AP Biology Unit 6 Terms
1/75
Earn XP
Description and Tags
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No study sessions yet.
76 Terms
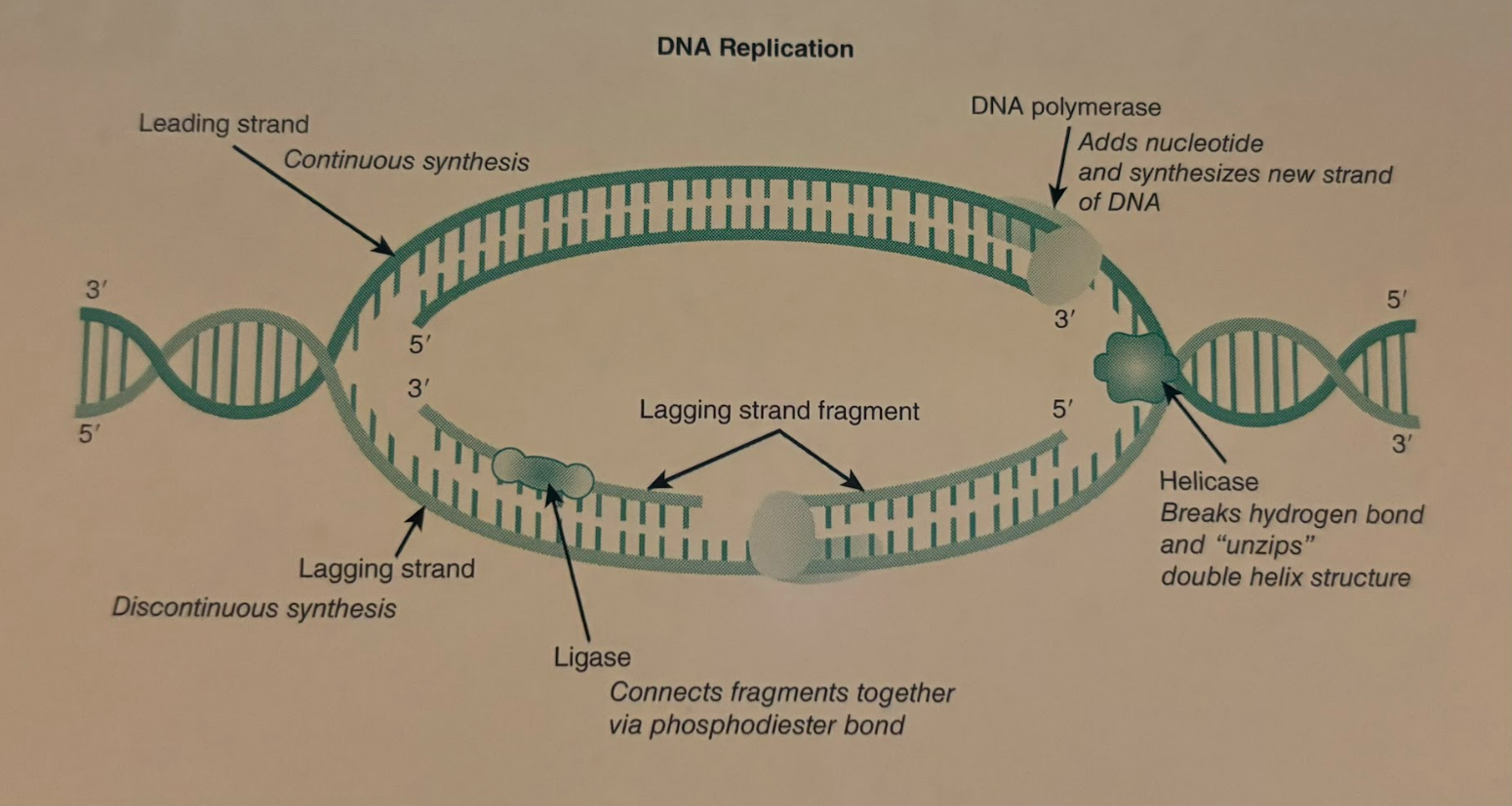
Helicase
The enzyme that unwinds the DNA double helix at the origin of replication site in preparation for DNA replication.
Origin of Replication
Also called “ori sites.” A sequence of DNA at which replication is initiated. Origins of replication tend to have a higher percentage of AT base pairs.
Topoisomerase
A protein that reduces some of the supercoiling, or torsional strain, that occurs as helicase unwinds the double helix.
DNA Polymerase
The enzyme that adds new DNA nucleotides the the 3’hydroxl group on a growing DNA strand. A necessary enzyme for DNA replication to occur.
Ligase
An enzyme that joins fragments of DNA during lagging strand replication.
Okazaki Fragments
Short fragments of DNA produced during the discontinuous replication of the lagging strand of DNA.
Leading Strand Replication
Leading strand replication occurs continuously in the 5’ to 3’ direction on one strand of a DNA molecule.
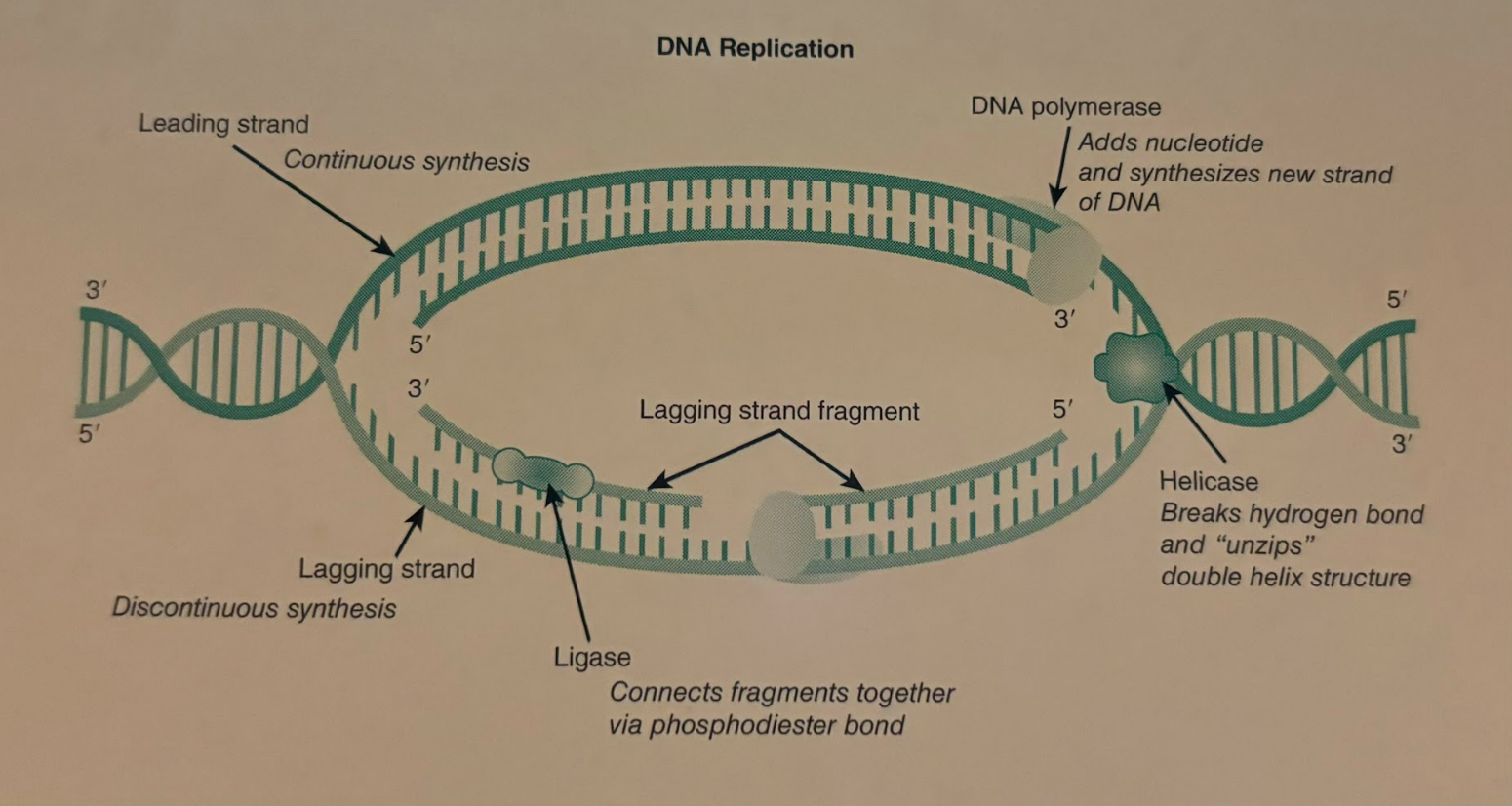
Lagging Strand Replication
Because the two strands of the DNA double helix are antiparallel, lagging strand replication occurs discontinuously in the 5’ to 3’ direction on one of the DNA strands. Short DNA fragments are produced and are eventually joined together by DNA ligase.
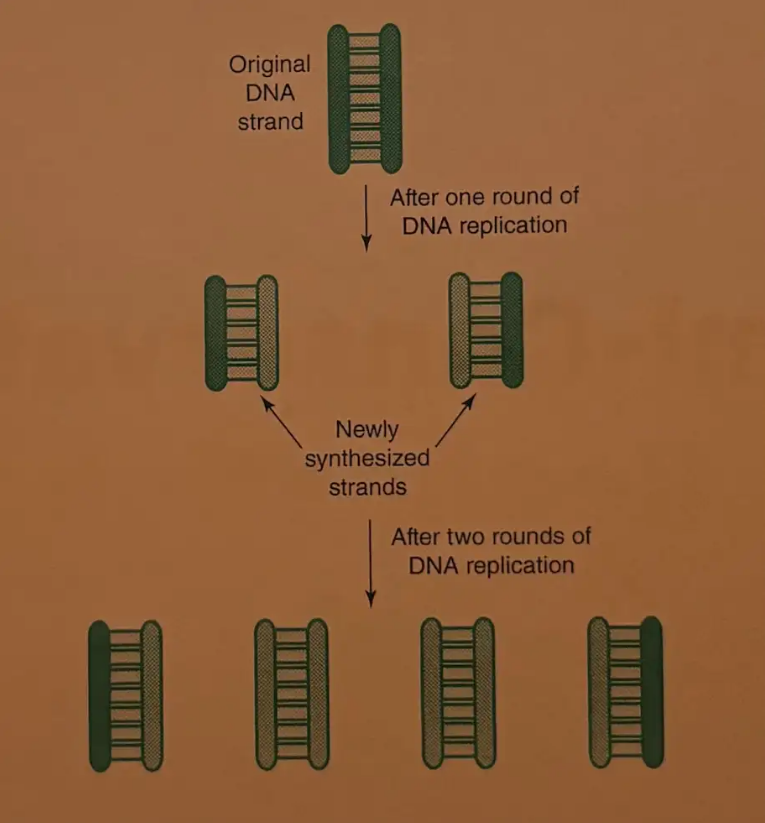
Semi-Conservative Replication
Each new DNA strand created by DNA replication contains one original template strand and one newly synthesized strand. This is semi-conservative replication.
Telomeres
Short, repetitive sequences at the ends of linear chromosomes. During each round of DNA replication of linear chromosomes, telomerase becomes shorter.
Transcription
The process in which the genetic information in a sequence of DNA nucleotides is copied into newly synthesized RNA molecules.
RNA Polymerase
The enzyme that transcribes a DNA sequence into RNA molecules.
mRNA
Messengers RNA is single-stranded and carries information from DNA to the ribosome. mRNA contains three base pair sequences called codons, which are complementary to the DNA base pair sequence. These codons will specify specific amino acids during translation.
tRNA
Transfer RNA folds into a three-dimensional structure that acts as an adapter molecule during translation. One end of the tRNA will bind to a specific amino acid, while the other end of the tRNA contains an anticodon, which will pair with the appropriate mRNA codon at the ribosome during translation.
rRNA
Ribosomal RNA folds into a three-dimensional structure. rRNA and proteins from ribosomes that perform translation. The three-dimensional rRNA acts as a ribozyme, catalyzing the reactions needed in translation.
Codons
Three nucleotide units in mRNA that specify a particular amino acid.
Anticodons
Three nucleotide units in tRNA that are complementary to codons in corresponding mRNA molecules.
Promoter
The noncoding DNA sequence upstream from a gene to which RNA polymerase can bind, starting transcription of the gene.
TATA Box
A repetitive sequence of thymine and adenine nucleotides that is within the promoter sequence and it recognized by RNA polymerase.
Transcription Factors
Proteins that help RNA polymerase bind to the promoter sequence and begin transcription.
Pre-mRNA
The initial mRNA transcript formed in eukaryotic cells that needs to be modified before it can leave the nucleus and be translated. Pre-mRNAs contain introns.
RNA Processing
In eukaryotes, the processes involved in preparing a pre-mRNA molecule for export as a mature mRNA to the ribosome.
Mature mRNA
In eukaryotes, the mRNA transcript that has been modified and is ready to leave the nucleus to be translated. Introns have been removed from mature mRNAs, a 3’ poly-A tail has been added, and a 5’ GTP cap is added to the mRNA to form the mature mRNA.
Introns
Noncoding RNA sequenced that are removed from eukaryotic mRNA transcripts before translation.
Exons
Protein coding sequences in eukaryotic mRNA transcripts that will be translated.
5’ GTP Cap
The addition of a guanine nucleotide to the 5’ end of a eukaryotic mRNA transcript. This ‘5 GTP cap allows the mature mRNA to leave the nucleus and helps the initiation of translation at the ribosome.
3’ Poly-A Tail
A sequence of adenine nucleotides that is added to the 3’ end of eukaryotic mRNAs. The 3’ poly-A tail helps precent degradation of the mRNA transcript.
Spliceosomes
Structures made of small nuclear RNAs (snRNAs) and small nuclear ribonucleoproteins (snRNPs) that remove the introns from pre-mRNAs and splice together the exons to form the mature mRNA transcript in eukaryotes.
snRNA
Small nuclear RNA molecules that make up part of the spliceosomes.
miRNA
MicroRNA that functions in gene regulation and RNA silencing.
RNA Silencing
The process by which small microRNA binds to mRNAs, forming a double-stranded RNA, preventing the translation of the mRNA.
snRNPs
Small nuclear ribonucleoproteins that make up part of the spliceosomes.
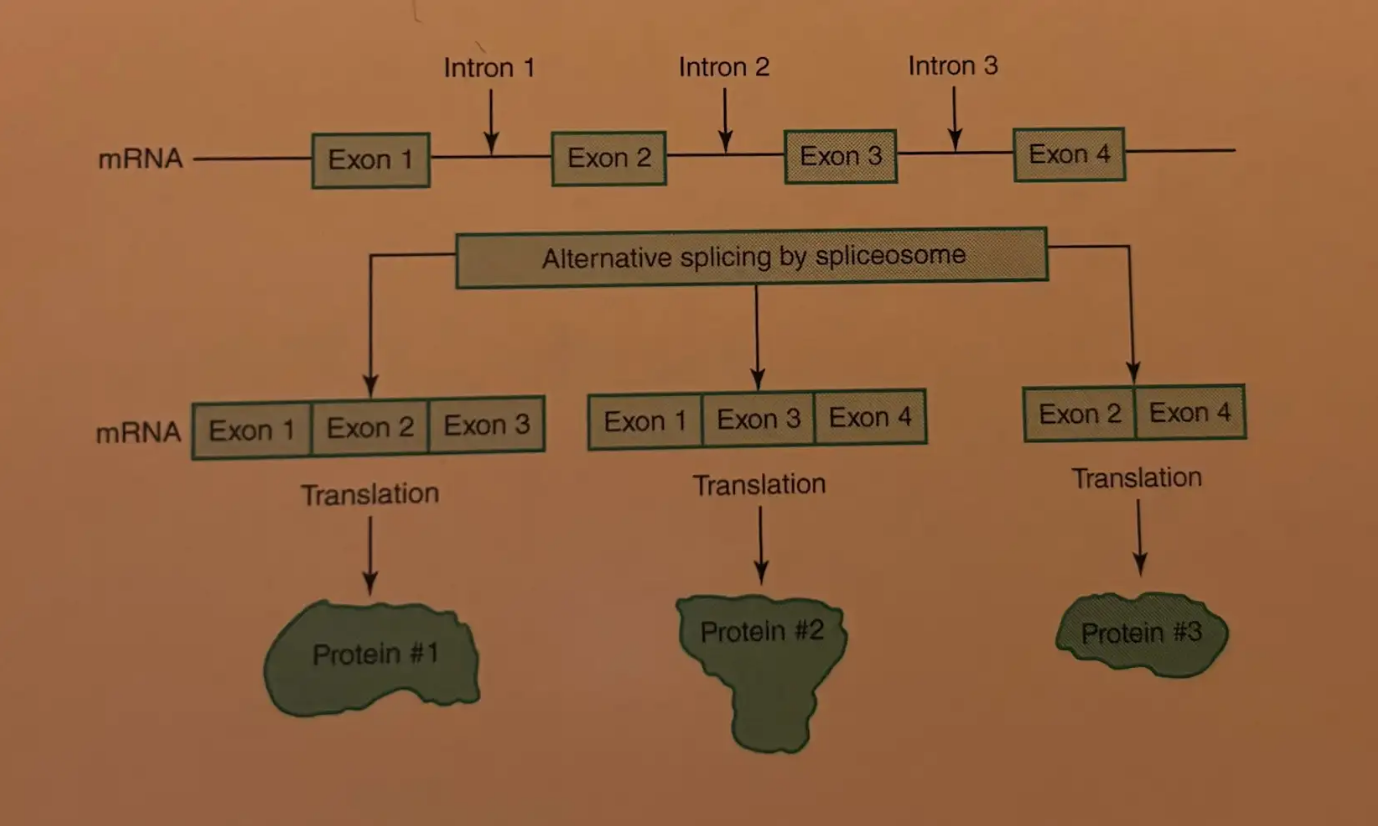
Alternative Splicing
In eukaryotes, the joining of exons in different combinations to create multiple RNA transcripts from the same gene.
Translation
The process by which a protein in synthesized based on the information provided by mRNA. Translation occurs in ribosomes, which may be in the cytoplasm, or in eukaryotes, attached to rough endoplasmic reticulum.
Polyribosomes
Found in prokaryotes only, this refers to multiple ribosomes translating an mRNA.
Start Codon
The first codon in an mRNA translated by the ribosome. The start codon (AUG) binds to rRNA in the ribosome, stimulating the start of transcription.
Stop Codon
The codon in mRNA that indicated the end of the polypeptide chain. The stop codon does not code for an amino acid. When ribosome reached the stop codon, a protein release factor binds to the ribosome, terminating translation.
Retroviruses
Viruses that contain RNA as their genetic material. Retroviruses also contain the enzyme reverse transcriptase.
Reverse Transcriptase
The enzyme in a retrovirus that makes a DNA copy of the retrovirus RNA. This DNA is then inserted into the host cell’s chromosome.
Regulatory Proteins
Proteins that can turn on or turn off genes by binding to a specific nucleotide sequences.
Regulatory Sequences
Noncoding nucleotide sequences to which regulatory proteins bind, affecting gene expression.
Operon
A cluster of genes with a common function under the control of a common promoter. Operons contain regulatory sequences, genes for regulatory sequences, and genes for structural proteins (which are responsible for the function of the operon).

Operators
Noncoding regulatory sequences that serve as binding sites for repressor proteins.
Structural Genes
Coding sequences that contain the genetic code for the proteins required to perform the function of the operon.
Inducible Operons
Operons that typically have a catabolic function (digesting molecules) and are turned off unless the appropriate inducer molecule is present. The repressor of inducible operons cannot bind to the operator if the inducer molecule is present.
Inducer
Molecule that binds to the repressor protein, changing its shape so that it can no longer bind to the operator sequence. This allows RNA polymerase to access the promoter sequence. This allows RNA polymerase to access the promoter sequence and begin transcribing the structural genes of the operon.
Repressor
A regulatory protein that binds to the operator sequence, preventing RNA polymerase from transcribing the genes in the operon.
Repressible Operons
Operons that usually have an anabolic function (synthesizing molecules) and are turned on unless the corepressor molecule is present. The product of the operon typically functions as the corepressor, allowing the cell to stop transcription of the operon when an abundance of the product is already present.
Corepressor
In repressible operons, the molecule that binds to the repressor protein, changing the shape of the repressor protein so that it can bind to the operator sequence, preventing transcription of the operon. The product of a repressible operon typically functions as the corepressor.
Regulatory Switches
Noncoding sequences to which activator proteins or repressor proteins may bind.
Enhancers
Regulatory switches to which activator proteins or transcription factors may bind.
Silencers
Regulatory switches to which repressor proteins may bind.
Activators
Regulatory proteins that bind to enhancer sequences, stimulating transcription of genes, thus upregulating gene expression.
Transcription Factors
Regulatory proteins that help RNA polymerase bind to promoter sequences, stimulating the start of transcription.
Mediators
Regulatory proteins that serve as “connectors” between other regulatory proteins and allow regulatory proteins to communicate.
Epigenetic Changes
Reversible modifications to the nucleotides of the DNA sequence, such as methylation and acetylation, that affect gene expression. Some epigenetic changes may be passed down from parents to offspring.
Methylation
Attachment of methyl groups to DNA nucleotides, affecting the expression of the DNA sequence. Often more methylation of DNA leads to lower expression of the DNA sequence.
Acetylation
Attachment of acetyl groups to histone proteins, affecting the expression of the DNA that is wrapped around the histones. Often more acetylation of histones leads to greater expression of the DNA sequence.
Histones
Proteins around which DNA is packaged into chromosomes.
Euchromatin
DNA in chromosomes that is more loosely wound around the histone proteins, is more accessible to RNA polymerase, and usually results in more expression of the genes within the euchromatin.
Heterochromatin
DNA in chromosomes that is more tightly wound around the histone proteins, is less accessible to RNA polymerase, and results in reduced expression of genes within the heterochromatin.
siRNAs
Small interfering RNA molecules are single-stranded RNAs that bind to complementary mRNA molecules, forming double-stranded RNA (dsRNA) molecules. Enzymes in the cell detect and destroy these dsRNA molecules, resulting in no translation of the targeted mRNA molecules and reduced gene expression.
Differential Gene Expression
The biochemical processes that determine which genes are transcribed and translated in different cells, giving each cell its unique characteristics.
Mutations
Changes in the genetic material of an organism.
Horizontal Transmission
The transfer of genetic material between organisms that are members of the same generation (not parent to offspring). Transformation, transduction, and conjugation are all potential methods of horizontal transmission of genetic material.
Transformation
The uptake and expression of naked foreign DNA by a cell.
Transduction
The transmission of DNA between cells by viral vectors.
Conjugation
The transmission of DNA through direct cell-to-cell contact, typically through a structure called pilus.
Transposition
The movement of DNA between chromosomes or within a chromosome. The DNA sequenced involved are sometimes referred to as “jumping genes.”
Bacterial Transformation
The process of introducing foreign DNA into bacterial cells and the expression of the foreign DNA in the bacteria.
Recombinant DNA
DNA that has been recombined from different source organisms.
Restriction Endonucleases
Enzymes that cut DNA at specific palindromic sequenced (also know as restriction enzymes).
DNA Ligases
Enzymes that connect DNA fragments by catalyzing the formation of phosphodiester bonds between the nucleotides.
Gel Electrophoresis
A technique that uses a gel (typically agarose or polyacrylamide) to separate DNA fragments by size. Smaller DNA fragments will migrate further in the gel than larger DNA fragments.
Polymerase Chain Reaction (PCR)
A technique that uses heat-stable DNA polymerases and specific primer sequences to make multiple copies of specific DNA sequences.
CRISPR-Cas9
A gene editing tool used in bacteria to destroy the DNA of invading viruses. CRISPR-Cas9 can be used in the lab to edit gene sequences.