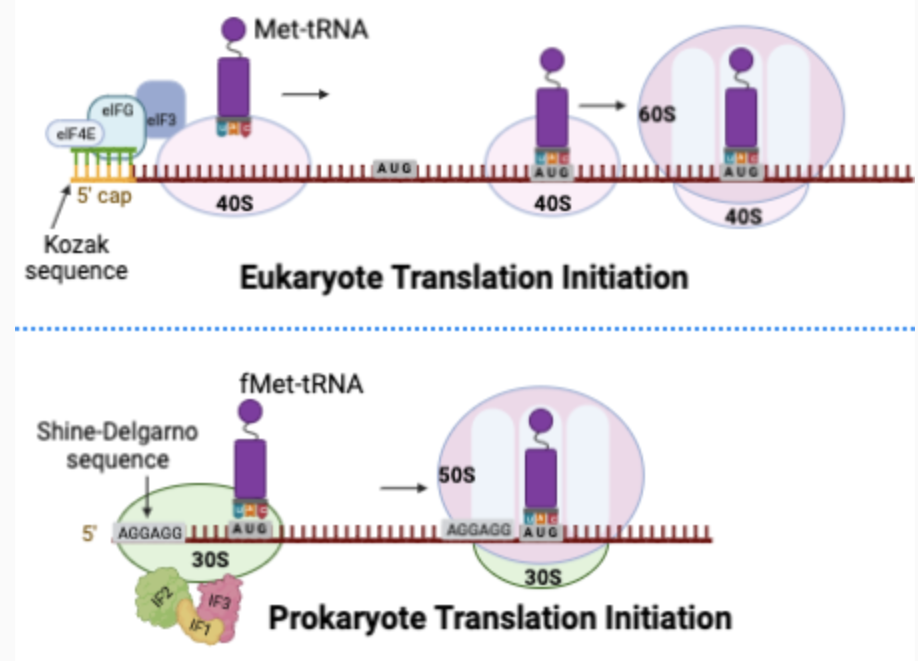
Prokaryotic vs Eukaryotic Translation
Prokaryotes = ribosomes recognize a sequence called the Shine-Dalgarno sequence, where they bind to initiate translation
Eukaryotes = ribosomal subunits recognize and bind to the 5’ cap
In eukaryotes there are many more protein factors involved (there are 3 Ifs in prokaryotes and at least 10 in eukaryotes)
In eukaryotes, the poly A tail likely increases rate of re-initiation
mRNA
the code that dictates the order of amino acids in a protein
Amino acyl site
a part of the tRNA
carries a specific amino acid based on the anti-codon site. A tRNA molecule can only care one specific amino acid