Cell Biology
1/224
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
225 Terms
What is a Cell??
A membrane with biomolecules that react to stimuli.
Structure of a Microscope
Contains an eyepiece that gives another 10x magnification.
An objective sense that sets magnification and focus.
A light source and a condenser that project light specifically on the specimen.
Fluorescence Microscopy
This is a form of microscopy where you isolate the specific colors of the specimen.
Electron Microscopy
This is another form of microscopy where an electron g*** to fire high speed electrons at a specimen which goes beyond the resolution of light.
How does EM work??
Biological tissues are stained with heavy metals because their high proton number increases electron scattering, improving contrast in electron micrographs.
Advantages & Disadvantages of EM
Advantages:
- Superior resolution
-Can visualise the whole cell
- Huge magnification
Disadvantages:
- Requires fixation of cells (so cells must be dead)
- Only small pieces of tissue can be imaged
- Time consuming
Endoplasmic Reticulum (mention the areas of the ER).
Rough ER where ribosomes are made.
Smooth ER (storage calcium, etc).
Tubules for increasing SA for ribosomes.
Key function is to make proteins for organelles and outisde of cell.
Inside is cytosolic side and outside is luminal
Nucleus in the EM
The nucleus is the largest organelle
We look at it on the microscope, we have dark spots which mean that more proteins signalling DNA is more active or RNA is presence in more abundance.
Golgi Apparatus
Protein modification and protein sorting is the key function
Fold the proteins by modifying the amino acids.
The proteins are taken up the cis side and then modification takes place.
Then they are released via the trans side, after which the protein can be sent to the target molecule.
How are functions in cells separated??
By large enzyme complexes (both in eukaryotes and prokaryotes) or by compartmentalisation of enzymes in organelles, which are surrounded by a membrane.
Organelles
Organelles are surrounded my membranes.
All organelles get proteins from ribosomes.
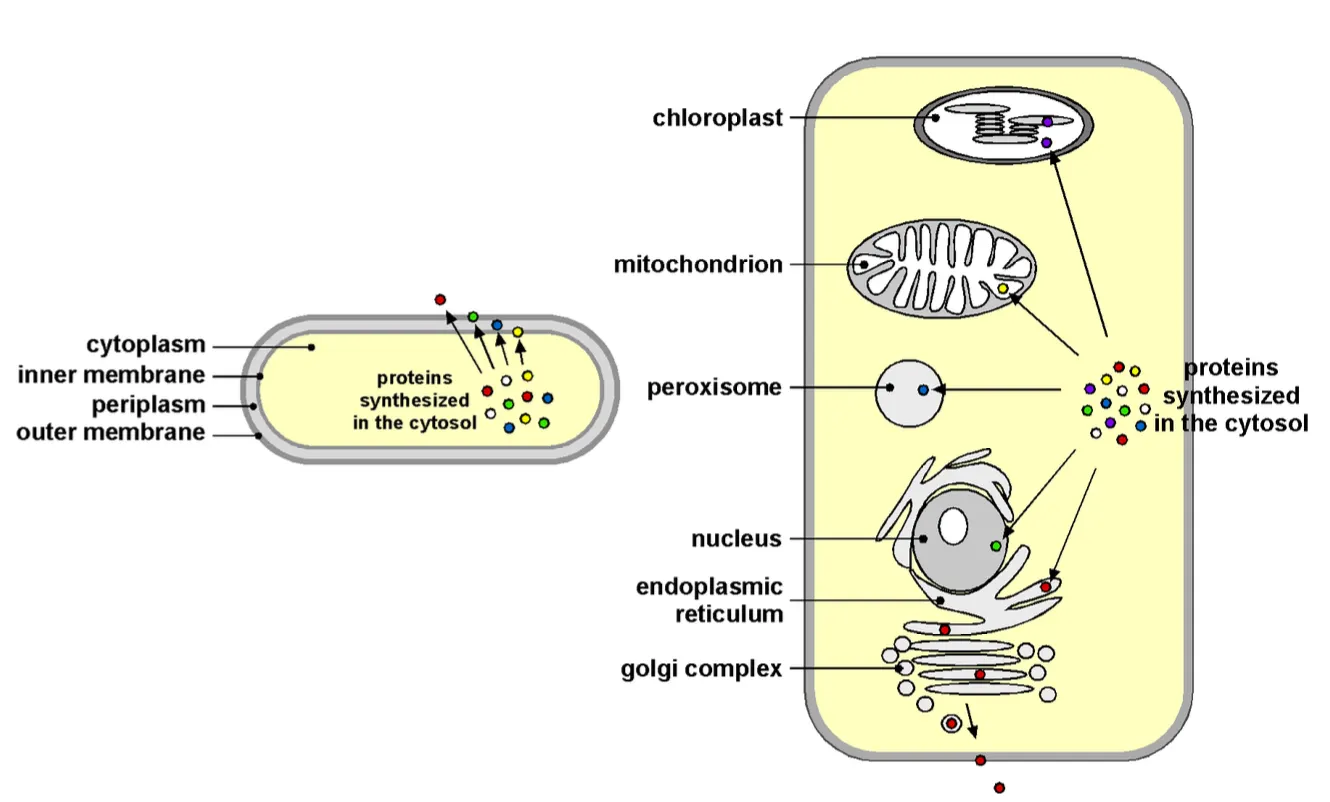
Organelles in Bacteria
Bacteria (prokaryotes) have no organelles in the cytosol but still proteins enter via the outer membrane, which is why there are also mechanisms to get the proteins in the cell.
Nucleus Function
DNA and RNA synthesis.
Cytoplasm
Overall this is a jelly matrix of proteins and contains Cytosol (protein synthesis/degradation) + organelles (specific functions).
What are the different functions taking place in the cytoplasm vs in the cytosol??
Cytoplasm:
- Transport
-Protein synthesis
Cytosol:
-Protein- breakdown
-Signal transduction
- Membrane fusion
-Ionic homeostasis
Cytoskeleton
Form and Transport
General Characteristics of Organelles
Have a lumen and a membrane
The cells are polarised,
Specific lipids and proteins are present in the lumen and membranes.
The abundance of an organelle will be different depending on a cell like muscle cells that have many mitochondria (ERs for gland cells because they actively secrete).
They also have the same position and function in all cells (for example that they are polarised and will have a specific apical and basal side helping with organisation of the cell also)
General Overview of Organelles
Precursors of eukaryotic organisms
Simple microorganisms, resembling present-day archaea, which had a plasma membrane but no internal membranes.
Explain how the organelles formed via Invaginations.
Golgi apparatus, Endosomes, and lysosomes most likely originated by invagination of the plasma membrane,
→ they together form what we call the endomembrane system
Nuclear envelope: partial invaginations
ER membrane: completely pinched off
Evolution of mitochondria and its Evidence
Theory that an anaerobic eukaryotic cell engulfed an aerobic bacterium, in a process called endosymbiosis.
Then over generation it developed into a mitochondria as the plasma membrane of the bacteria was degraded and eventually we were left with a mitochondria with a double membrane.
Evidence of this is that the mitochondria and chloroplast both have their own genomes and membranes and also their ribosomes are the same as bacteria. They also do not participate the the vesicular traffic of a cell.
Evolution of Chloroplast
Same goes for chloroplasts as the same as the mitochondria.
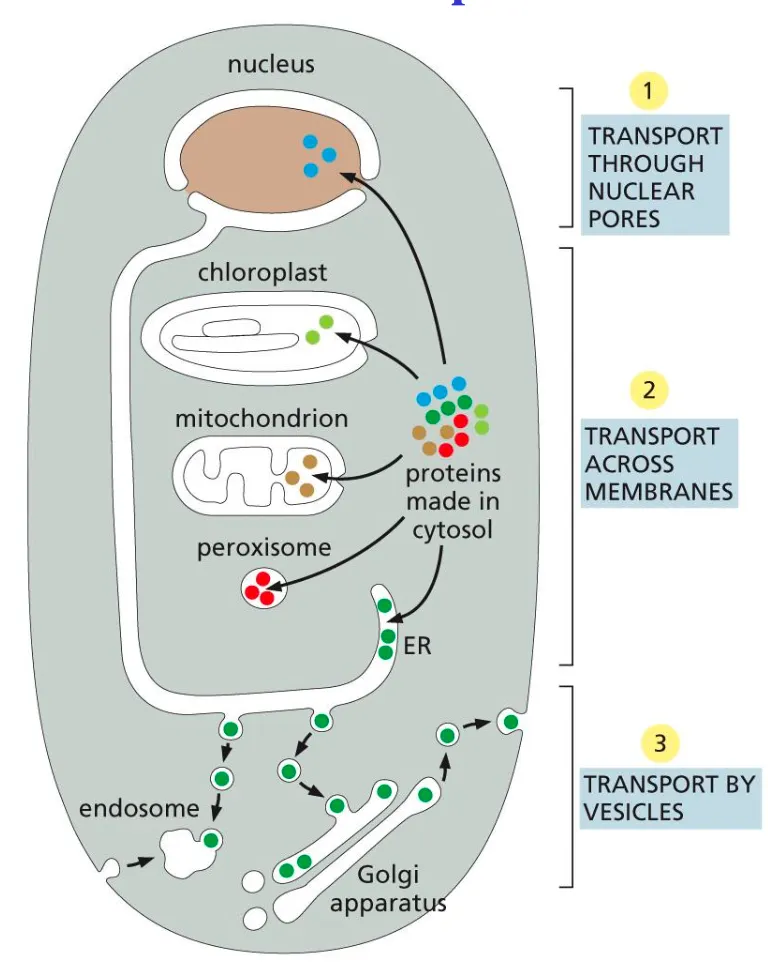
3 main transport mechanisms
Large pores: cytosol to nucleus (gated transport)
Narrow channels: Cytosol to ER/mitochondria/chloroplast/peroxisome
Vesicles: ER to GA to endosome/lysosome/ PM
General Mechanism of Transport Route in Cell
First molecules go out the nucleus via the nuclear pores then across narrow channels (transport membranes) and finally they undergo vesicle transport and can be exocytosed out the cell.
Signal Sequences
These are sequences in the genetic code which are 3-30 amino acids long and are used to target specific proteins to specific organelles. They are usually recognised by a receptor and are used to target the protein to the destination.
They are necessary and sufficient for sorting
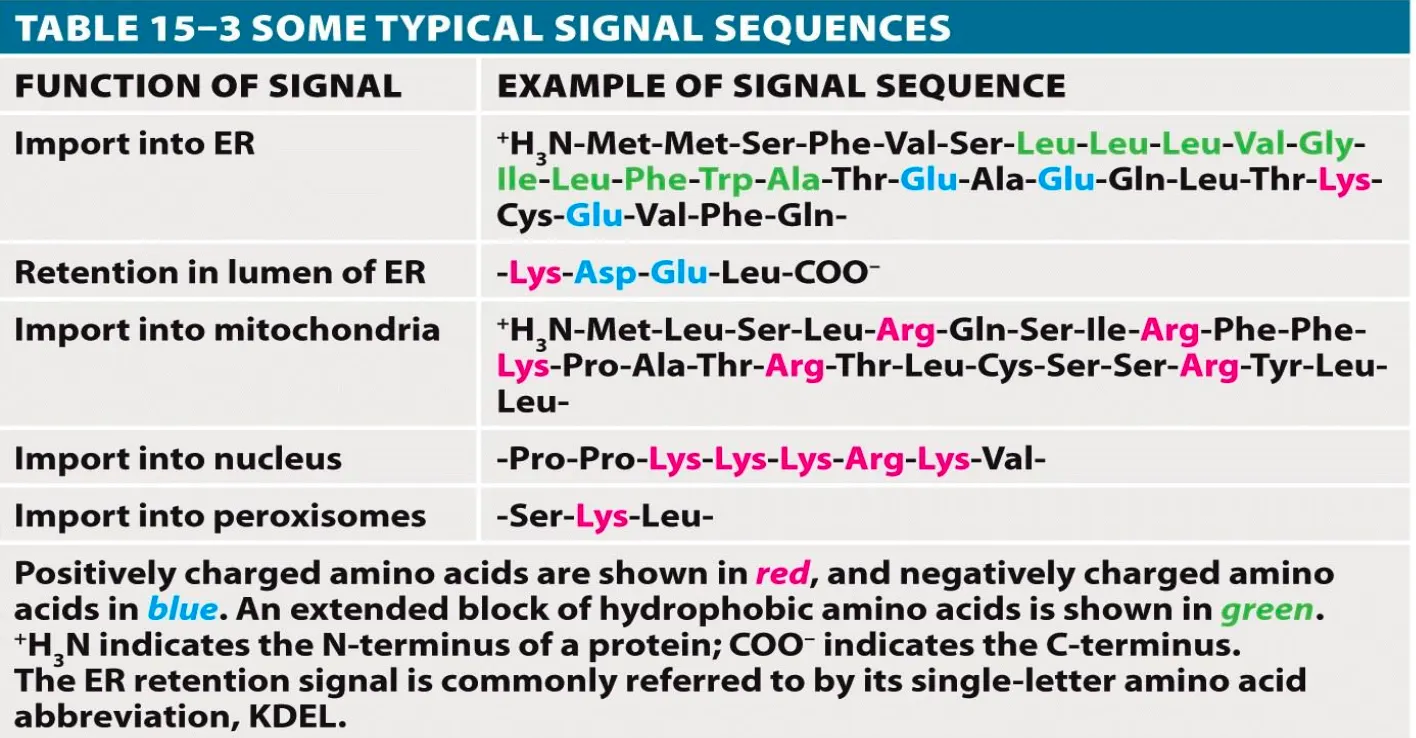
Experiment to Understand Signal Sequences
You need to know that if you add a signal sequence to a protein to be targeted to the ER that protein will go there so it is key that the correct proteins have the correct target sequence. That can negatively interfere in the process. Experiments have been done to see that is a ER tag is added to an cytosolic protein the protein will go to the ER and not the cytoplasm.
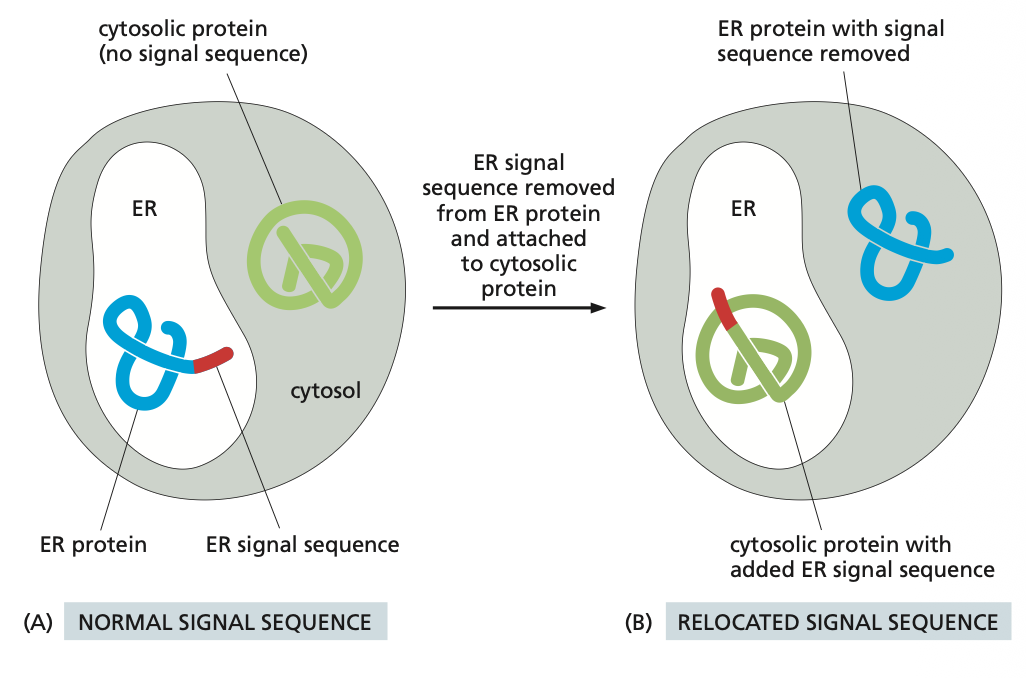

1. Cytosol → Nucleus
To import a protein from the cytosol to the nucleus we first have the prospective nuclear protein which has a nuclear localisation signal, which indicates that a protein needs to be transported into the nucleus. This signal is received by a nuclear import receptor, which binds to the signal.
The signal sequence must be positive as it must undergo an electrostatic interaction.
The transported proteins are folded.
Now this complex can be taken in the nucleus. This is done with help of the cystolic fibers, which help the proteins into the cell, which has a inter membrane space such as a gel like mesh network of nuclear proteins.
There is a nuclear basket which prevents molecules leaving that are not meant to.
Once the protein is in the nucleus, the receptor detaches and the protein can be transported to where it needs to be.
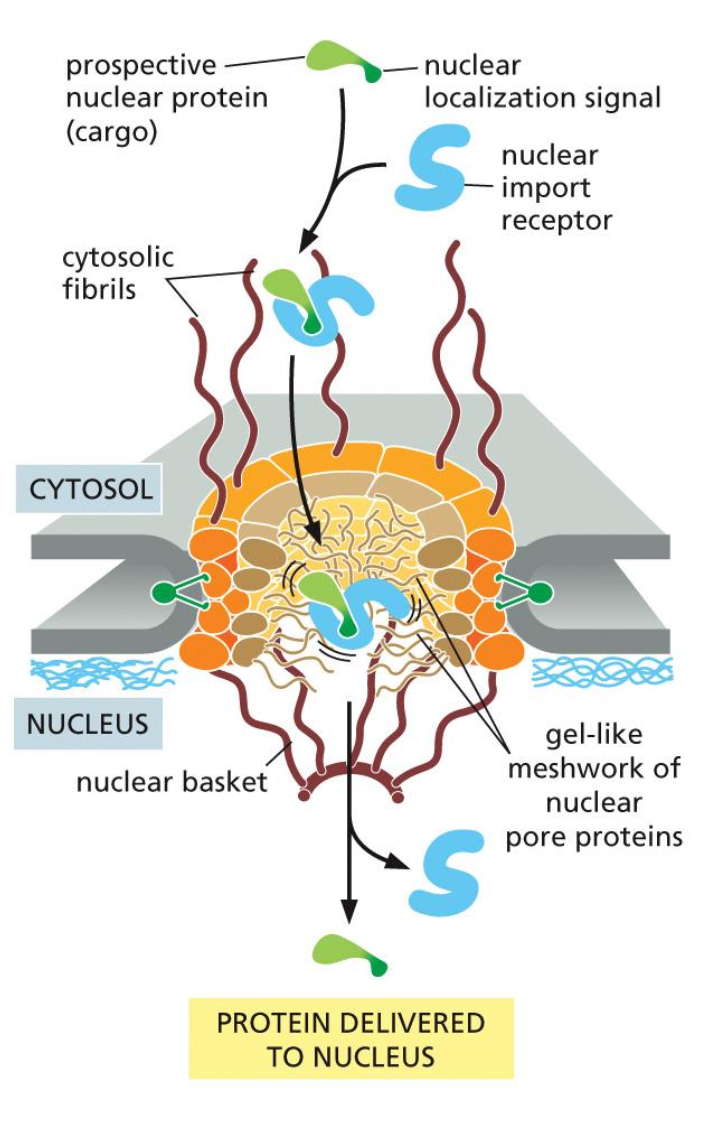
Mitochondria
powerhouse of the cell and undergoes ATP synthesis.
derived from the bacterial cells and also contains the same ribosomes and DNA.
some of the proteins are synthesised in the mitochondrion and some of them in the cytosol.
they also contain the following subcomponents: inner and outer membrane and lumen is the matrix.
Chaperone Proteins
Chaperone proteins can keep a protein unfolded but can also assist in the folding of a protein to a certain conformation.
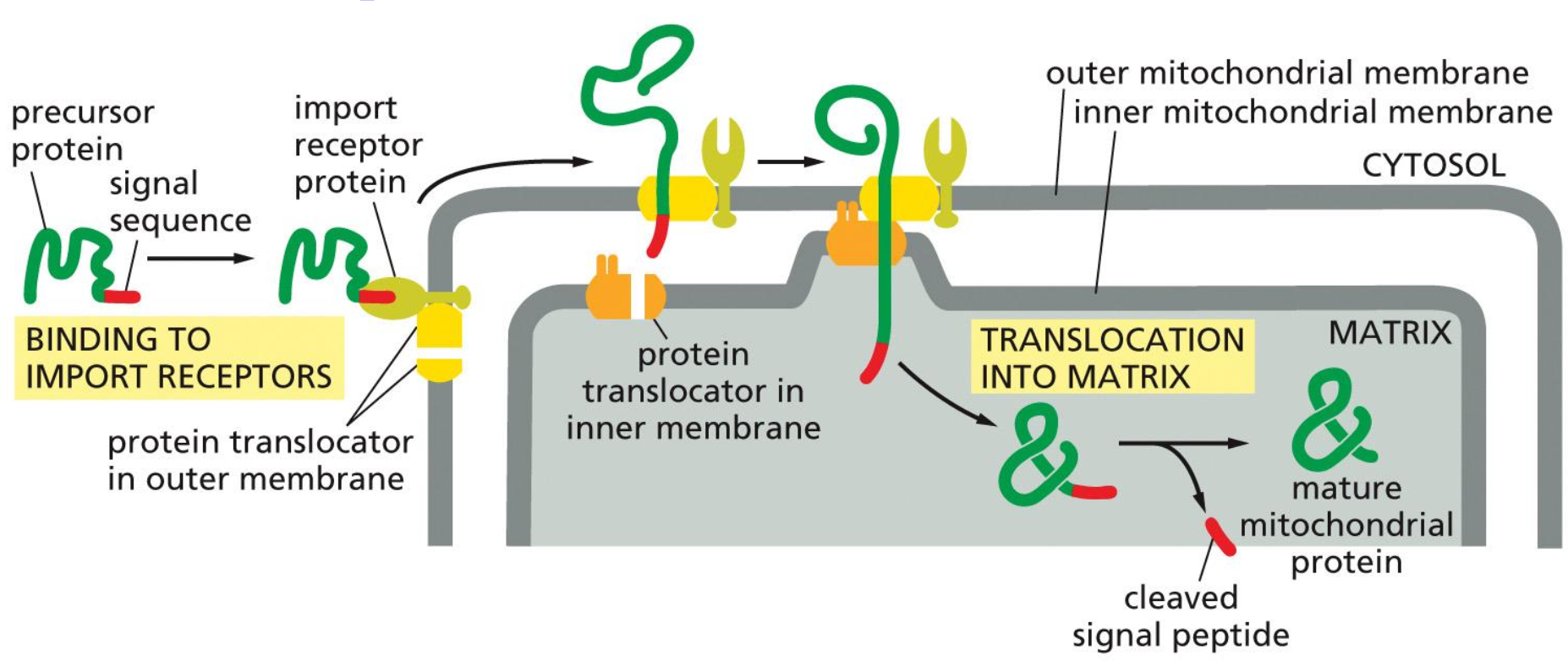
Import into mitochondria
So in this process we have the protein which is binded to the signal sequence in the cytoplasm, which then binds to chaperone proteins.
Eventually the signal sequence will bind to the import receptor protein and translocation occurs at the contact site. Following this process the protein can bind to the matrix. chaperones, which help in pulling the protein through and folding it into the correct shape.
Then the signal peptide is cleaved and we have the mature mitochondrial protein.
→ This is a post-transcriptional process.

Signal sequence for transport into the mitochondria.
As of the properties of the signal sequence: Also has the signal sequence which have positive charges that are not clustered like the nucleus ones. These ones are amphipathic and have an alpha helix. This will also be accepted by a certain receptor proteins.
Most mitochondrial and chloroplast proteins are coded for in the nucleus and usually have a signal sequence at their N-terminus that allows them to enter their specific organelle.
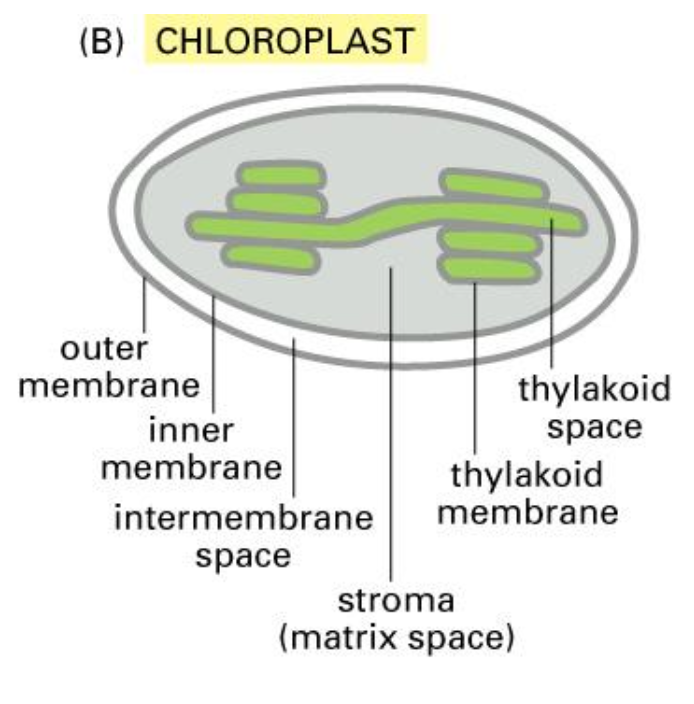
Chloroplast
chloroplast has same structure (exception of thylakoids).
lumen is called the stroma.
Note that an extra signal sequence is required to transport a protein to the chloroplast and it also requires a different targeting mechanism.
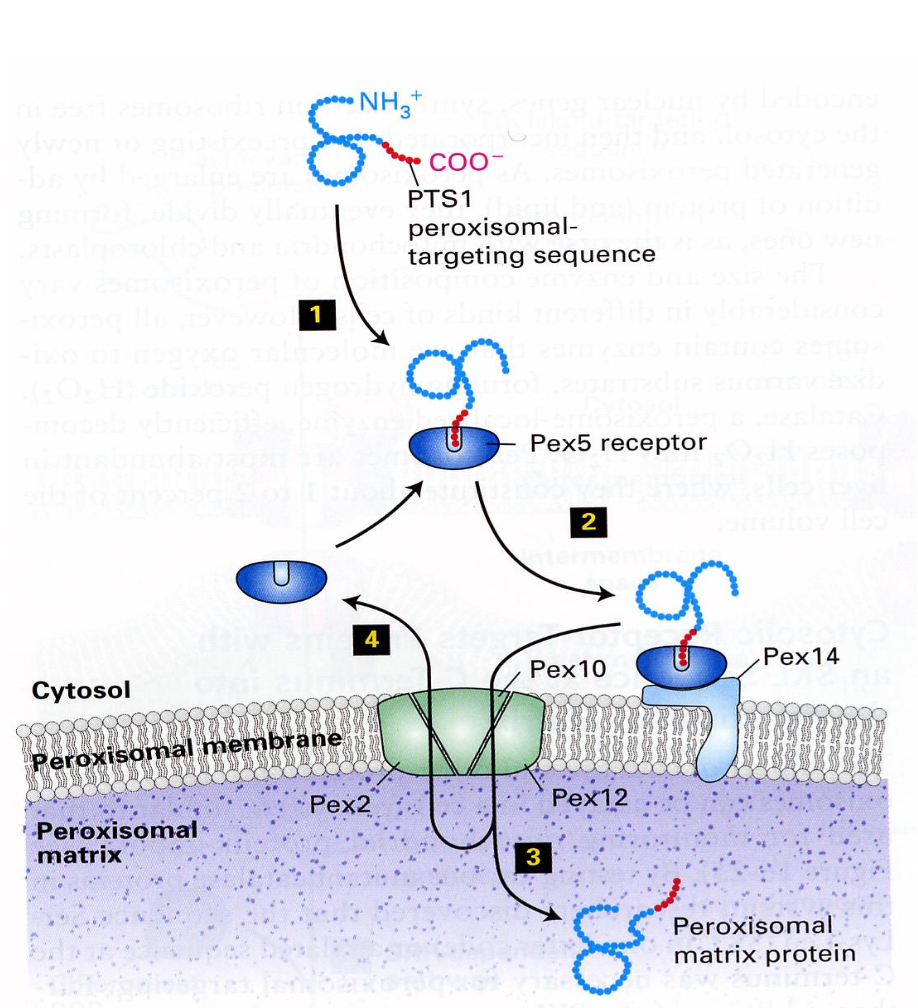
Peroxisomes
detoxification and oxidation of fatty acids
has a single membrane and has no DNA or ribosomes.
peroxidase abundant protein in cell.
evolved from invaginations.

3. Cytoplasm → Peroxisomes
Peroxisomal Import: synthesis with PTS, binding to the receptor in cytosol and binding to receptor in membrane, translocation through membrane channel and cytosolic receptor turns to cytosol. Protein transport in folded form.
The narrow channel needs to temporarily open up to let the folded protein to enter the matrix.
Signal sequence at the C-terminus, protein is completely synthesised.
Endoplasmic Reticulum
The ER is a continuous network and we have two types the rough ER and the smooth one. The rER is responsible for protein synthesis and has many ribosomes present and the sER is responsible for specialised functions (detoxification, production of steroid hormones, storage of Ca).
Once proteins are in the ER they can no longer be transported out alone they will instead be ferried by transport vesicles from organelle to organelle within the endomembrane system, or to the plasma membrane.
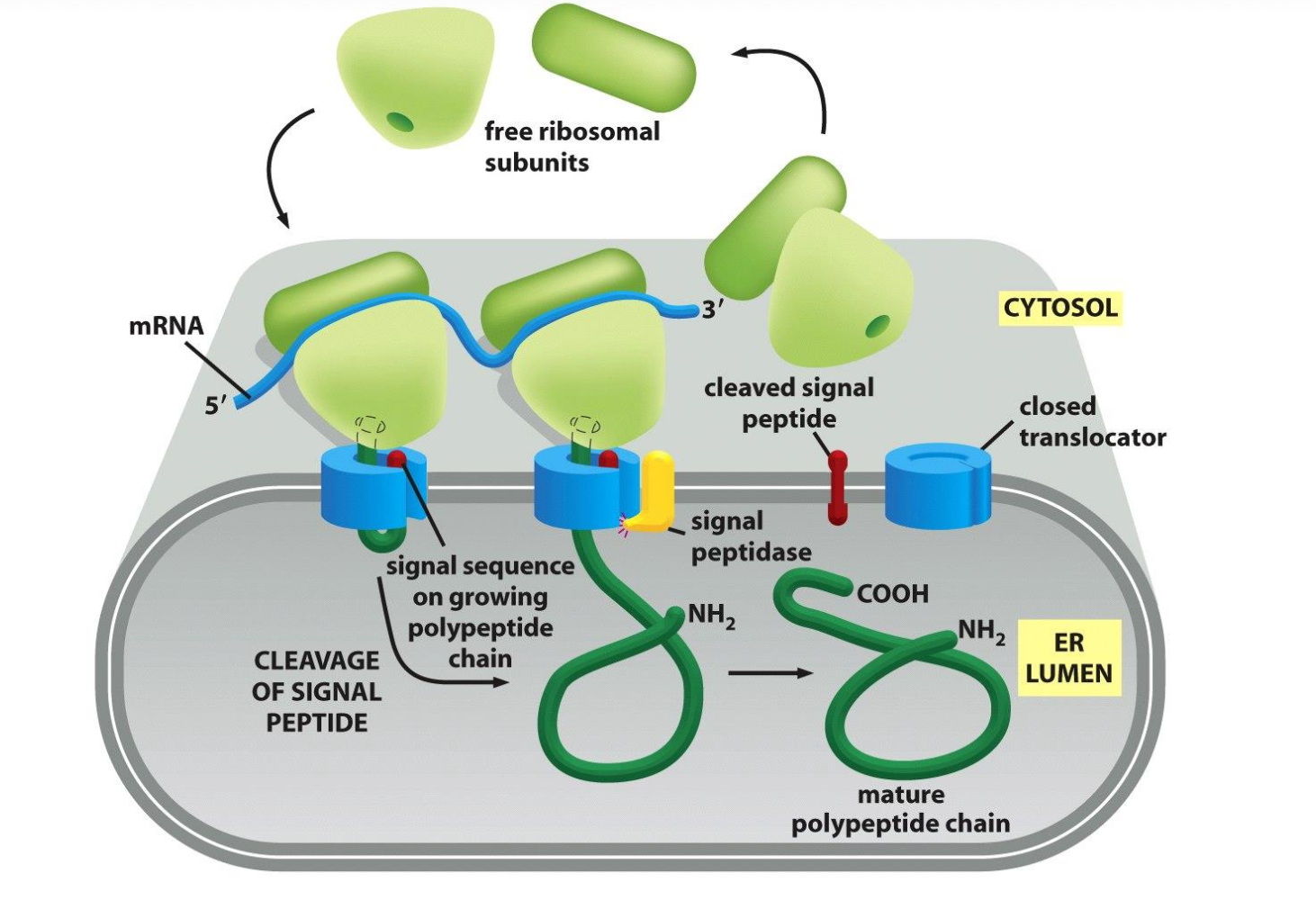
In order for proteins to be transported along the ER membrane the ribosomes have to be on the ER membrane so that the process of protein synthesis and transport across the ER can happen together. That is why the ribosomes coat the rER.
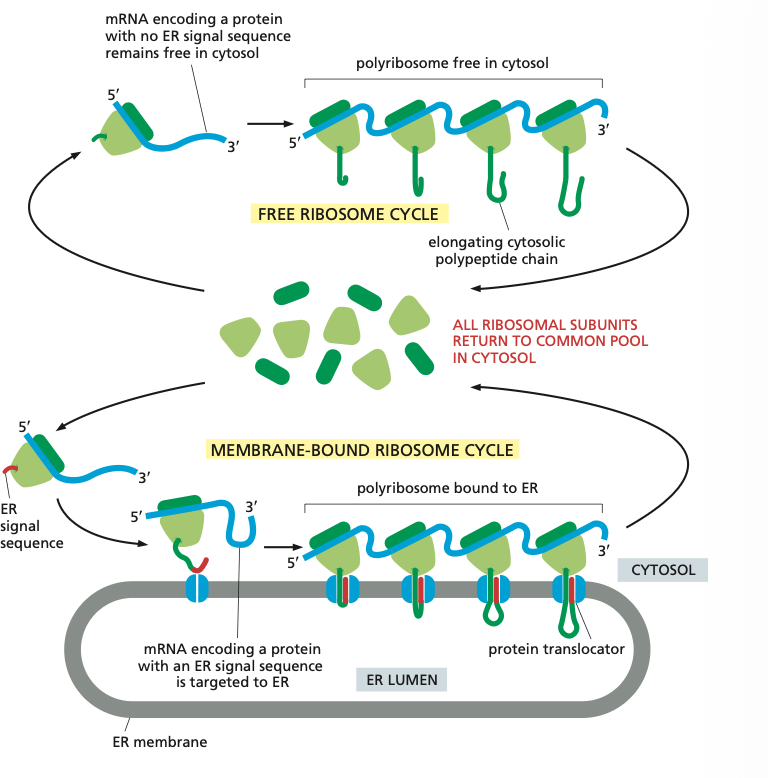
What is the difference between free ribosomes and membrane-bound ribosomes?
Free ribosomes:
- make cytosolic proteins
- proteins for the mitochondria/chloroplast/peroxisome and nuclear proteins
- post-translational process
Membrane-bound ribosomes:
- All the proteins of the secretory pathway (ER/GA/lysosomes/endosomes/PM)
- co-translational process
- Targeting sequence on mRNA determines if a protein is free or membrane-bound
Membrane bound ribosomes
Attached to the cytosolic side of the ER membrane.
ER signal sequence it is made by the membrane bound ribosomes so it can directly be taken up by the ER membrane.
Elongation of each polypeptide provides the thrust needed to push the growing chain through the ER membrane.
The membrane bound ribosomes form a polyribosome which is as mentioned bound to the ER.
Describe the signal sequence for ER?
- hydrophobic
- N terminal with hydrophobic core
- directs transport to ER and is cleaved in ER
functions:
- targeting to ER via binding of SRP
- opening of translocation channel
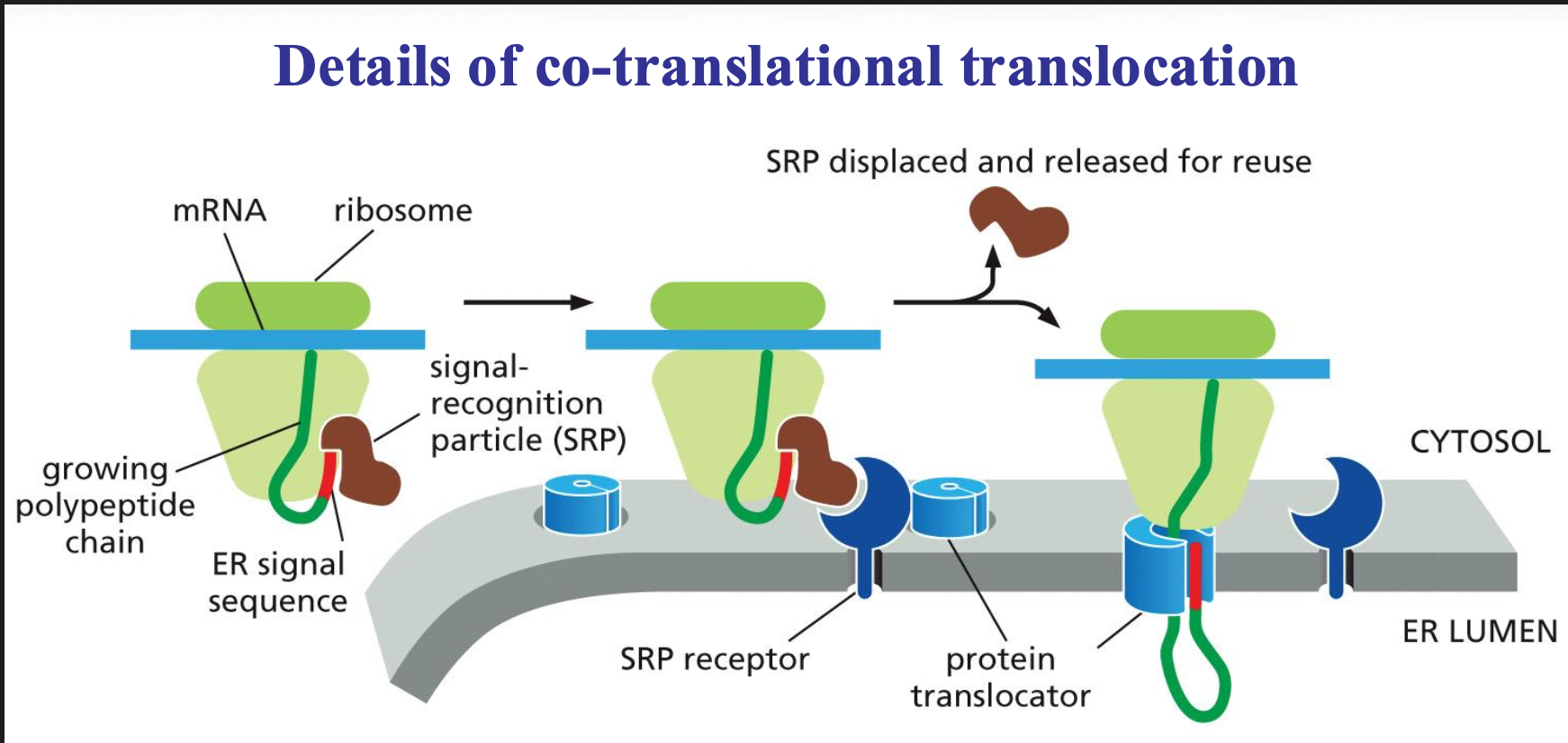
What is the signal recognition particle (SRP)?
It is a ribonucleoprotein complex that consists of 6 proteins and RNA
- binds ss of nascent (just coming into existence) proteins and this causes translation arrest and binds to SRP receptor
Co-translational vs Post translational translocation
There are also differences in the translocation obviously we have co translational translocation (membrane bound) which is when the translation of the protein and translocation takes place at the same time and post translational translocation (free ribosomal) is when the strand is translated first and then translocated into the membrane.
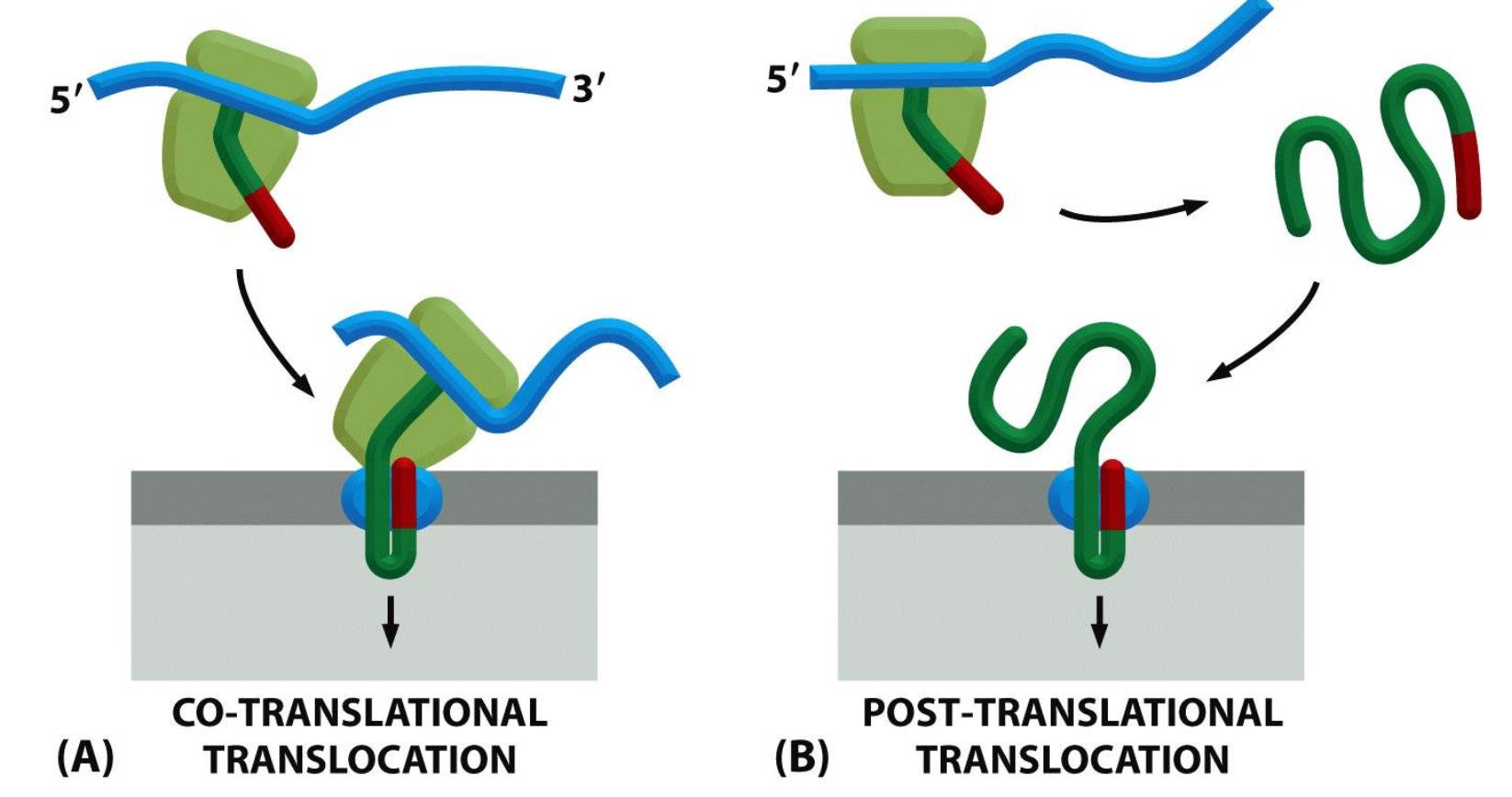
Co-translational translocation Process
Two molecules the SRP molecule and the SRP receptor.
The SRP molecule binds to the ER signal sequence and to the ribosome, and in that state can slow down (pause) the translation of the sequence. This will be slowed down until the SRP molecule can interact with the receptor.
Once they do bind the SRP is released and the sequence is passed on to a translocator in the ER membrane and protein synthesis can take place again. The sequence is then threaded through a channel in the translocator.
The ER signal sequence opens the translocator, guides the protein into the ER, is then cut off by signal peptidase, and the completed protein ends up free inside the ER lumen.
→ end stage of translocation: ss cleaved and degraded; mature protein folds and can be further sorted.
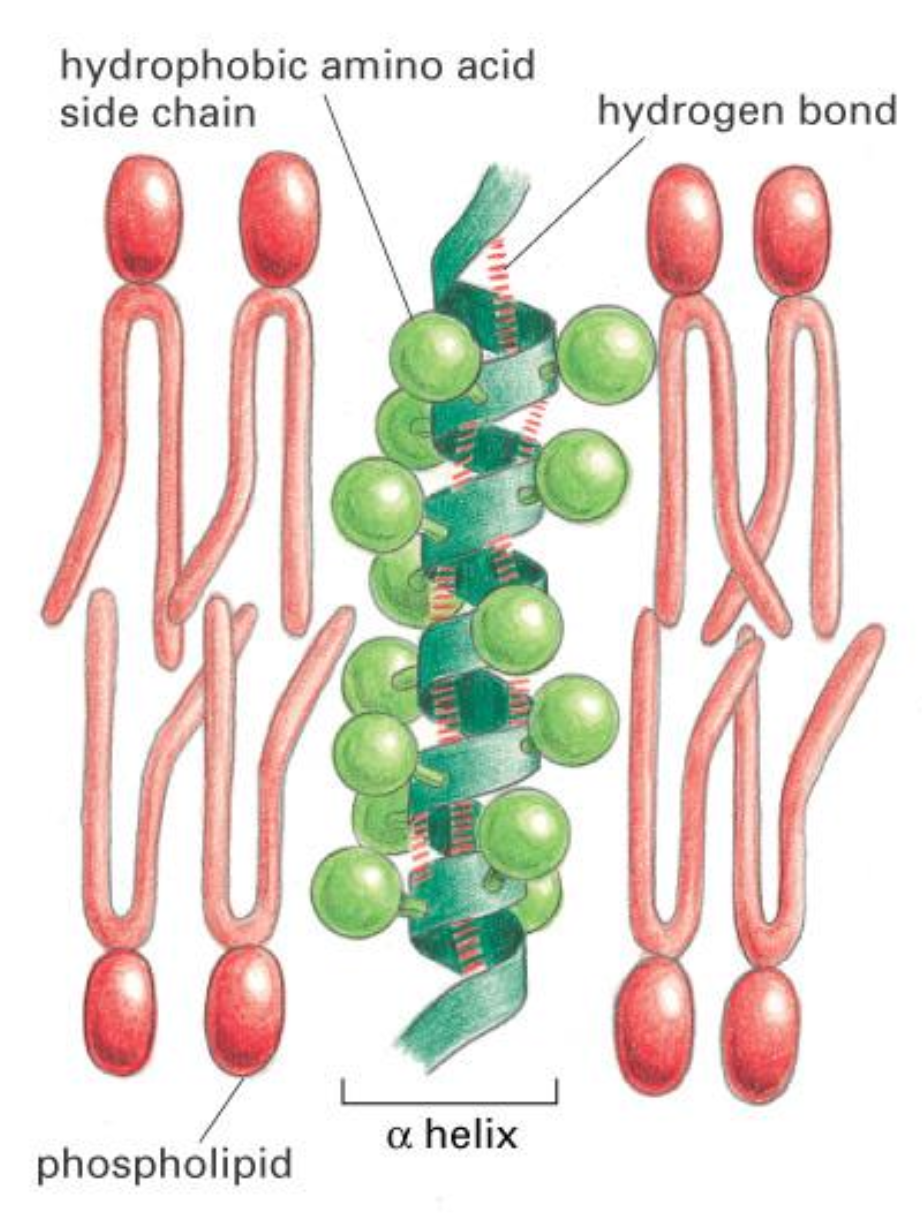
What are the different types of membrane (-associated) proteins?
Integral:
- Transmembrane: alpha helix, beta barrel
- Membrane associated
Peripheral membrane protein:
-Lipid linked
-Protein attached
Importance of membrane proteins:
30% of sequenced genes encode membrane proteins.
membrane proteins are involved in vital cellular processes (signal transduction, communication, transport)
70% of drug targets are membrane proteins
many diseases due to compromised folding of membrane proteins (Cystic Fibrosis, Alzheimer)
very few high resolution structures available
Transmembrane Helix
Transmembrane Proteins are anchored in lipids via hydrophobic alpha-helices.
How do proteins pass through the membrane???
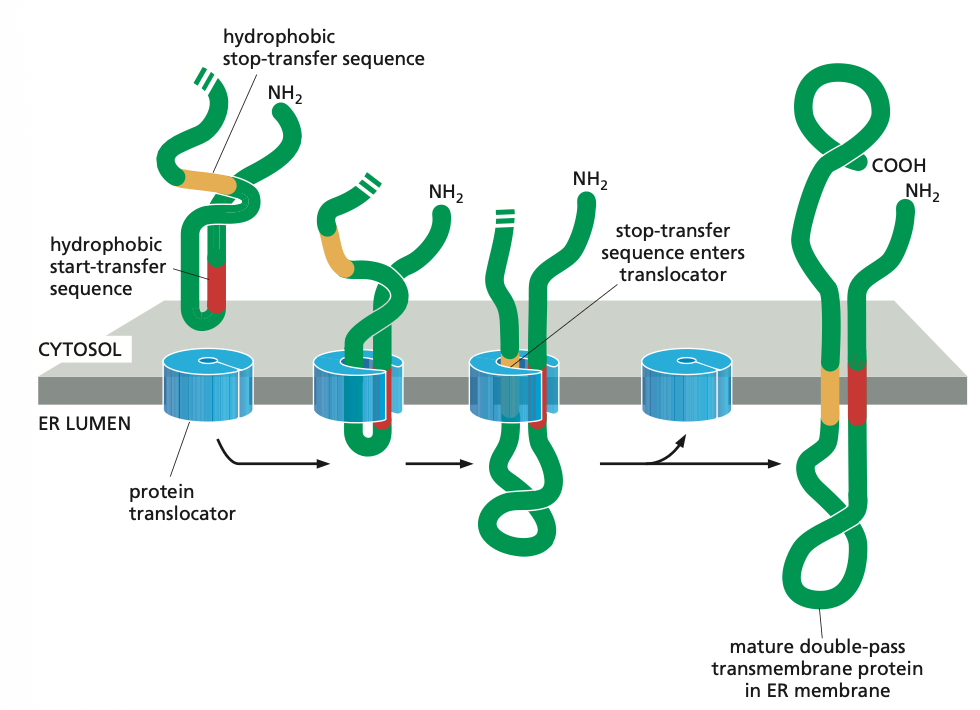
Some times proteins are not transported through the ER membrane but rather remain in the ER membrane. In this case some parts of the polypeptide chain must be translocated completely across the lipid bilayer, whereas other parts remain fixed within the membrane. We have two mechanisms for this one is the single pass and the other is the double pass.
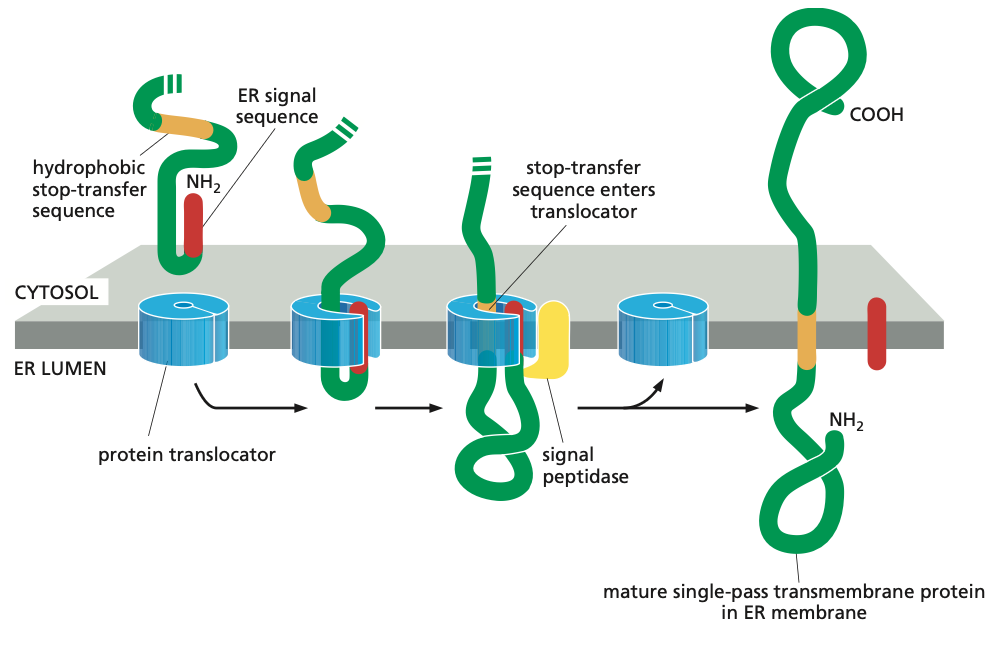
Single-Pass Mechanism
Such membrane proteins have a single membrane-spanning segment, the N-terminal signal sequence initiates translocation—as it does for a soluble protein. But the transfer process is then halted by an additional sequence of hydrophobic amino acids, a stop-transfer sequence.
Now the protein translocator can release the growing polypeptide into the lipid bilayer.
The N-terminal signal sequence is cleaved off, and the stop-transfer sequence remains in the bilayer, where it forms an α-helical membrane-spanning segment that anchors the protein in the membrane.
Now we have a membrane protein that has a fixed orientation, where the N-terminus is on the lumenal side of the lipid bilayer and the C-terminus on the cytosolic side.
Note also that the orientation of these proteins are never changed.
Single Step with no stop Sequence
From the PPT we know that it is also possible that there is no hydrophobic stop transfer sequence. In that case the charge of the N-terminus will determine what the cytosolic side is and which one is luminal.
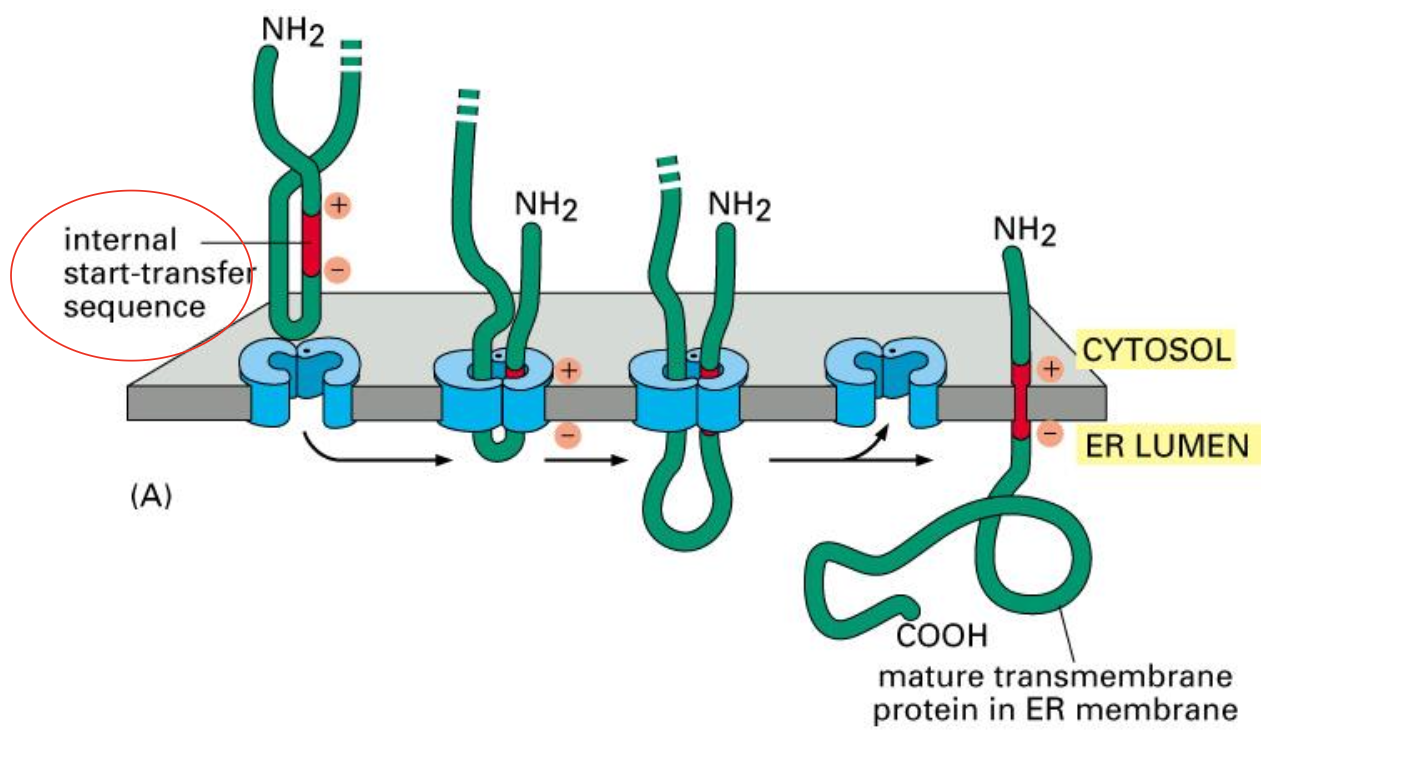
Insertion of a double-pass membrane protein
This internal sequence (red ) not only acts as a start- transfer signal, it also helps to anchor the final protein in the membrane.
Like the N-terminal ER signal sequence, the internal signal sequence is recognized by an SRP, which brings the ribosome to the ER membrane (not shown).
When a stop-transfer sequence (orange) enters the protein translocator, the translocator discharges both sequences into the lipid bilayer.
Neither the start-transfer nor the stop-transfer sequence is cleaved off, and the entire polypeptide chain remains anchored in the membrane as a double- pass transmembrane protein.
Proteins that span the membrane more than twice contain additional pairs of start- and stop-transfer sequences, and the same process is repeated for each pair.
Fate of proteins that have entered the ER
Proteins either stay in the ER via retention signals or they are routed via secretory pathways via vesicles (vesicular transport).
Roadmap of Vesicular Transport (Endocytic Pathway or Secretory Pathway)
Vesicular traffic can either be through an Endocytic Pathway or a Secretory Pathway.
Endocytic Pathway is responsible for the ingestion and degradation of extracellular molecules, which move materials from the plasma membrane, through endosomes, to lysosomes.
Secretory Pathway follows the synthesis of proteins on the ER membrane and their entry into the ER, and it leads through the Golgi apparatus to the cell surface; at the Golgi apparatus, a side branch leads off through endosomes to lysosomes.
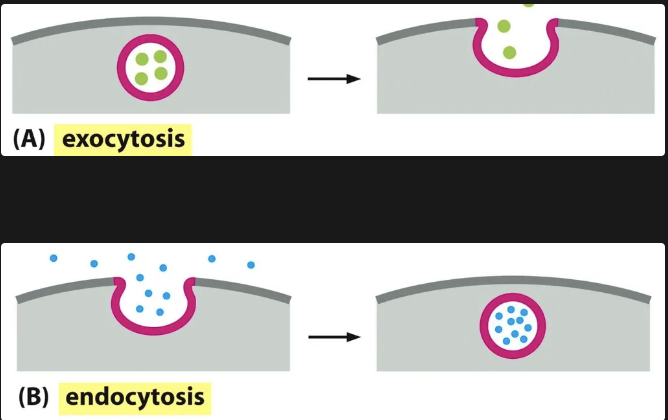
Endocytosis vs Exocytosis
When we speak of the secretory pathways we need to understand the movement through endocytosis and exocytosis. Vesicular transport allows materials to exit or enter the cell.
During exocytosis, a vesicle fuses with the plasma membrane, releasing its content to the cell’s surroundings
During endocytosis, extracellular materials are captured by vesicles that bud inward from the plasma membrane and are carried into the cell.
General Characteristics of Vesicle Budding
Both soluble and insoluble molecules can be transported and different types of vesicles are used to ensure the right molecules are brought to the right organelles. This transport is also well-regulated and specific.
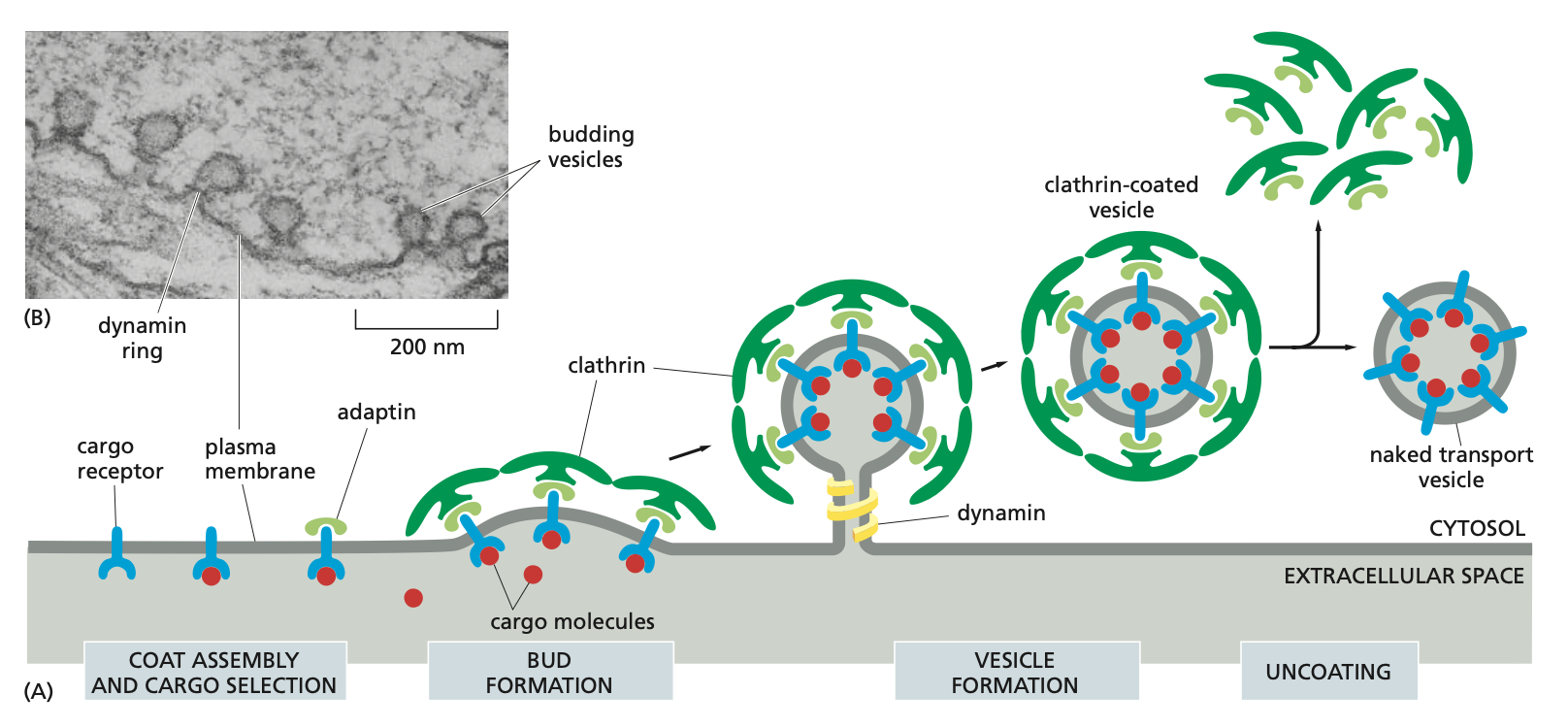
Vesicle Budding
Vesicles that bud off membranes usually have coats which remain intact until the vesicle reaches the destined organelle after which they are broken down.
The coat serves at least two functions: it helps shape the membrane into a bud (triskelions) and it captures molecules for onward transport.
Process if vesicle budding
1. The coat gives shape to the vesicle
Clathrin (example of coat) consists of triskelions --> football-like structures that pinch off from donor membrane
2. The coat selects cargo (how dynamin, adaptin, and clathrin work together)
This is how molecules are transported into the cell via endocytosis.
The cargo receptors together with the the cargo molecule bind to the adaptin which are then further bound to the clathrin molecules.
This is where the bud formation takes place and the dynamin binds around the membrane. Once it is activated with GTP is pinches the membrane and then buds off the membrane.
Once the clathrin coated vesicle is formed the clathrin are released in a process called clathrins and then the naked transport vesicle is ready.
→ Also note that there are different kinds of adaptins which depend on the membrane that it is pinching (donor membrane).
Specificity of transport (where do the naked vesicles go)
Naked vesicles are transported along the cell via motor proteins across the cytoskeleton.
However specific molecules need to be recognised by the correct organelles and there are specific mechanisms that allow the process.
The identification process depends on Rab proteins which are on the membranes of the vesicles and the corresponding tethering proteins on the organelle membranes.
The coding system of matching Rab and tethering proteins helps to ensure that transport vesicles fuse only with the correct membrane.
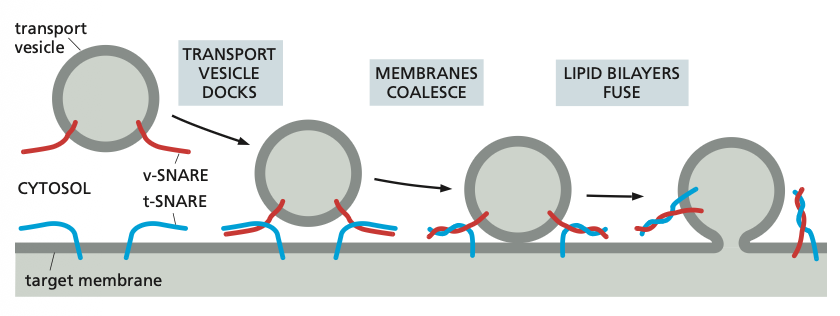
Additional recognition is provided by a family of transmembrane proteins called SNAREs.
SNAREs on the vesicle (called v-SNAREs) interact with complementary SNAREs on the target membrane (called t-SNAREs), firmly docking the vesicle in place.
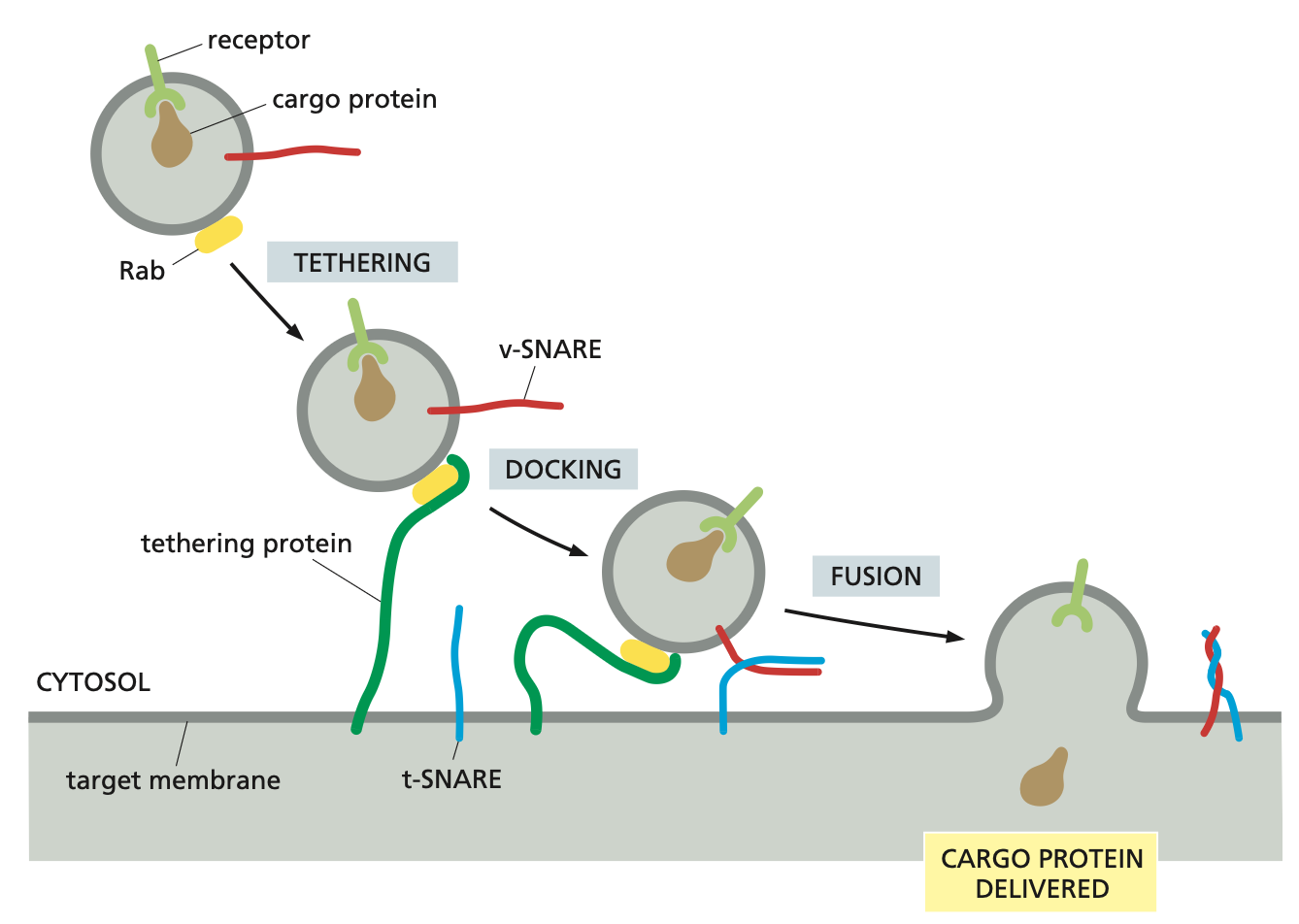
SNARE proteins
SNAREs on the vesicle (called v-SNAREs) interact with complementary SNAREs on the target membrane (called t-SNAREs), firmly docking the vesicle in place.
For vesicle fusion, the SNARE proteins themselves catalyse the process: when fusion is triggered, the v-SNAREs and t-SNAREs wrap around each other tightly, thereby acting like a winch that pulls the two lipid bilayers into close proximity.
After fusion, the SNAREs are pried apart so that they can be used again.
Secretory Pathways
Secretory pathways begin at the ER → Golgi Apparatus → Plasma Membrane → external environment. This all takes place via vesicles.
During transport there are the following processes: modification, quality control, sorting. This way we ensure that the correct proteins are processed and sorted in the correct organelles.
Trimming and processing of sugars continues in ER and GA --> structure, protection, folding and sorting
Modification
Occurs co-translationally in the ER lumen. A preassembled branched oligosaccharide is transferred intact from the lipid carrier dolichol phosphate to an asparagine residue in the consensus sequence Asn–X–Ser/Thr by oligosaccharyl transferase. Sugars are trimmed and further processed in the ER and Golgi. Glycosylation aids protein folding, protects against degradation, and contributes to quality control and sorting.
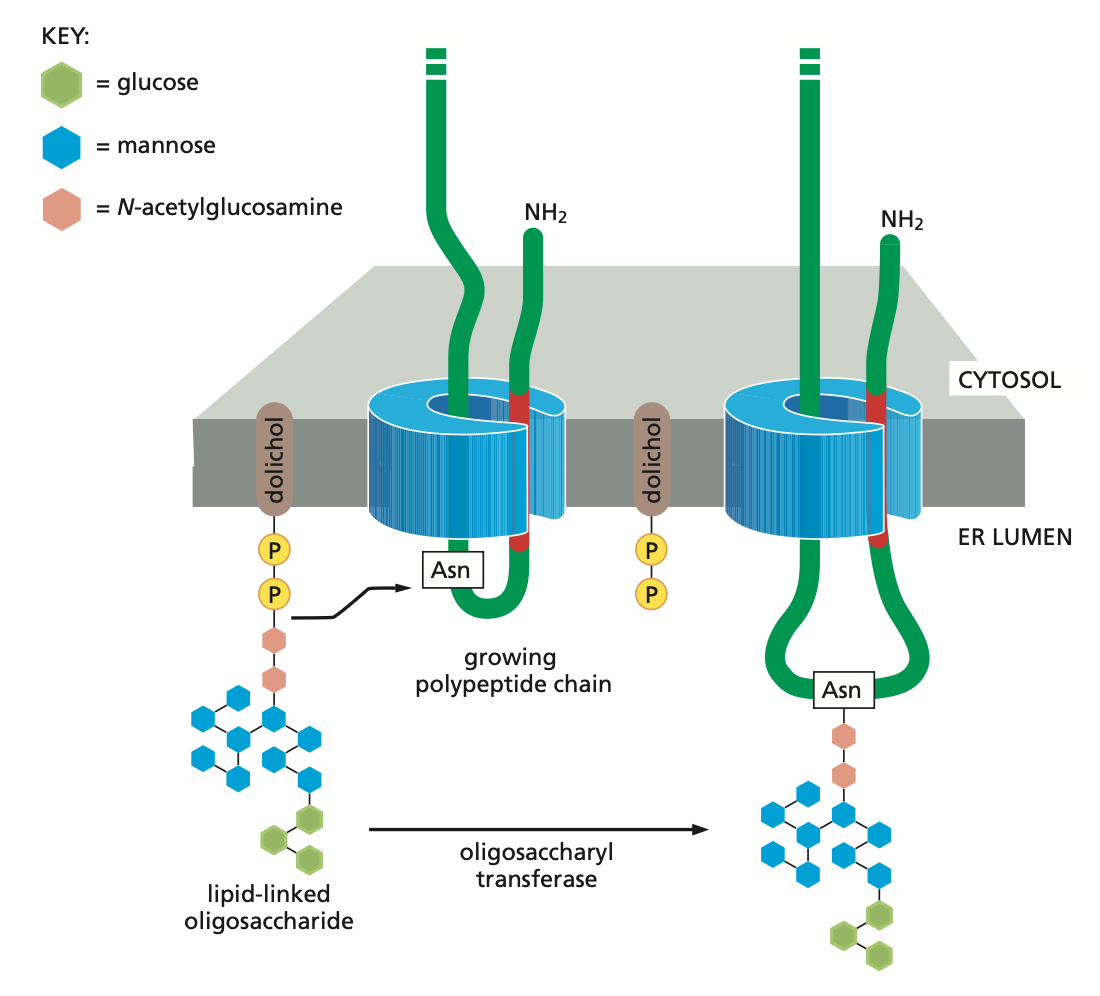
Quality control
Only correctly folded proteins can leave the ER
Unfolded proteins remain complexed with chaperones --> folding or transfer to cytosol for degradation
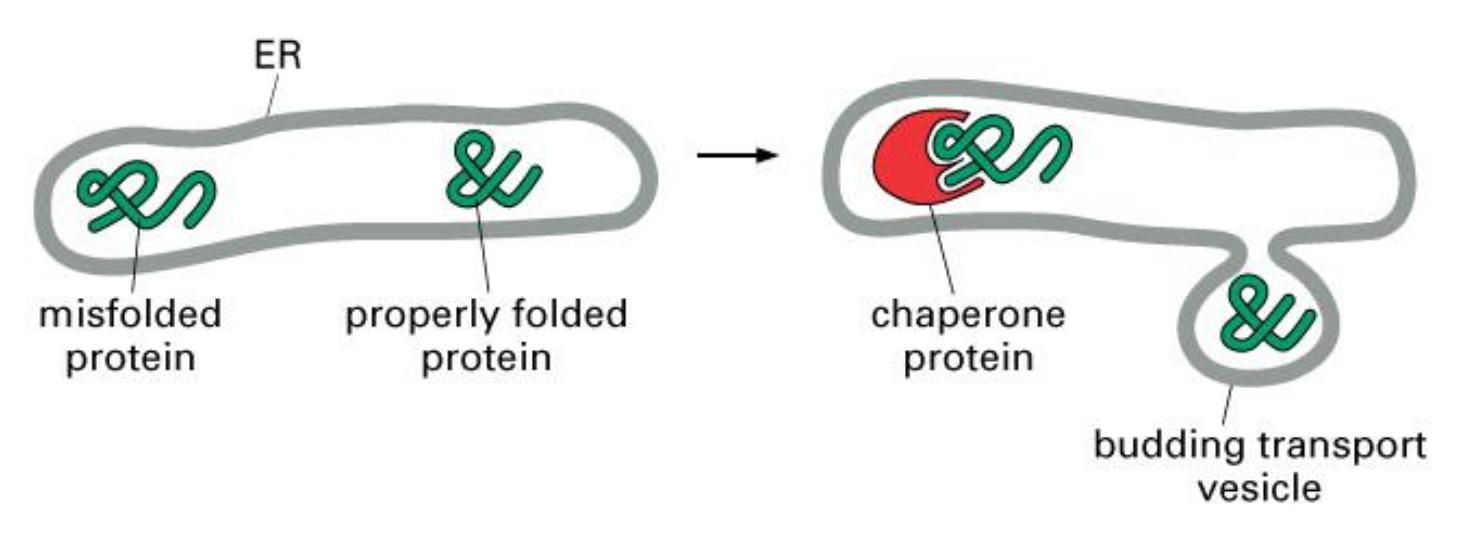
Unfolded Proteins
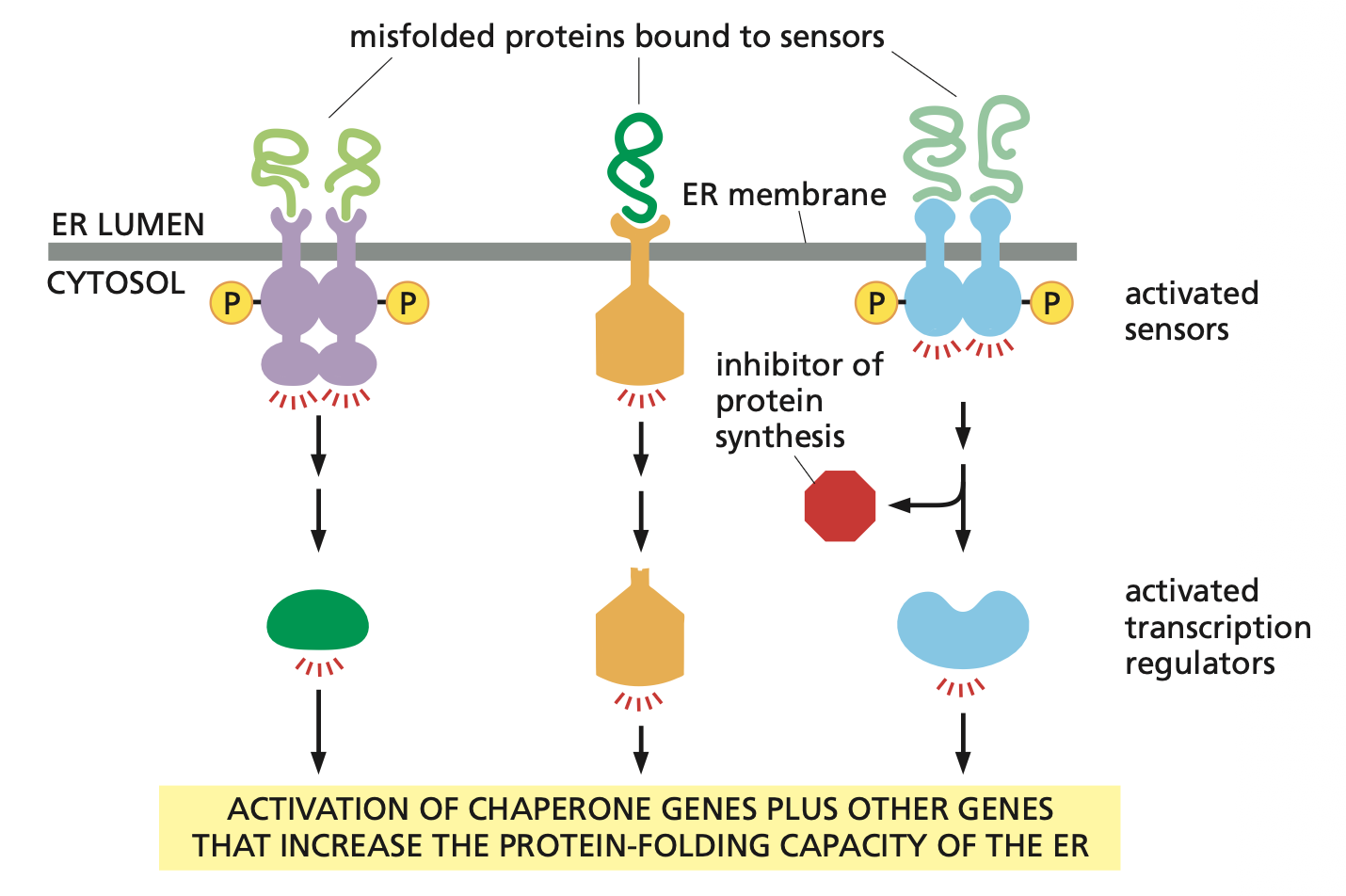
Accumulation of misfolded proteins in the ER lumen triggers an unfolded protein response (UPR).
The misfolded proteins are recognized by several types of transmembrane sensor proteins in the ER membrane, each of which activates a different component of the UPR. Some sensors stimulate the production of transcription regulators that activate genes encoding chaperones or other proteins involved in ER quality control. Another sensor can also inhibit protein synthesis, reducing the flow of proteins through the ER
Sorting
Proteins that have entered the ER can either stay there or move on towards the GA
Proteins “move on” by default; they need special mechanisms to stay in the ER
A. Aggregation or interaction with chaperones (prevents packaging in vesicles)
B. Retention in ER via recognition of retention signal in the protein (KDEL or KKXX)

Retrieval of ER resident proteins, mechanism, part I
ER proteins sometimes leave the ER by mistake in COPII vesicles.
In the Golgi, the more acidic pH allows the KDEL receptor to bind these escaped ER-resident proteins.
They are then packaged into COPI vesicles and transported back to the ER, where the neutral pH causes them to be released.
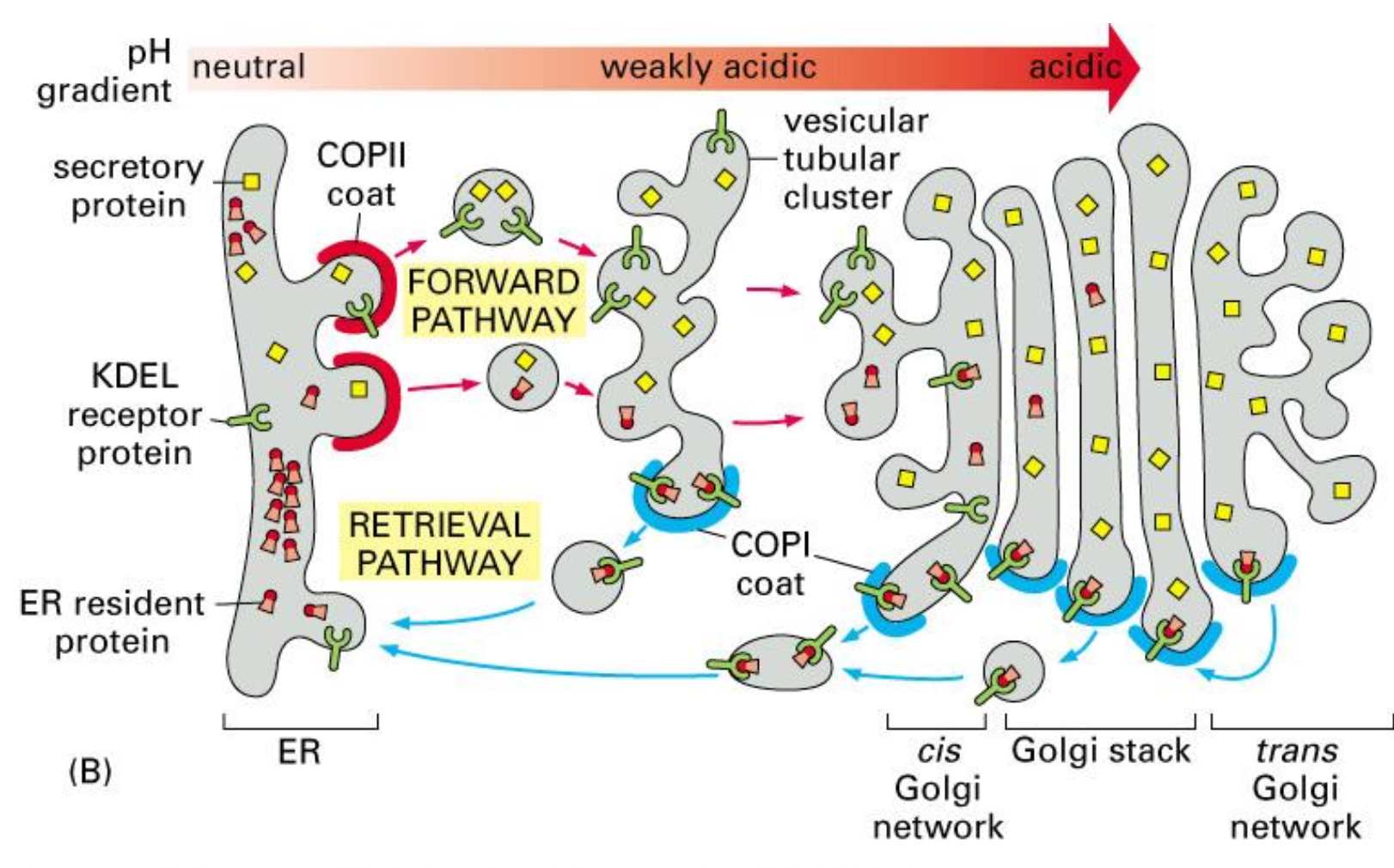
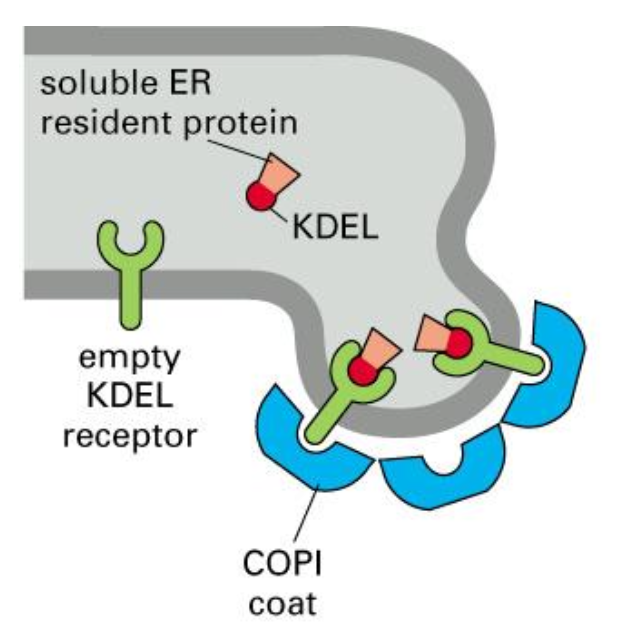
Retrieval of ER resident proteins, mechanism, part II
All ER resident proteins have the retention signal; if they “ escape” to the GA, they are recognised by a receptor and shuttled back to the ER
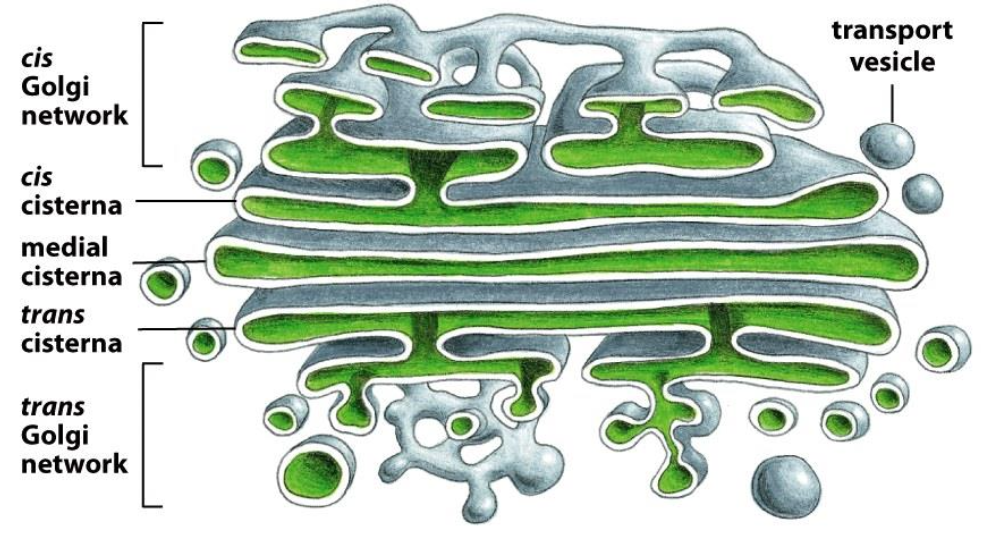
The Golgi Apparatus
Stacks of cisternae and tubules
Golgi Apparatus is polarised (cis-side faces the ER, trans-side faces the plasma membrane)
Function: Modification of proteins, primarily trimming of sugars (glycosylation).
The Golgi apparatus is functionally compartmentalized into CGN, cis, medial, trans cisternae, and TGN.
CGN and TGN are sorting stations (entry and exit points).
The cis → medial → trans cisternae contain different enzymes, each performing specific steps of glycosylation(mannose removal, GlcNAc addition, Gal addition, NANA addition, sulfation).
Proteins move between compartments by vesicle trafficking.
Together, the cisternae act as the “powerplant” of glycosylation.
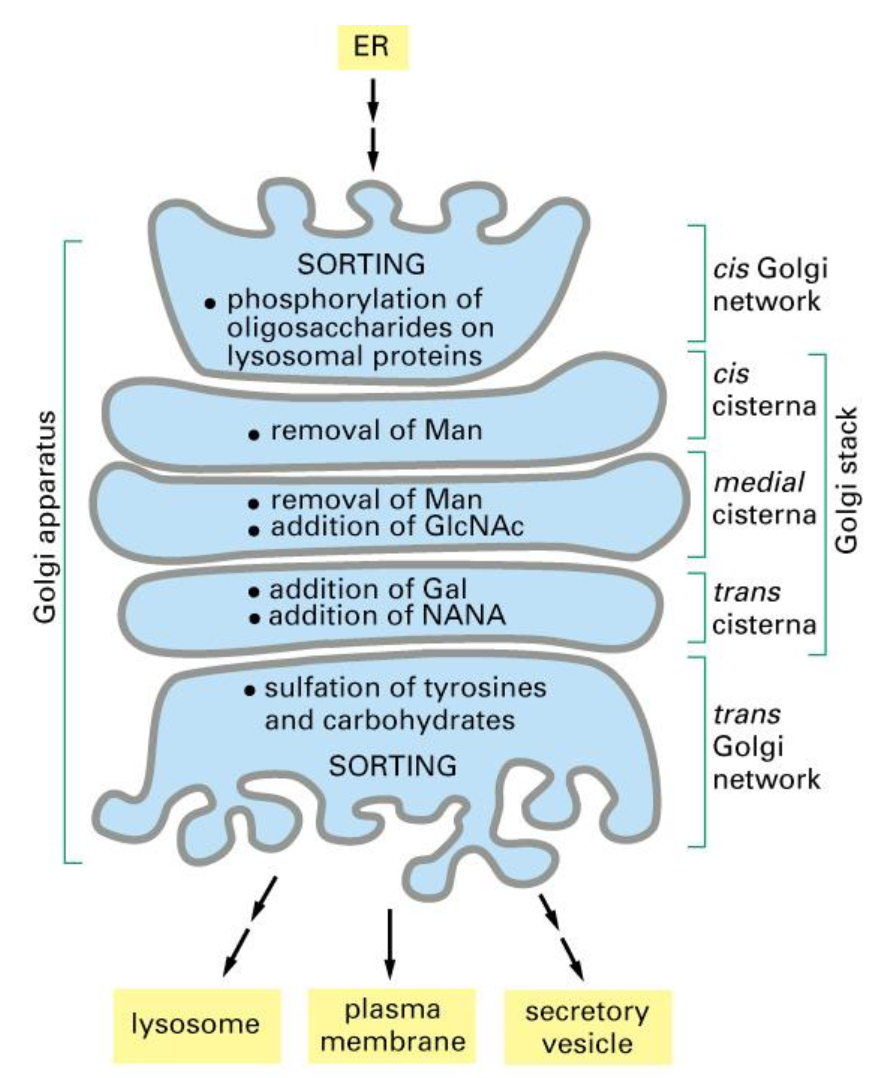
Two pathways of exocytosis
Constitutive secretion is a default process, which is present in all cells and it includes the permanent flow of materials to the membrane which are exocytosed out and counteracts endocytosis.
Regulated secretion is when the flow is dependant on a stimulus and is only present in specialised cells. This secretion depends on the extracellular signal that binds on the receptor on the membrane after which the exocytosis can occur.
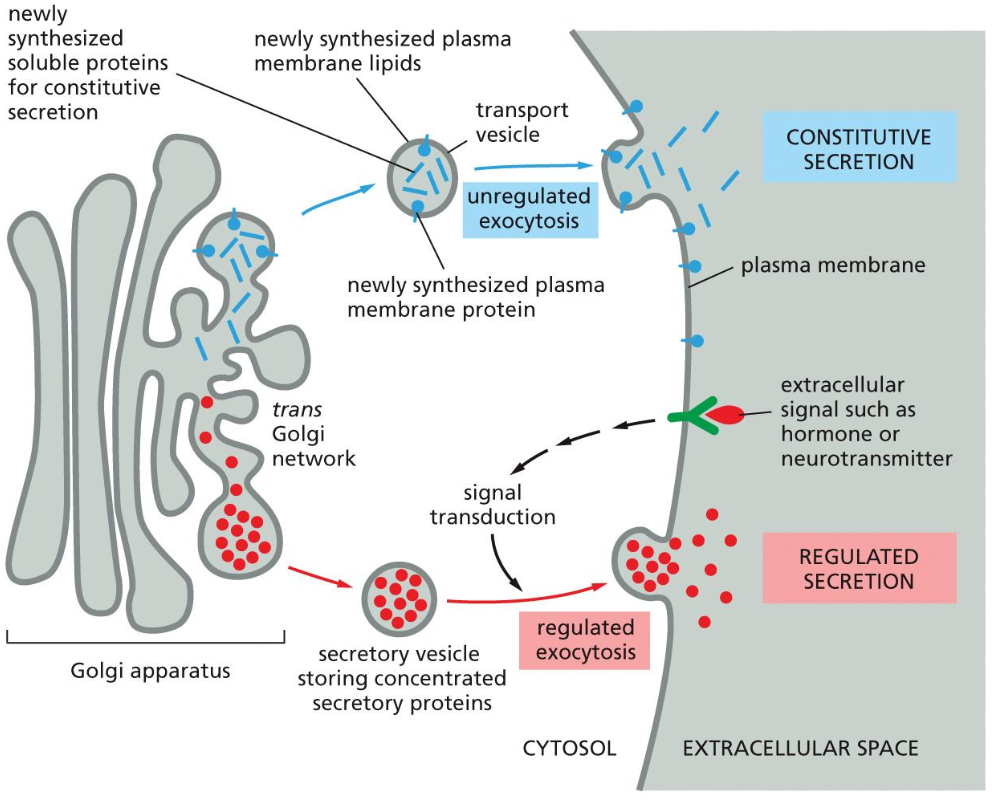
Endocytosis and degradation
There are two means of endocytosis and degradation. The first one being phagocytosis and the other Pinocytosis. Pinocytosis is also broken down in ordinary pinocytosis and receptor mediated endocytosis (RME).
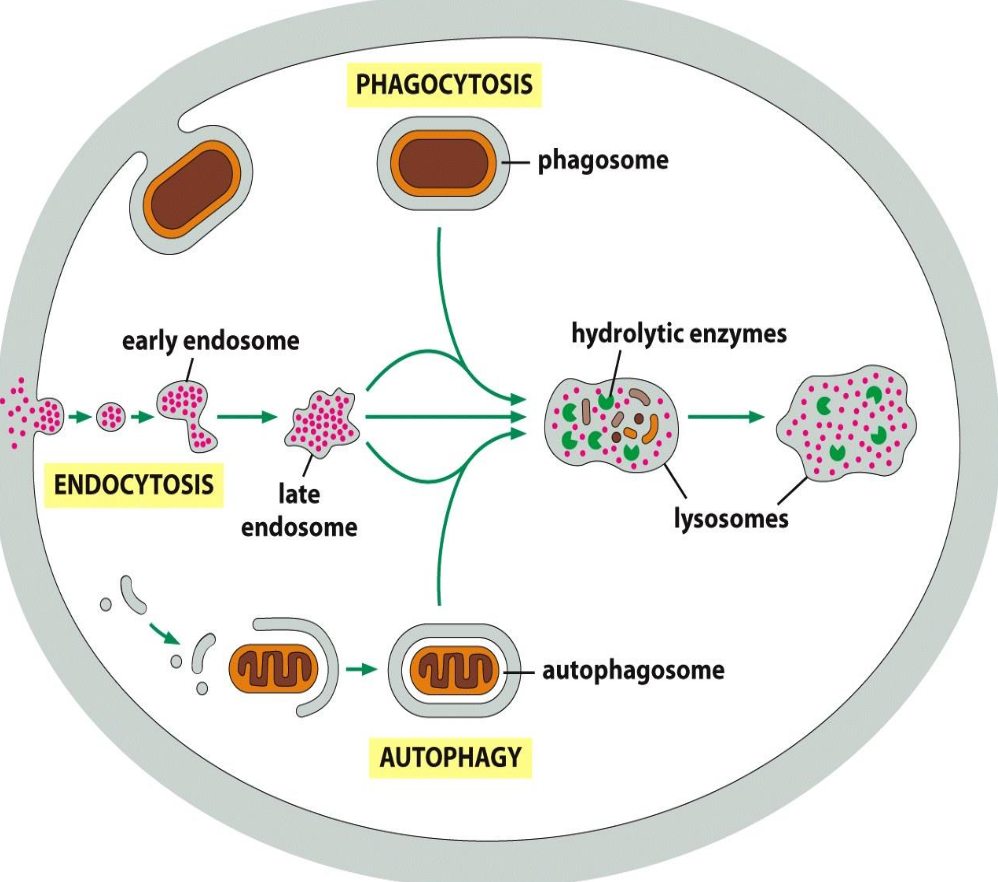
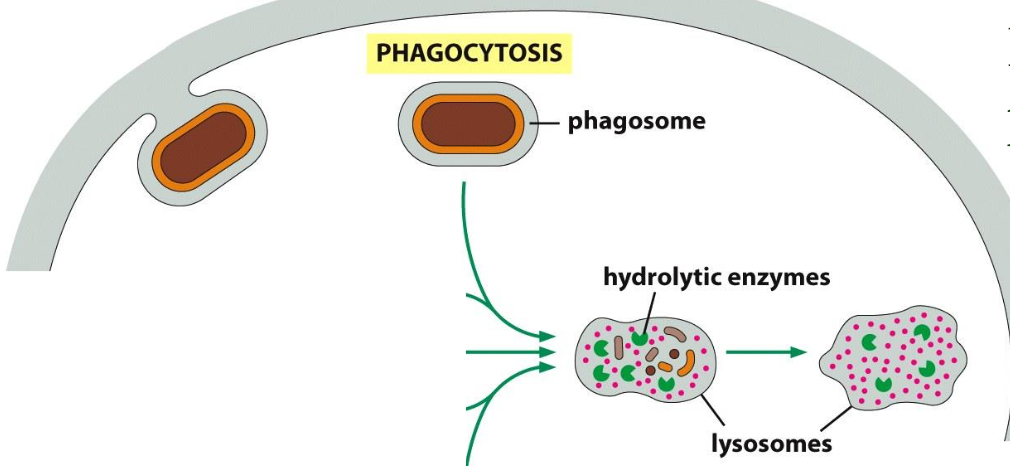
Phagocytosis
Defined as the uptake of large molecules.
Uptake of bacteria (defense mechanism) or cellular debris; by specialised cells (macrophages)
Particles usually bind to receptors (f.i. for antibodies) before uptake (”triggered process”)
Uptake --> large endocytic vesicles (phagosomes) --> fusion with lysosomes --> degradation
Pinocytosis
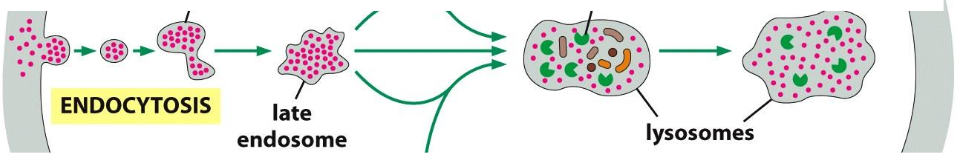
Defined as Ordinary pinocytosisuptake of solutes (small stuff)
Uptake via clathrin coated pit-->clathrin coated vesicle --> uncoating--> fusion with early endosome --> late endosome--> lysosome
Ordinary pinocytosis
Indiscriminate continuous uptake --> retrieval of membrane material.
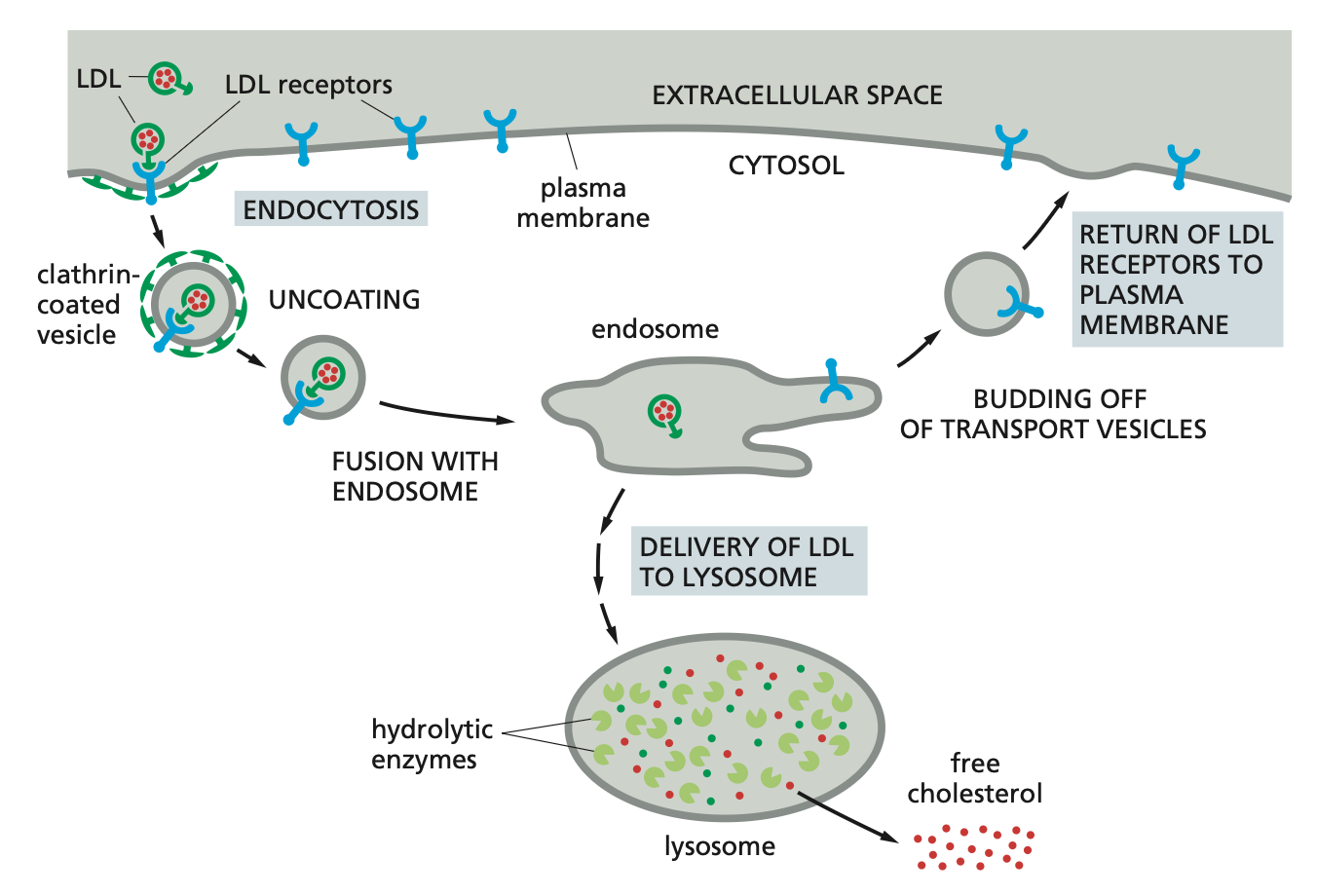
Receptor-mediated endocytosis (RME)
Specific binding of macromolecules to receptors --> accumulation in clathrin coated pits --> uptake.
The macromolecules bind to complementary receptors on the cell surface and enter the cell as receptor–macromolecule complexes in clathrin-coated vesicles. This process, called receptor-mediated endocytosis.
Example: Cholesterol in blood in LDL-->binds LDL receptor -->concentration in coated pit --> uptake -->endosomes -->
dissociation of LDL- receptor complex by low pH --> transfer to lysosome --> LDL is degraded/receptor returns to PM
Endosomes
Early endosomes --> fusions to form larger (late) endosomes --> develop into or fuse with lysosomes
Gradual decrease in pH (lowest in lysosomes)
Receptors are often released at low pH --> recycling, degradation or transcytosis
Cargo always goes to lysosome for degradation
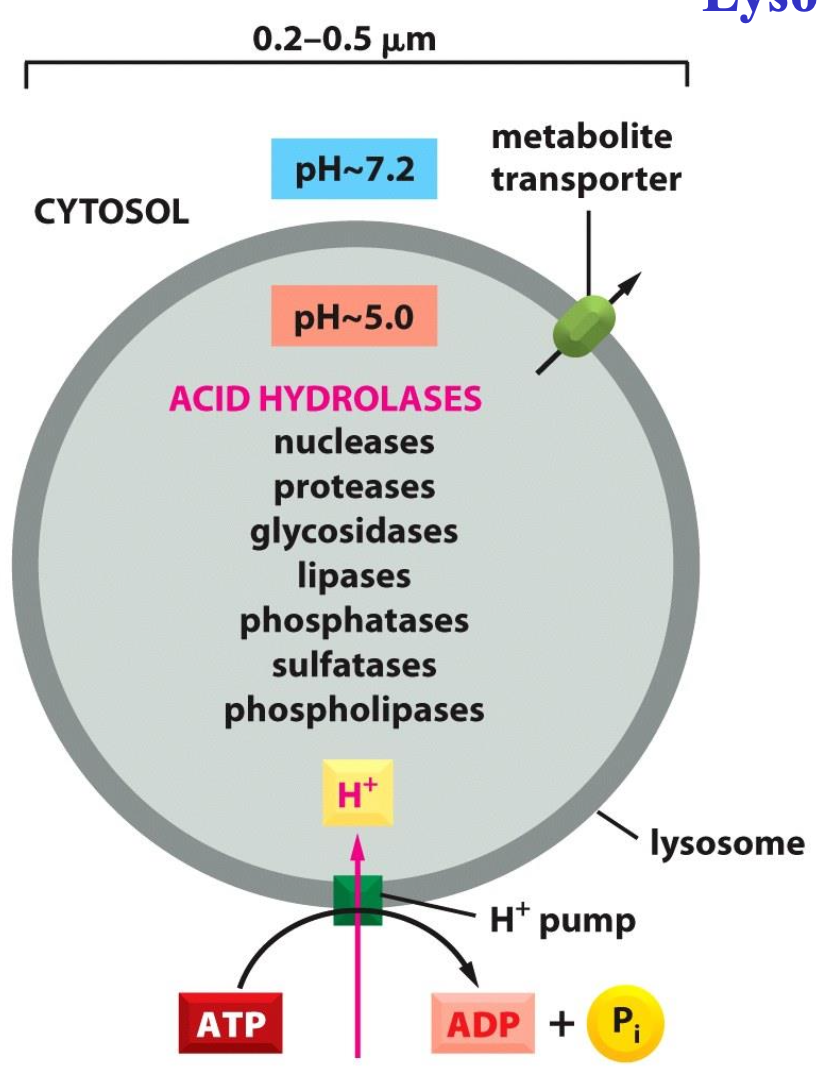
Lysosomes
Degradation of macromolecules by acid hydrolases these hydrolases have low pH optimum
One membrane:
protects cytosol
contains many transporters
contains ATPase
A lysosome contains a large variety of hydrolytic enzymes, which are only active under acidic conditions. The lumen of the lysosome is maintained at an acidic pH by an ATP- driven H+ pump in the membrane that hydrolyses ATP to pump H+ into the lumen.
4 entry pathways for lysosomes
Pinocytosis/endocytosis
Phagocytosis
Autophagy: Cells have an additional pathway that supplies materials to lysosomes; this pathway, called autophagy, is used to degrade obsolete parts of the cell.
Vesicular traffic from GA - how the lysosome gets degrading enzymes
Vesicular traffic from GA to lysosomes
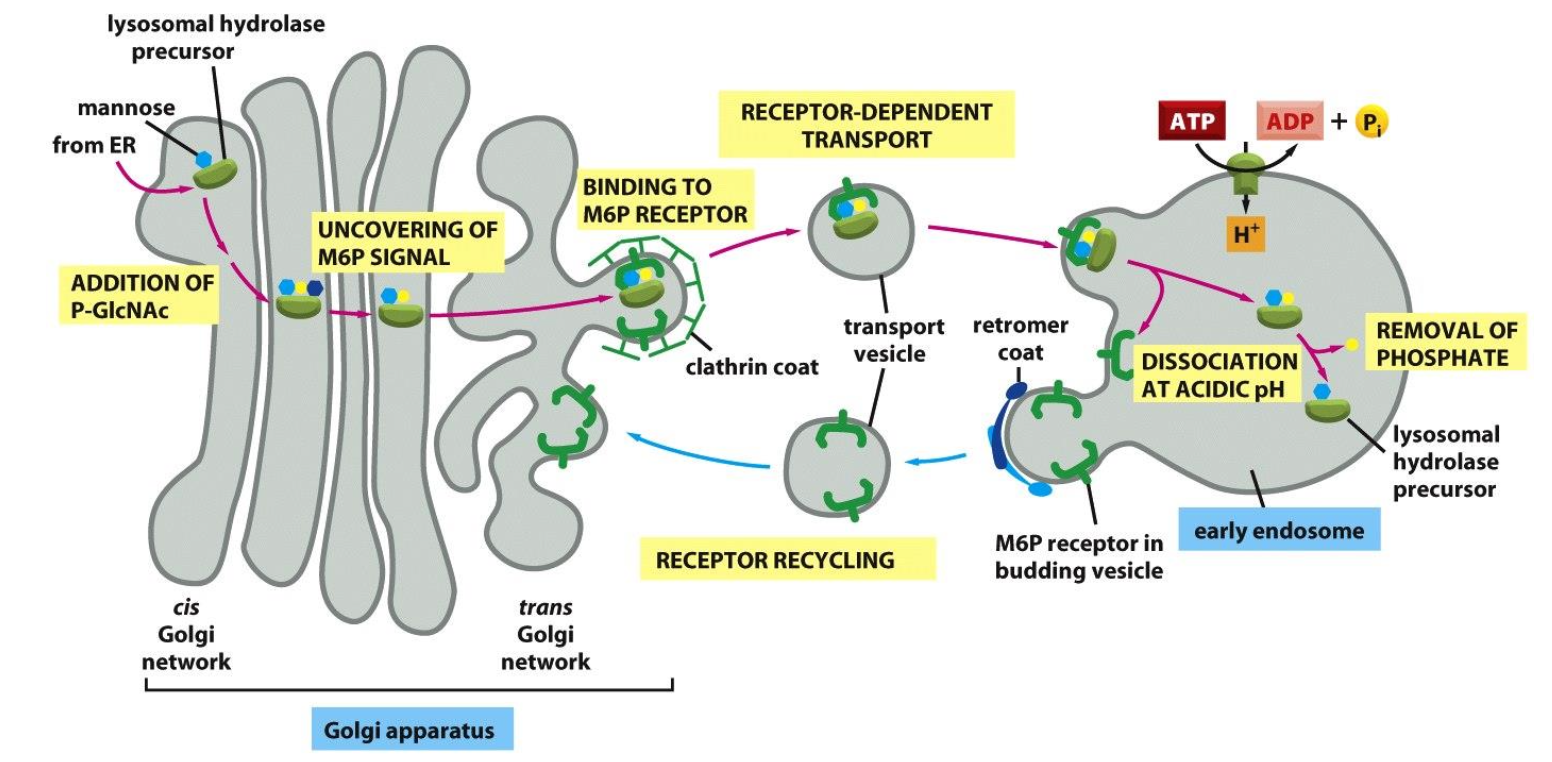
Lysosomal hydrolases get a mannose-6-phosphate (M6P) tag in the cis-Golgi.
M6P-tagged enzymes bind M6P receptors in the trans-Golgi and are packed into clathrin-coated vesicles.
Vesicles fuse with early endosomes; low pH releases the enzymes, which continue to lysosomes and become active.
The empty M6P receptors are recycled back to the Golgi for reuse.
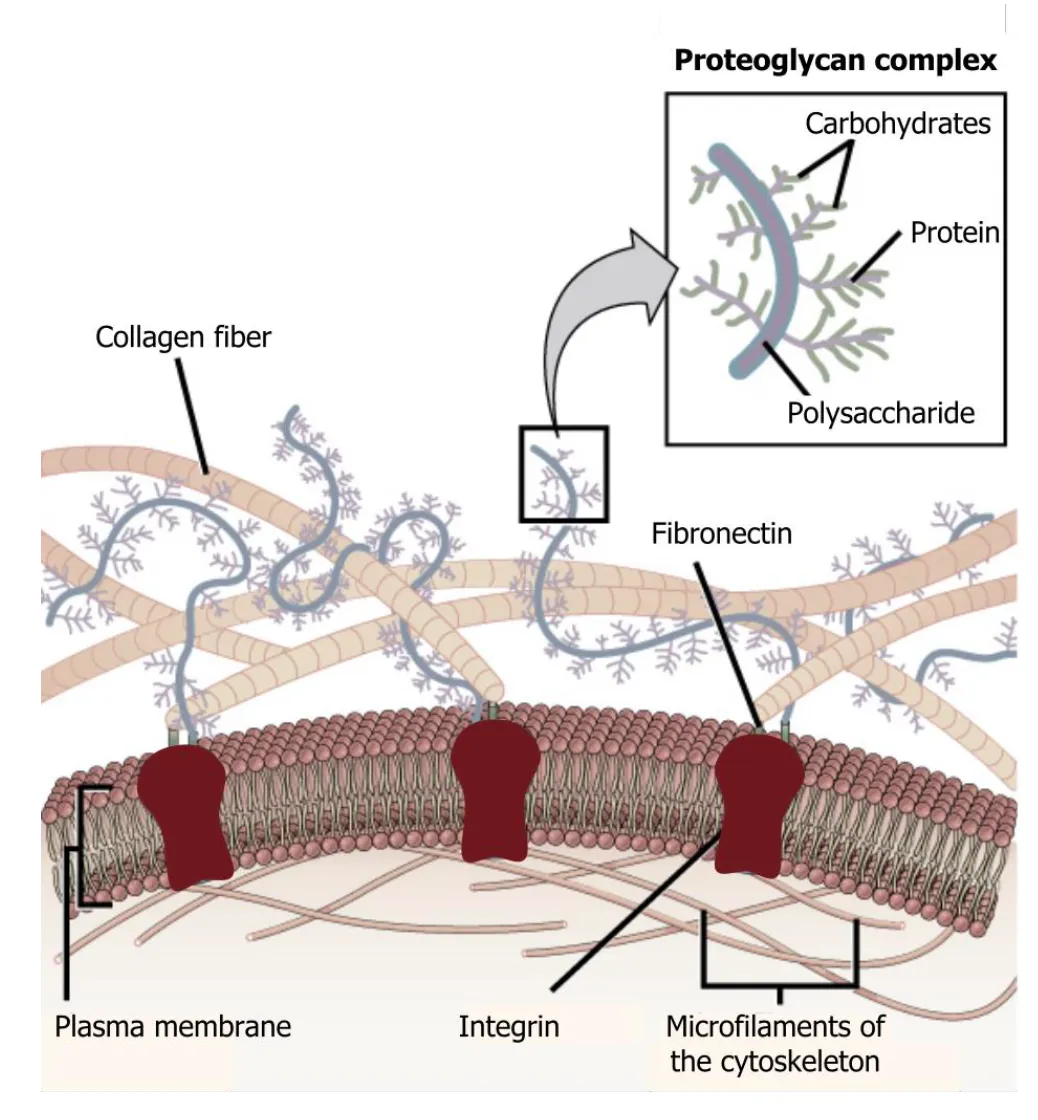
Explain how connective tissue is part of the extracellular matrix??
Bone is connective and consists of collagen and calcium deposites.
Extracellular matrix is connective tissue, what is outside the nucleus.
Explain how carbohydrates and collagen are part of extracellular matrix??
Carbohydrates are a big part of the matrix (proteoglycans) the other big part are collagen components.
Collagen gives rigid structure and resistance to pressure comes from the carbohydrates.
In bones you would need more collagen, for the structure.
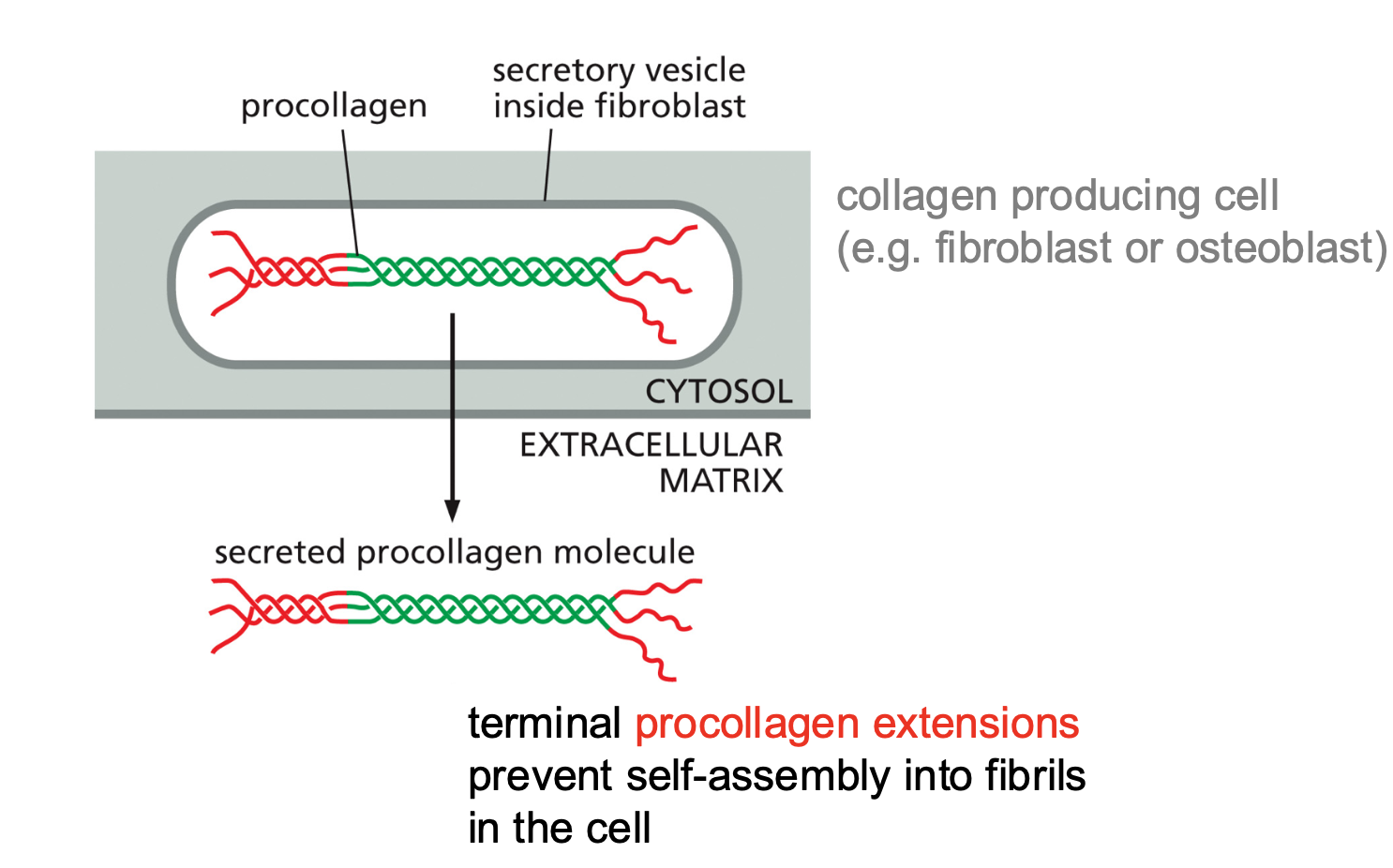
How do collagen fibers form?
Single collagen chain assembles together with other strands to become a triple stranded collagen molecules and when they interact they make up fibrils of collagen and these fibrils assemble into fibers.
What is the role of fibroblasts in the extracellular matrix?
Fibroblasts make most of collagen. You can see the fibrils being made in the ER and around the fibroblast you can also see the fibers that are assembled around the cell.
So the fibroblasts can tell where the collagen needs to be oriented. And also they know how the collagen is secreted and aligned.
What is the role of procollagen in the formation of collagen??
The fibers are first formed as pro collagen and they have red extensions, which prevent the fibers from self assembling. If these extensions do not exist then they would not be able to form properly.
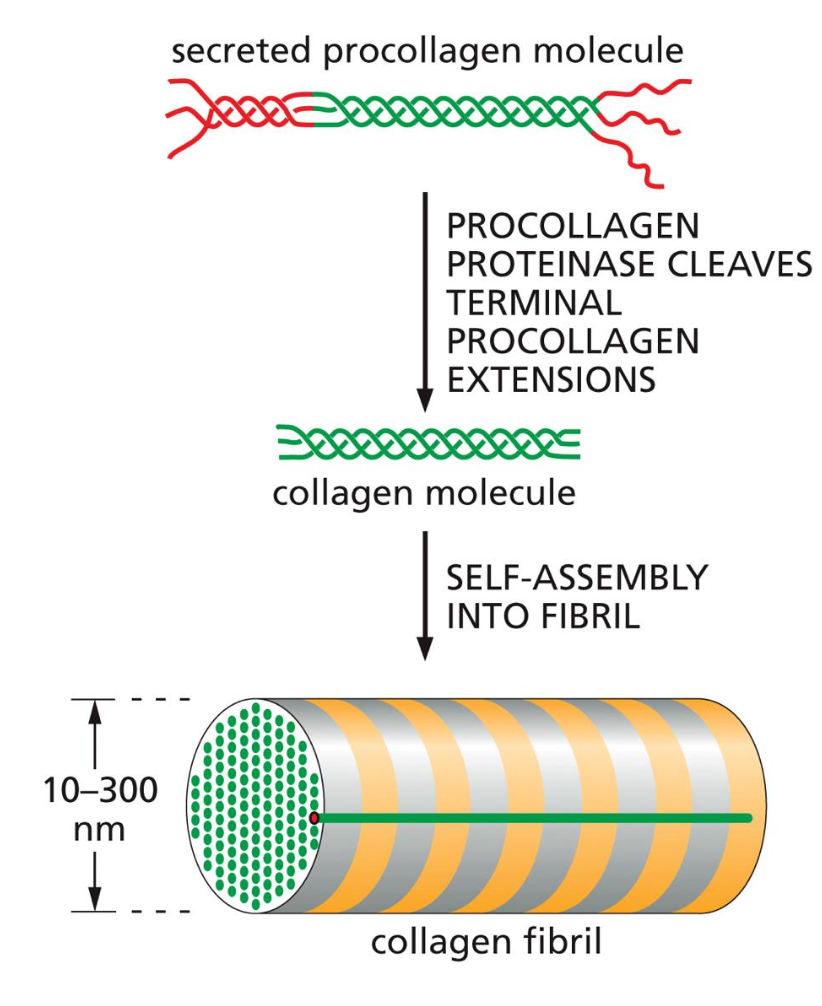
These extensions are then cleaved by protease cleavers and then the self-assembly into collagen fibers can happen.
pro collagen extensions are cleaved by proteinases in the extracellular space to allow self-assembly into collagen fibrils
What connects the intracellular cytoskeleton to EM
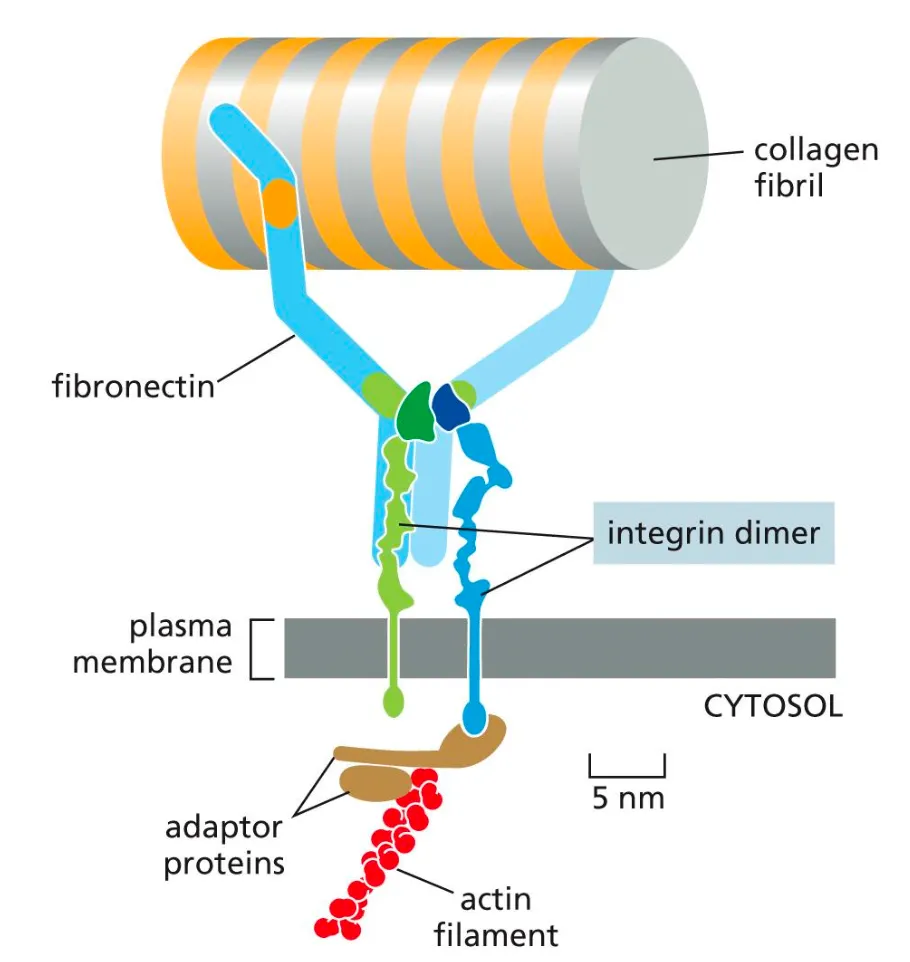
Integrins through fibronectins
Integrins
Integrins come together as dimers and they bind to the collagen fibers through fibronectins. And this way integrins can bind various adaptor proteins that can connect to the cytosplasm.
Integrins allow cells to grasp on and move through the extracellular matrix
Many integrins assembling together and allow cells to hold on to them and move around the matrix.
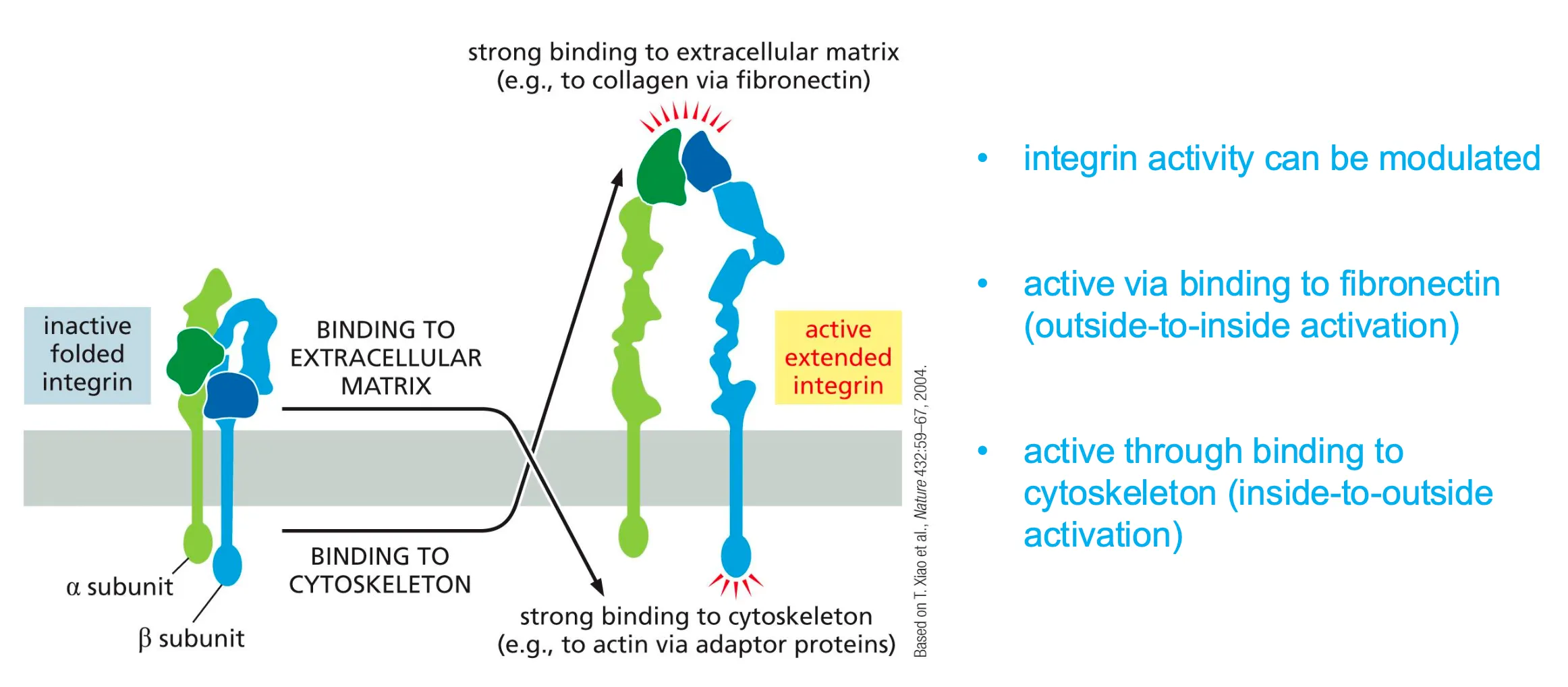
Integrins are in an inactive state and become activated via two states.
Bi-directional activation of integrin
When binding to either matrix or cytoskeleton and then unfolds to bind to the fibronectin.
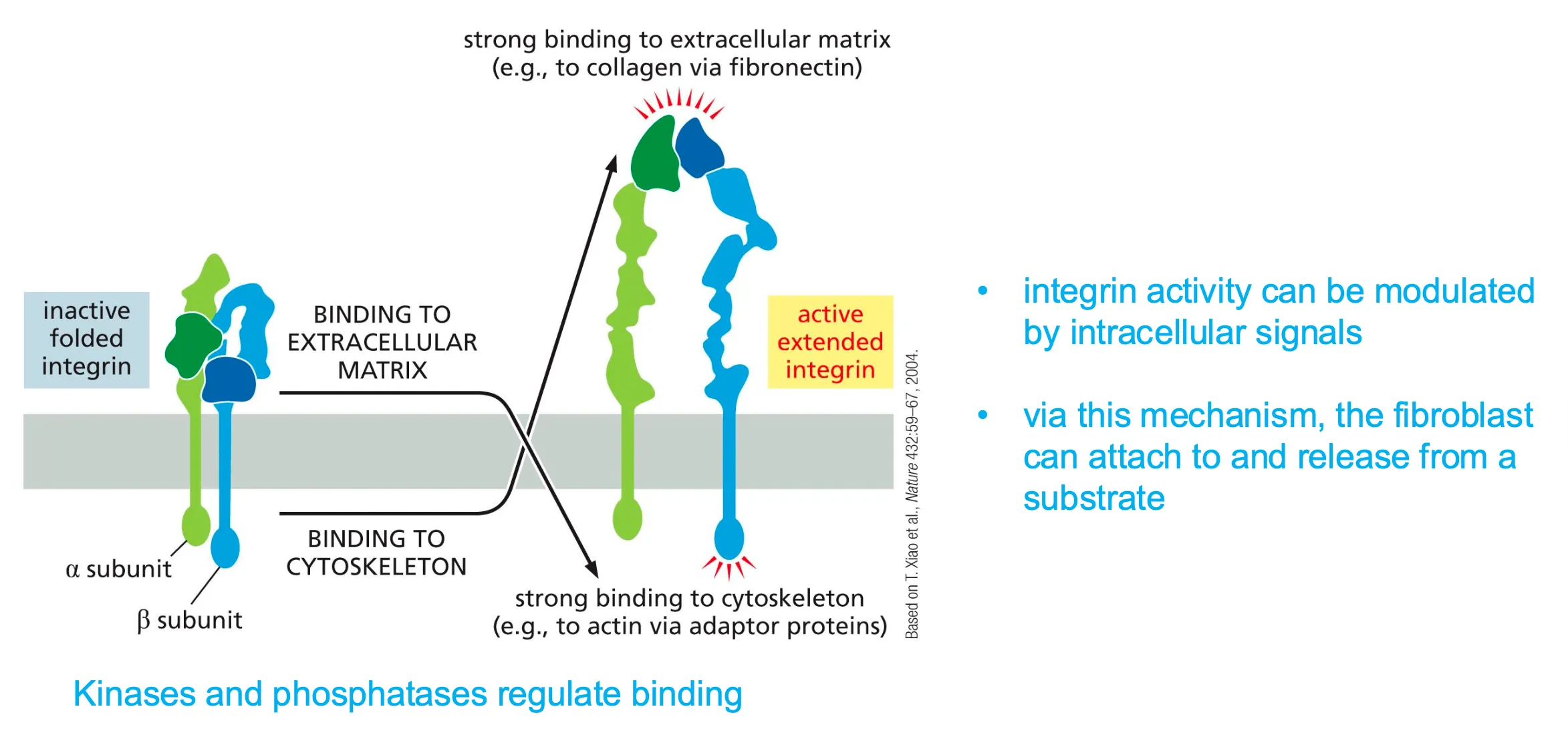
Modulation of integrin activation by intracellular signaling
Active via binding to fibronectin or to cytoskeleton
- binding unfolds interns
- activity can be modulated by intracellular signals
- via this mechanism a fibroblast can attach to and release from a substrate
- kinases and phosphates regulate binding
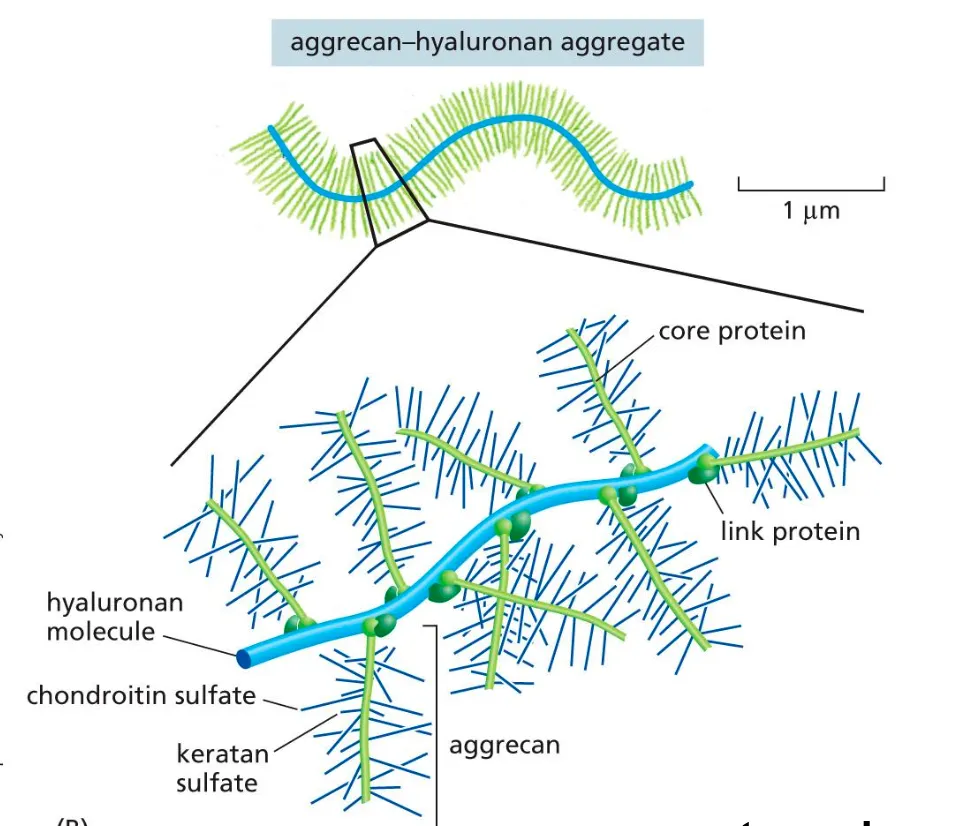
Glycosaminoglycans
Are extracellular matrix components that provide resistance to compression
Glycosaminoglycans are long, negatively charged polysaccharide chains.
Chains of glycosaminoglycans are often covalently linked to proteins forming proteoglycan.
The glycosaminoglycan hyaluronan consists of 2,000–25,000 disaccharides, which corresponds to polysaccharides with relative molecular masses of 106–107 and polymer lengths of 2–25 μm.
Variety of cell comes from the combinations of Glycosaminoglycans in terms of the side branches they have.
The charges also give osmotic pressure and soaks water up giving a rigid structure.
Proteoglycans
Proteoglycans and glycosaminoglycans can form extensive, brush-like structures
Proteoglycans are made of polysaccharides and proteins.
Functions of Proteoglycans
Structural support and hydration: GAGs form strongly hydrophilic gels that attract positive ions (Na+) that suck up large amounts of water
together with collagens this provides a rigid matrix that can resist compression and absorb shocks
help organize tissues by faciliating adhesion and arrange collagen fibers
these gels can also act as molecular barriers that restrict flow of molecules through the extracellular matrix
proteoglycans can bind growth factors and cytokines to stir cell migration through the extracellular space
How do tissues acquire and maintain their shape to function properly?
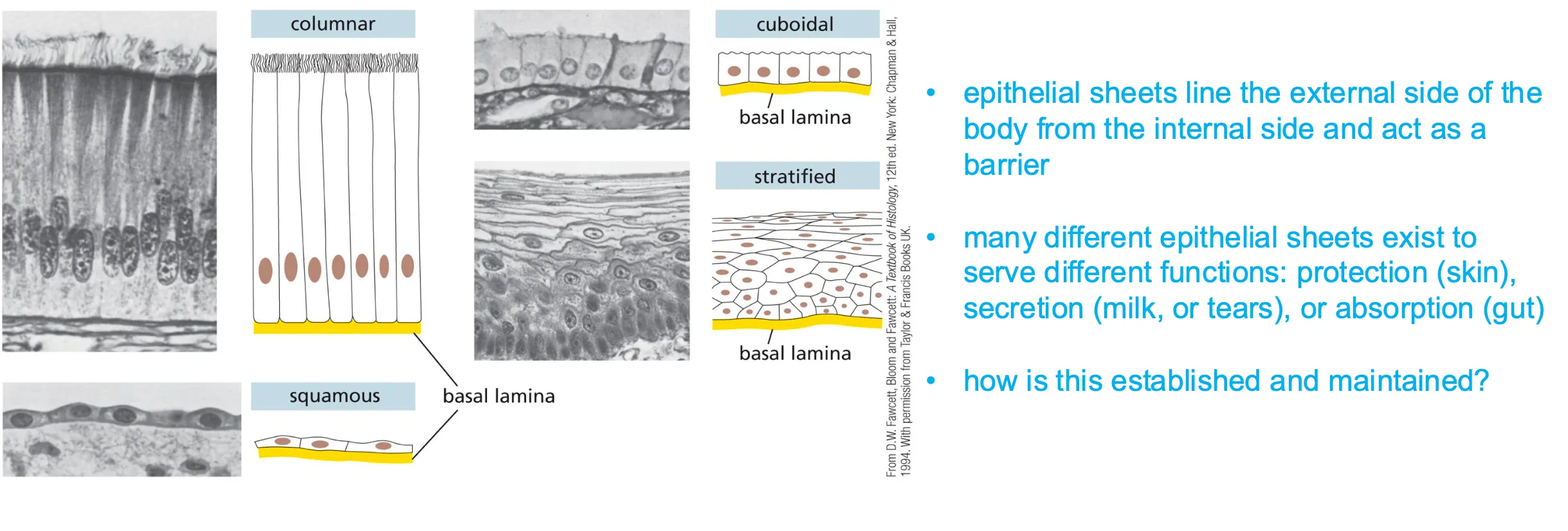
Sheets of cells have to be able to form different structures.
Epithelial sheets and cell junctions
All cells are aligned. And epithelial sheets have two sides apical and basal (has a rich extracellular matrix)
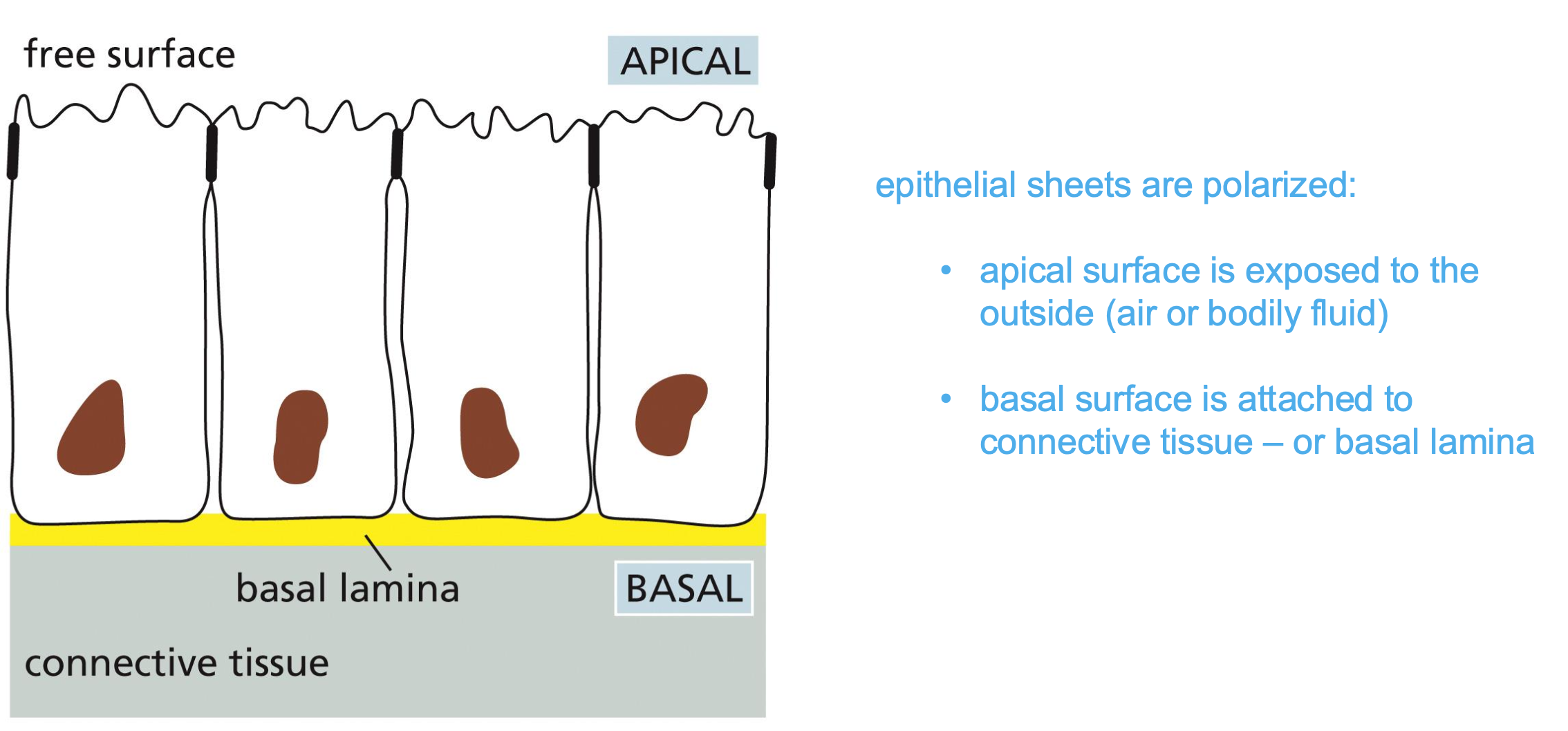
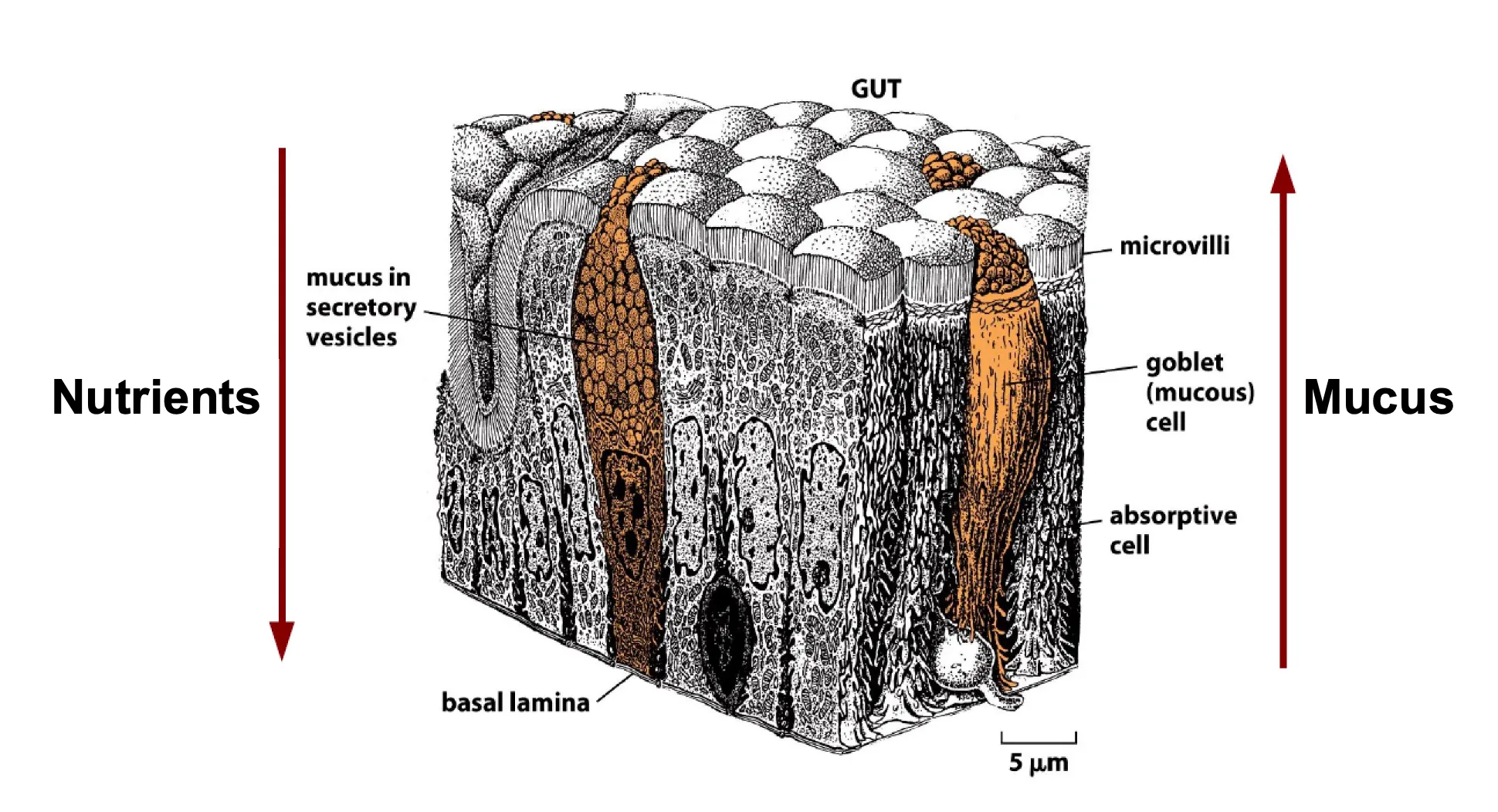
Polarisation of Epithelial Sheets
Both cell types are polarised but apical side has to take up nutrients which is why they have microvilli and goblet cells are polarised because they need to secrete substances out of the cell.
What do Different Types of Junctions do in the extracellular matrix?
Help in holding together the components of the extracellular matrix.
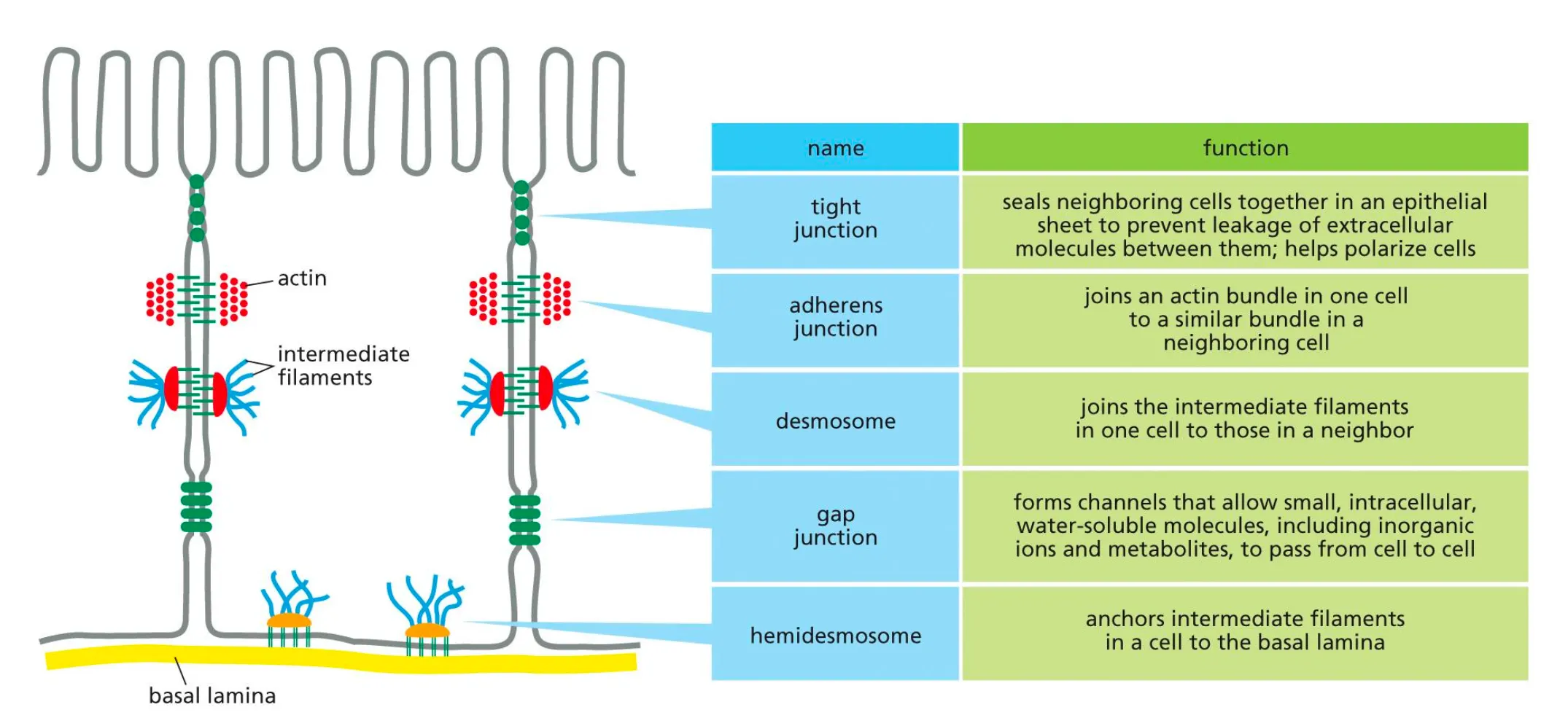
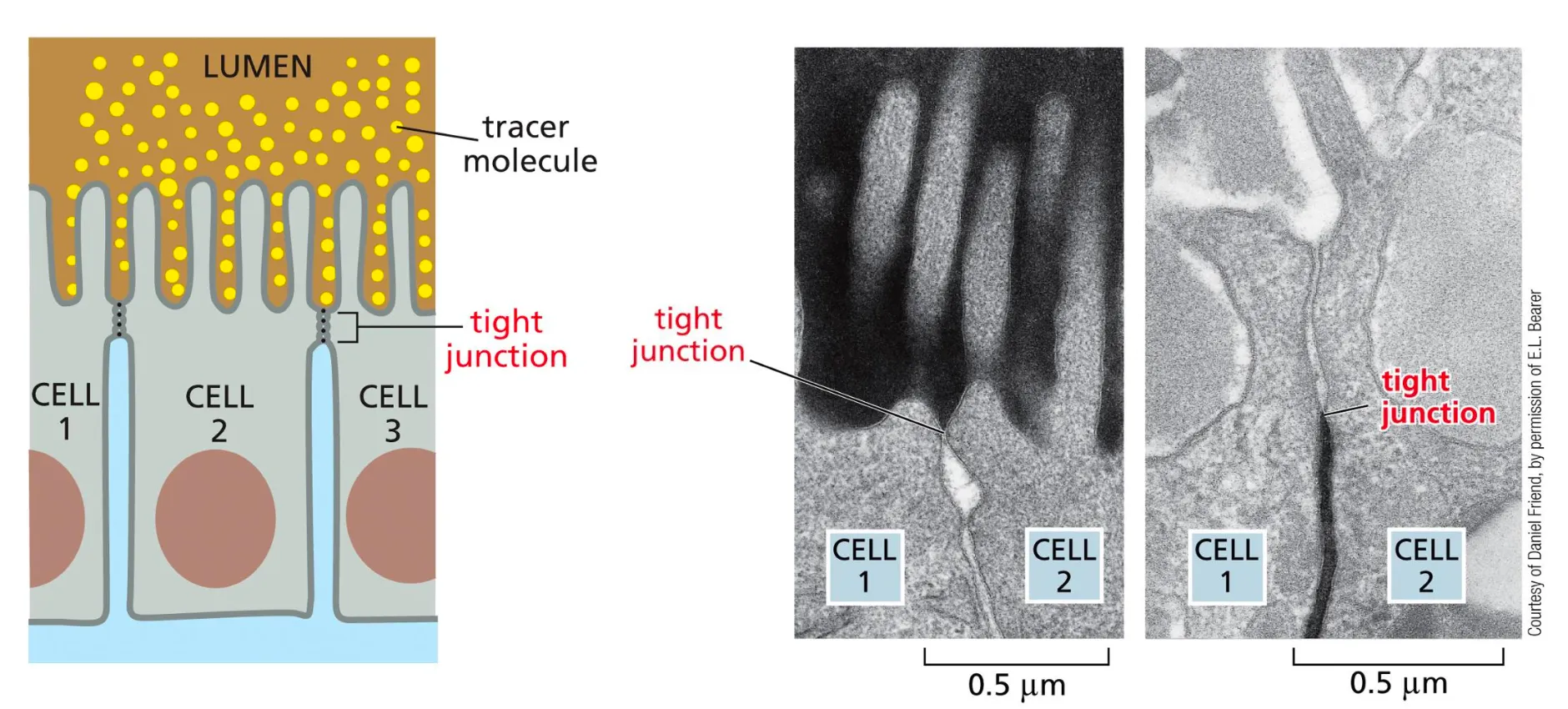
Tight Junctions
Tight Junctions form a tight barrier sealing the outside from the inside. Very tight and do not allow any substances through and they make a barrier between the external and internal world.
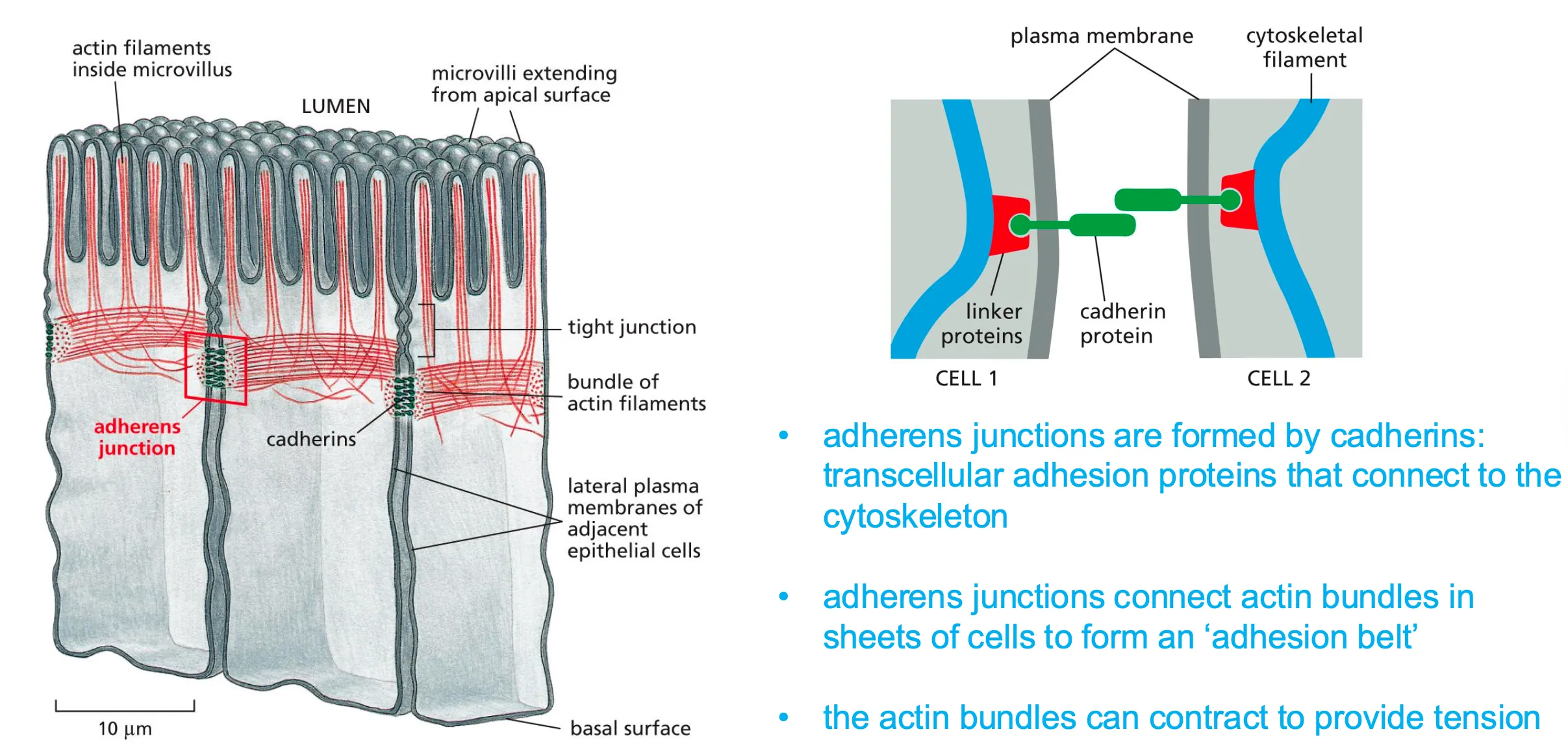
Adherens junctions
Connect the actin cytoskeleton in neighboring cells.
Bridges between cells and are formed by cadherins and main function is to form the intracellular bridges.
Adherens grab actin filaments which form sheets which form the belt that holds all the cells together.
Actin we know can exert a force but also allows the belt to expand and gives dynamic to the adherens.
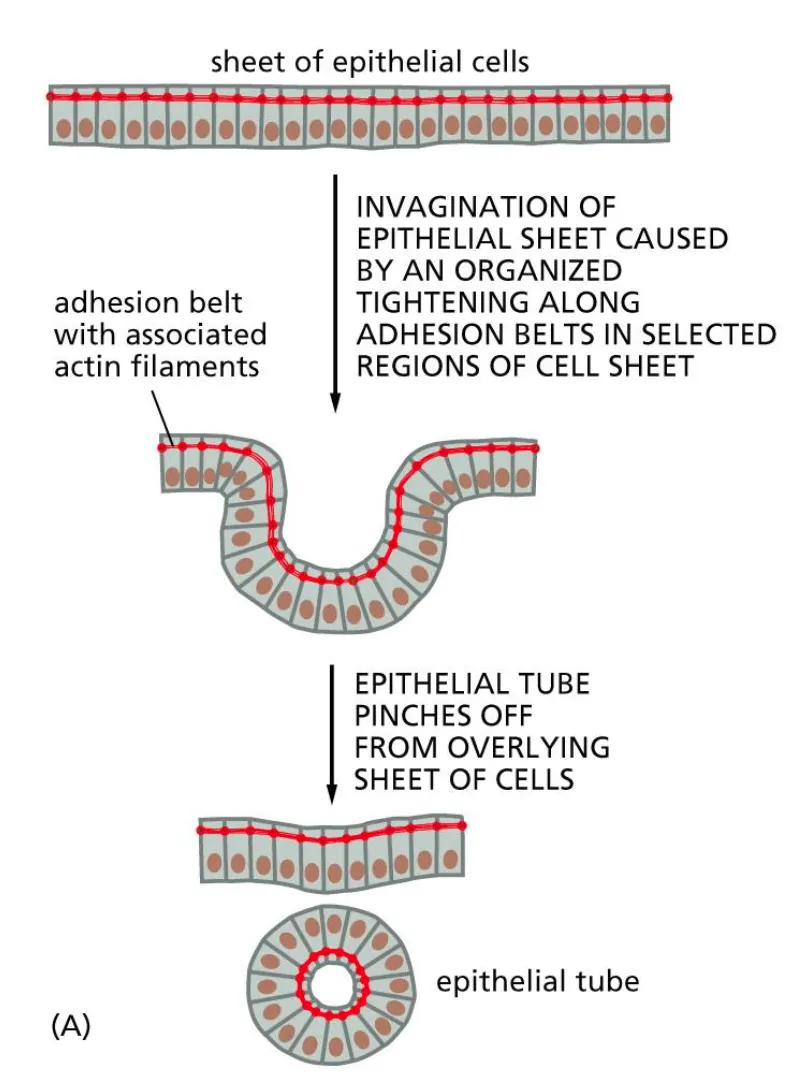
Adhesion belt
Contraction of the adhesion belt allows the formation of complex tissue structures.
Formation of cysts via invaginations.
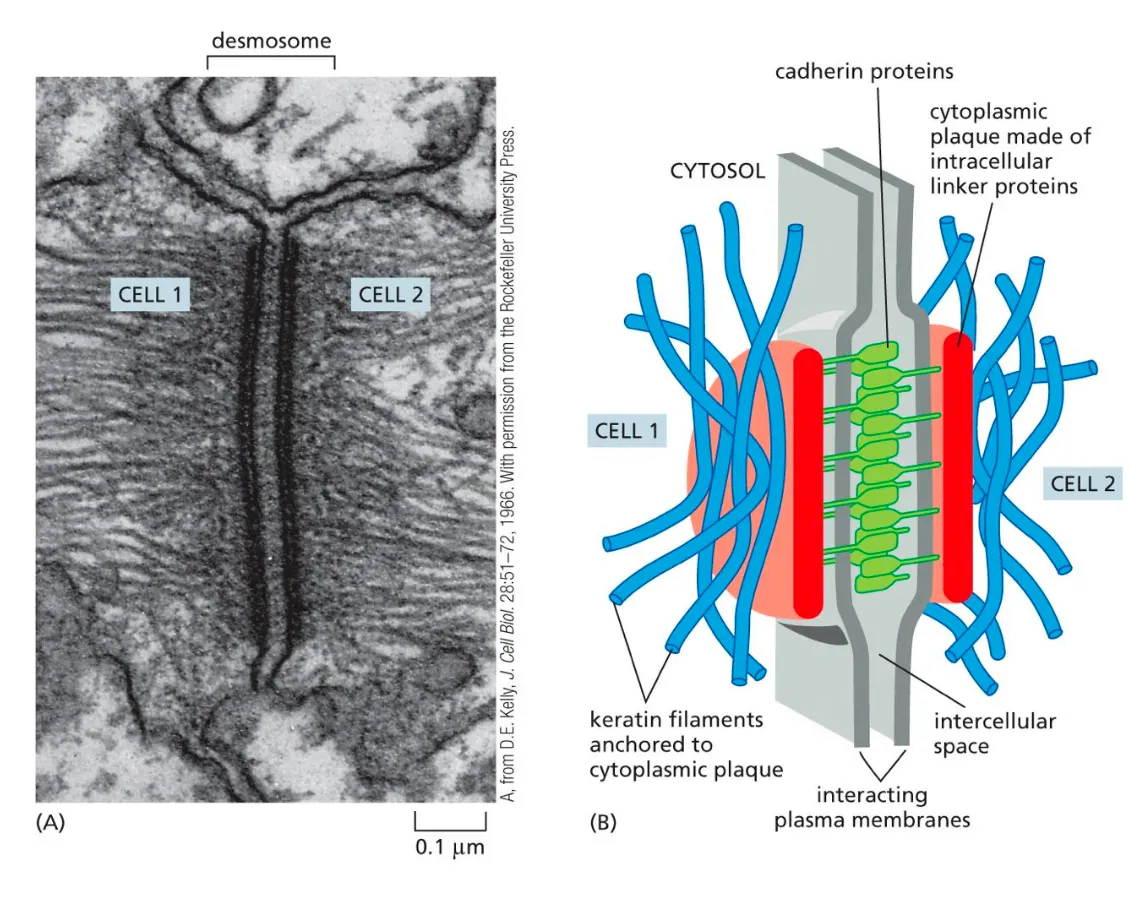
Desmosomes
Desmosomes connect intermediate filaments between neighboring cells
Connections are formed between the cells but now the cadherins are connected to intermediate filaments.
Desmosomes connect intermediate filaments in one cell to the ones in another cell to form a tight sheet of cells
Same function as adherens but is another example of how the junctions occur. (structure to form that tight sheet)
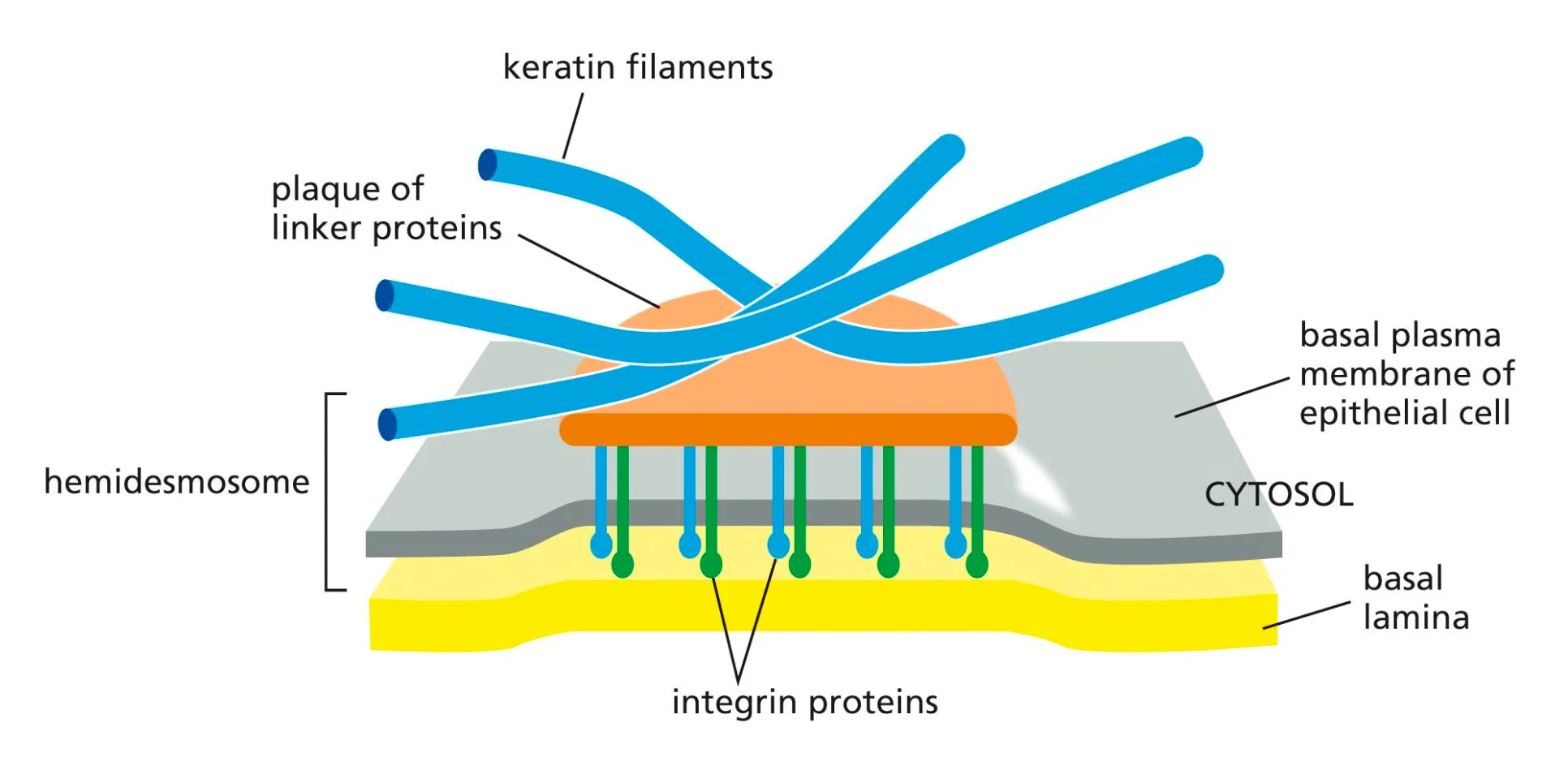
Hemidesmosomes
Connected to the basal lamina via integrin proteins.
Any mutations in these cells would mean the epithelial cell would not be connected to the basal lamina and you would not have polarity in that cel
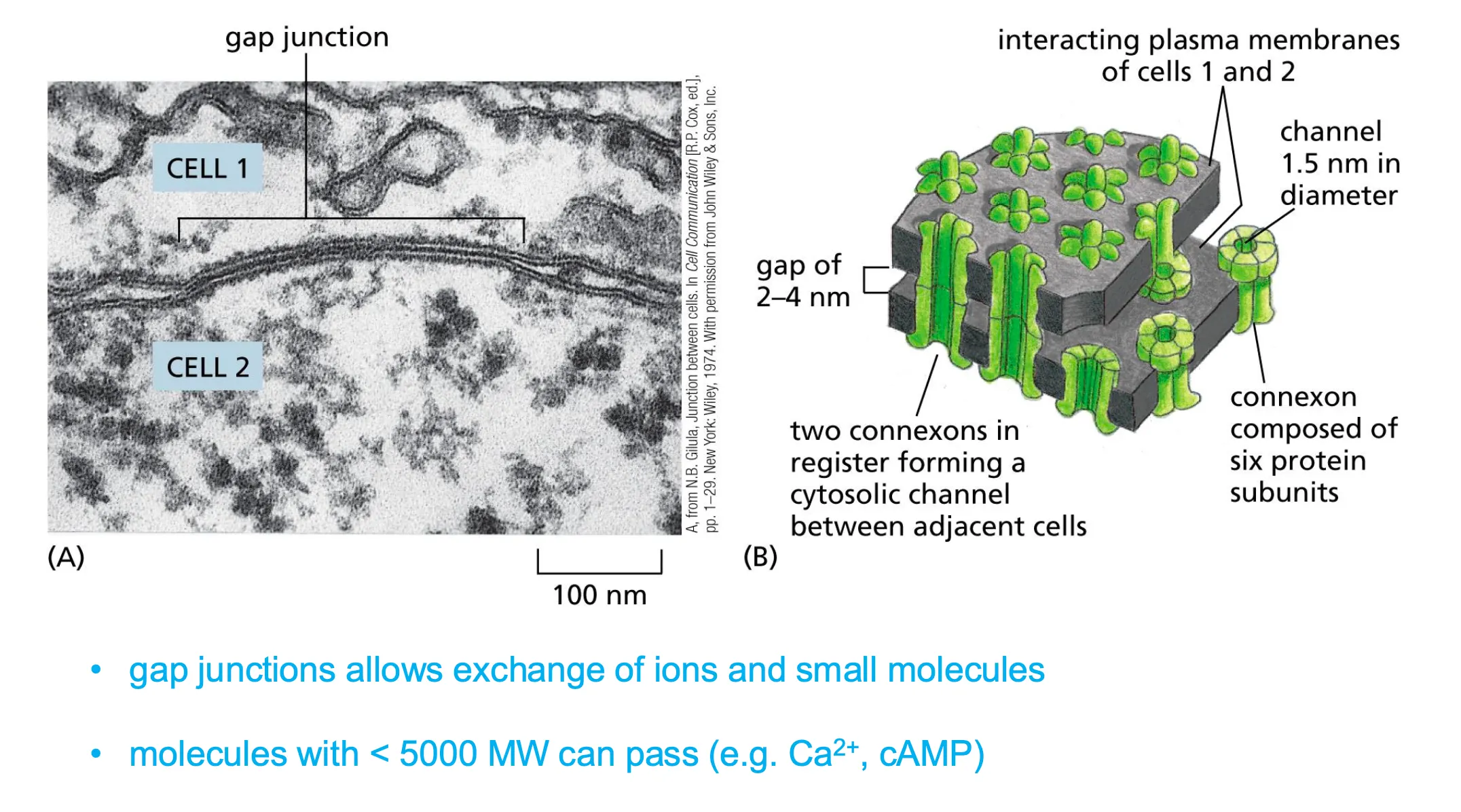
Gap junctions
Permeable for molecules.
Connexxins form connexons and form channels which need to be alligned between two cells so that we can transport molecules between two cell. alignment needs to be very precise.
As long as there is a specific molecular weight the molecules are allowed to diffuse through freely.
Connexons are also regulated and can open and close.
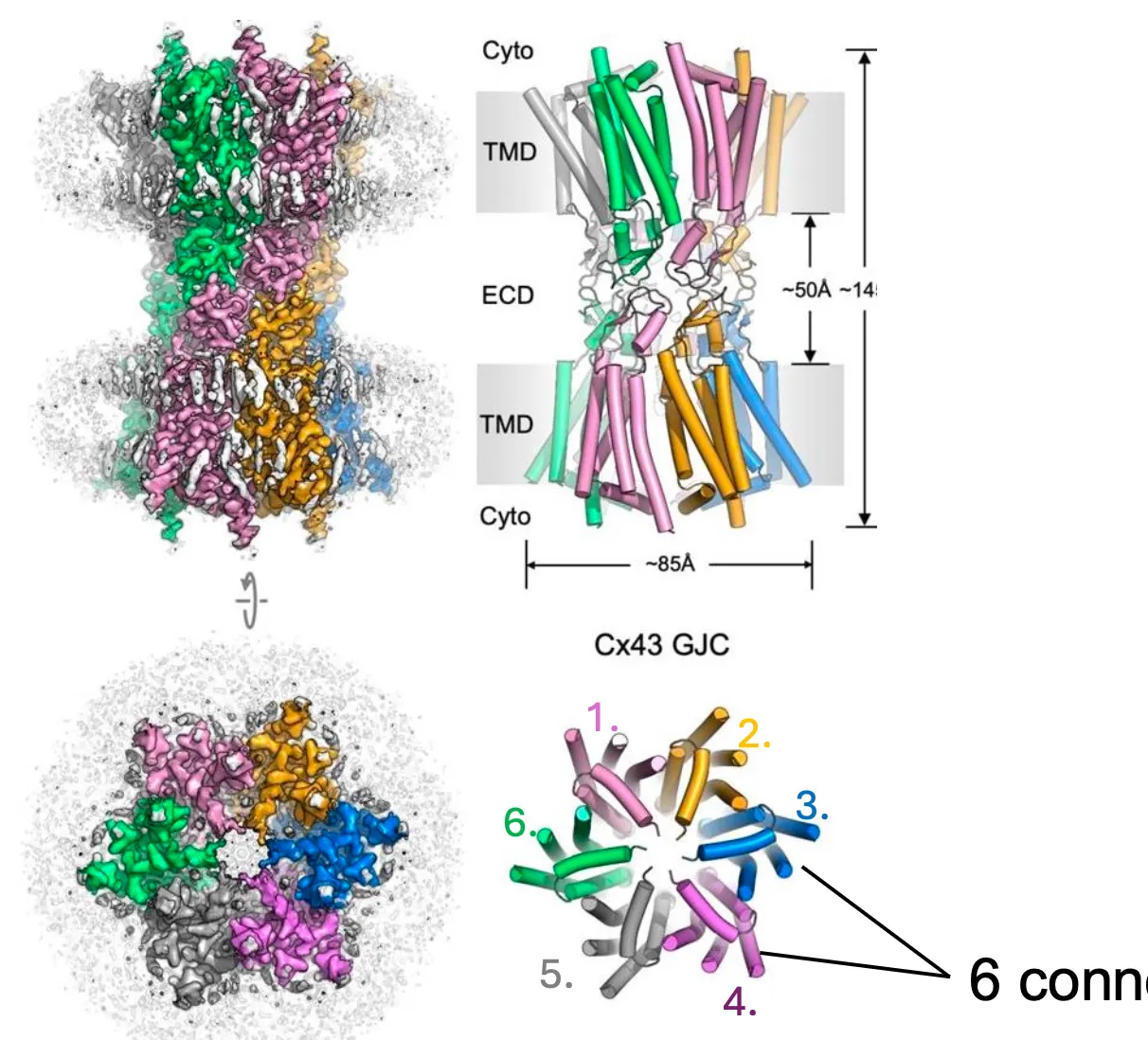
Gap Junctions structure
Gap junctions made of connexins.
And the 6 connexins align to form a pore called the connexon (gap junctions)
And then 2 connexons form a gap junction channel.
Usually they come in packages and so usually we have more than 100 gap junctions that connect the cells.
What are the 4 modes of signalling and please describe them also??
Endocrine (secretion of ligands via bloodstream and the ligand secreted is called a hormone which is produced in endocrine cells)
Paracrine (ligands diffuse directly through the extracellular matrix and act as local mediators and this can be seen in immune responses)
Neuronal (can generate quick electric impulses which are then converted to chemical signals and this is done via neurotransmitters)
Contact dependant (direct contact of cell between ligand and receptor which are embedded in each cell).
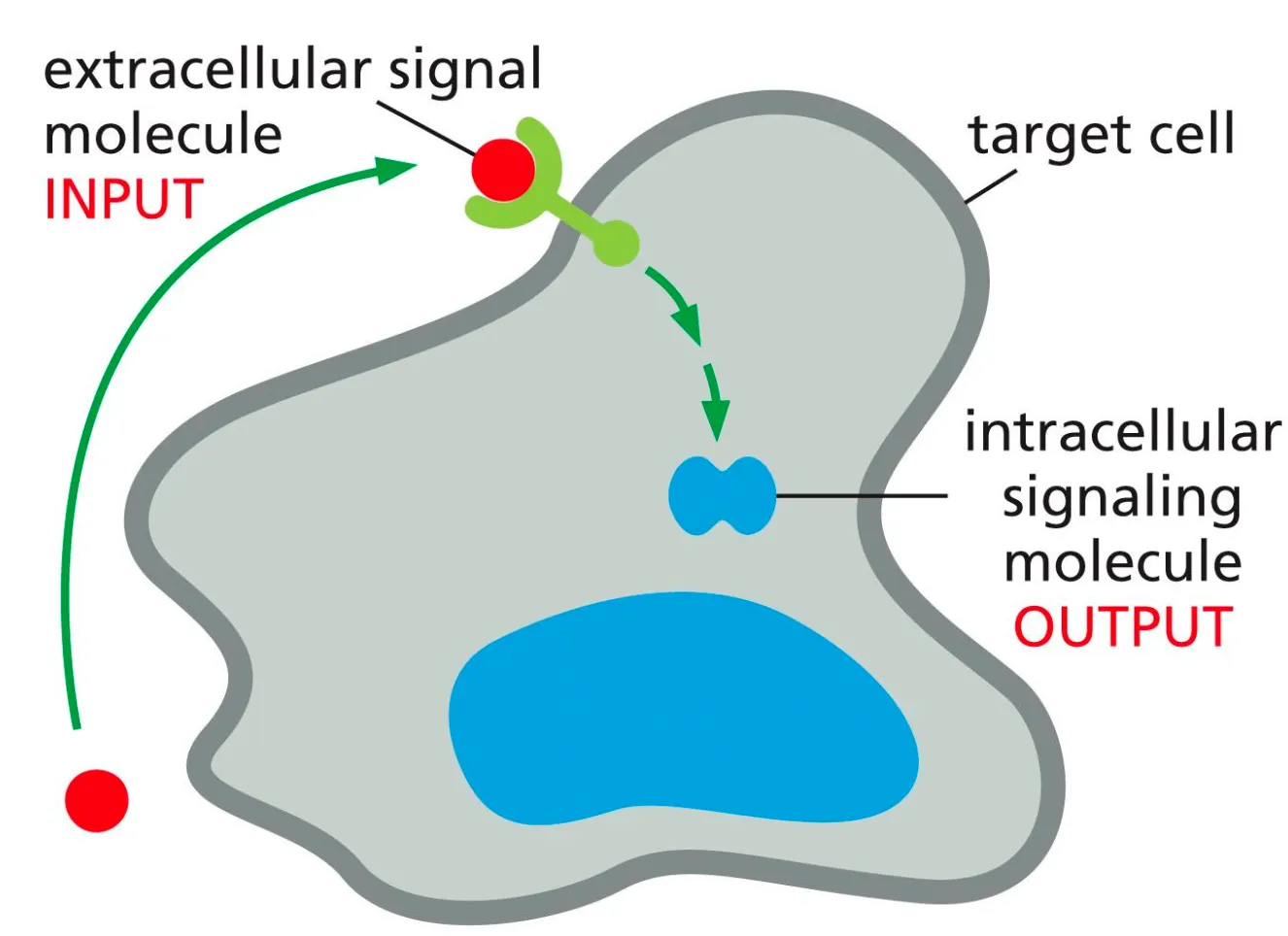
What is the general mechanism of Signal Molecules??
Target cells have proteins which are receptors and can recognise ligands. Signal transduction happens when the receptor on the target cell receives an incoming extracellular signal and then produces intracellular signalling molecules, which alter the cell behaviour.
The response of a cell to a certain signal depends on whether that cell has the given receptor for the ligand. Each receptor is only activated by a single ligand.
The reaction mechanism of a cell on the signal depends on which cascade of intracellular signals the cell-surface receptor can generate and how that effects the effector proteins.
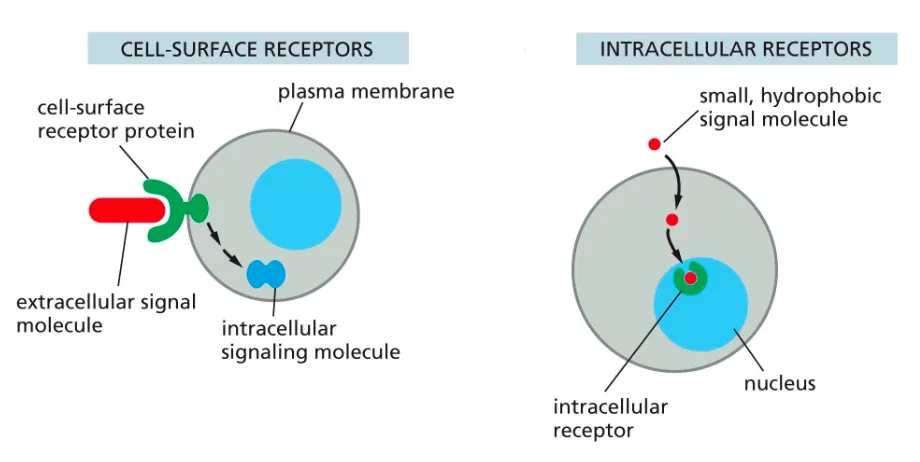
Define Receptors?? and what are the two types.
Receptors can be cell surface ones or intracellular receptors.
The intracellular receptors, are found in the cytosol of the cell and recognise hydrophobic and smaller molecules which are able to pass through the membrane.
Cell surface receptors help large and hydrophillic molecules get targeted and recognised in order for the intracellular events to take place.