Biology - DNA and Proteins
1/93
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
94 Terms
what process does prokaryoitc cells use the reporduce
binary fission
what process does eukaryoitc cells use the reporduce
mitosis and meiosis
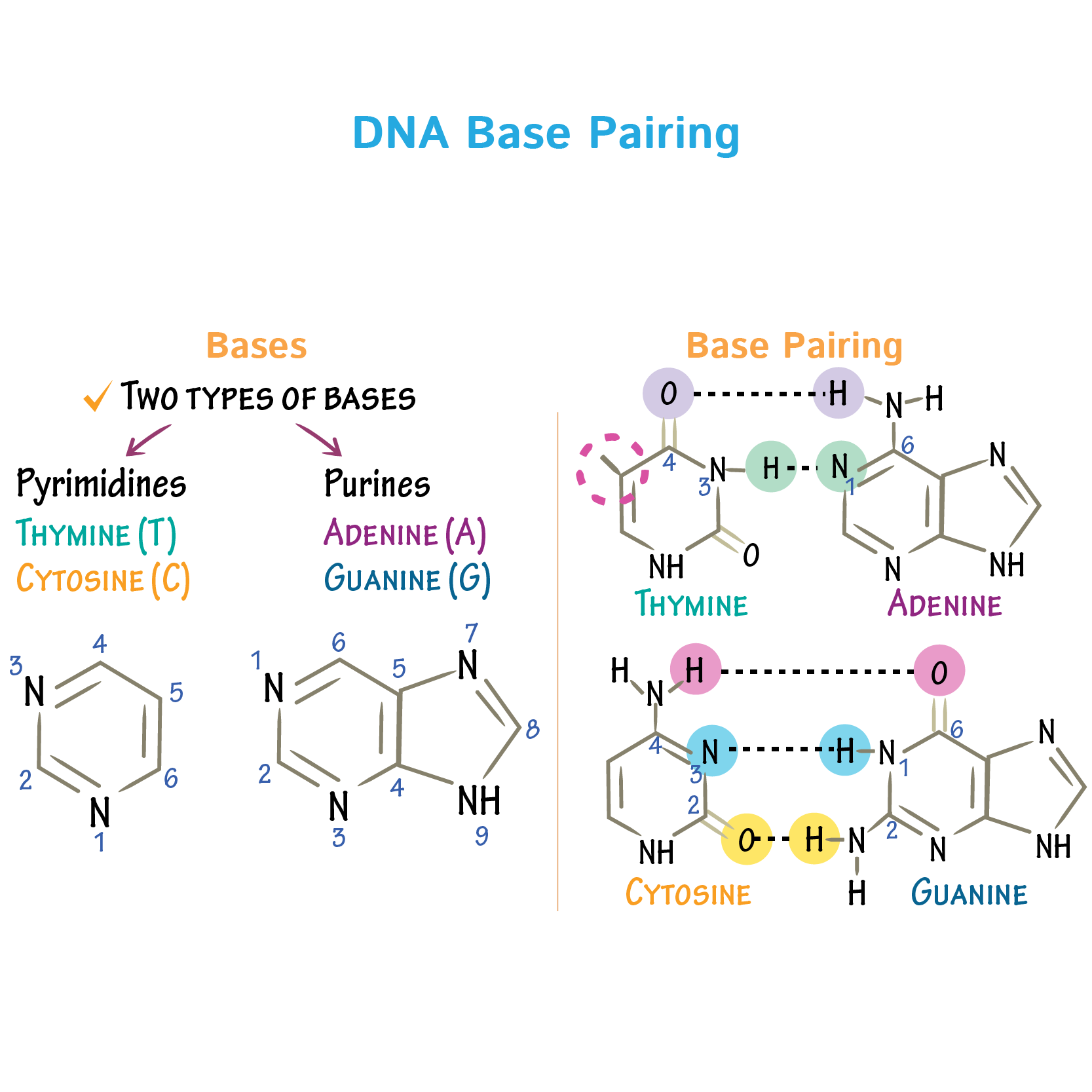
what nitrogenous bases are purines (one nitrogen ring)
adenine and guanine
what nitrogenous bases are pyrimidines ( two nitrogen rings)
thymine and cytosine (and uracil)
what is the bond holding the nitrogenous bases together
weak hydrgen bonds
what is the bond holding the nucelotides together
strong covalent bonds
why do only certain nitrogenous bases fit togther
the position of their atoms match up with eachother
what is the process of semi conservitaive replication
1. Unwinding: The DNA double helix unwinds with the help of enzyme helicase, creating two separated strands.
2. Primer Binding: Enzymes called primases add short RNA primers to each single strand of DNA. These primers provide a starting point for DNA synthesis.
3. DNA Synthesis: DNA polymerase enzymes start building new DNA strands by adding complementary nucleotides. They move along each strand in opposite directions:
- Leading Strand: DNA polymerase synthesizes this continuously in the 5' to 3' direction.
- Lagging Strand: DNA polymerase synthesizes this discontinuously in short sections called Okazaki fragments, each initiated by a primer.
4. Primer Removal and Fragment Joining: DNA polymerase removes the RNA primers and replaces them with DNA nucleotides. DNA ligase then joins the Okazaki fragments into a continuous strand.
5. Result: The process results in two identical DNA molecules, each consisting of one original strand and one newly synthesized strand.
This replication process ensures genetic information is accurately copied and passed on during cell division. It's crucial for maintaining genetic stability and diversity in living organisms.
when shown a table of percentages of nitrogenous bases in a DNA strand, what does it mean if the complementary bases do not match (or are close to matching)?
it is single stranded (ie. prokaryote DNA) and thus ther is no complementary pairing
what strand in DNA replication is the template strand? and what is the coding strand?
the template strand is the strand from the original DNA helix that the free nucleotides are attaching to
the coding strand is the other original DNA strand that the free nucleotides are a RNA copy of
is RNA double stranded
no it is single stranded
what are proteins
macromolecues composed of long chains of amino acids folded into unique shapes
what are the processes involved in gene expression
transcription and translation
do prokaryotic cells have introns
no so no splicing need to be done
what is the process of transcription
1. Initiation: RNA polymerase binds to a specific starting point on the DNA called the promoter causing the DNA to unwind. Helicase unzips the dna.
2. Elongation: RNA polymerase moves along the DNA, unwinding it and creating a complementary RNA strand. It adds RNA nucleotides that match the DNA template (A-U, T-A, C-G).
3. Termination: Transcription stops when RNA polymerase reaches a special sequence on the DNA that signals the end of the gene.
4. Processing (in eukaryotes): The initial RNA transcript undergoes modifications like adding a cap and tail, and removing non-coding sections (introns).
5. Export and Translation: The mature mRNA leaves the nucleus and is translated by ribosomes in the cytoplasm to produce proteins.
This process allows the genetic information stored in DNA to be converted into RNA, which then directs the synthesis of proteins in the cell.
what is the process of translation
1. Initiation: The mRNA attaches to a ribosome, and the ribosome finds the start codon (usually AUG).
2. Elongation: The ribosome reads the mRNA in sets of three letters (codons). Transfer RNA (tRNA) molecules bring amino acids to match each codon. The ribosome links these amino acids together to form a growing chain (polypeptide).
3. Termination: Translation stops when the ribosome encounters a stop codon (UAA, UAG, or UGA) on the mRNA. This signals the end of protein synthesis.
4. Protein Formation: The chain of amino acids (polypeptide) folds into its specific shape, determined by its sequence, and may undergo modifications to become a functional protein.
5. Function: The protein performs its specific job in the cell, such as enzymes catalyzing reactions, structural proteins giving shape to cells, or signaling proteins communicating between cells.
Translation is crucial for converting genetic instructions from mRNA into functional proteins that carry out essential tasks in the cell.
where does transcription and translation happen in eukaryotes? where is prokaryotes?
eukaryotes: nucleus → cytoplasm
prokaryotes: cytoplasm
what is the primary level of protein structure
the sequence of amino acids
what is the secondary level of protein structure
the 2D structure. weak hydrogen bonds that form between certain atoms to create a 2D shape. ie pleated sheet or helix
what is the teriary level of protein structure
3D shape. resulting from attractive forces including hydrgon bonds, hydrophobic interactions, covalent bonds and ionic bonds
what is the quaternary level of protein structure
the highest level of protein structure, where two or more polypeptide chains (subunits) come together to form a larger, functional protein complex.
what are the bonds tht hold together a polypeptide called?
peptide bonds
proteins are essential to cell structure and function
proteins are essential to cell structure and function
why is the 3D shape of a protein so important to its function
The three-dimensional shape of a protein is crucial for its function because:
1. Functionality: Each protein's shape is like a lock that fits a specific key (its substrate). This allows proteins to perform their jobs, such as speeding up chemical reactions (enzymes) or transporting molecules.
2. Specificity: Proteins recognize and interact with other molecules based on their shape. This specificity ensures that proteins can carry out precise tasks in cells without interference.
3. Stability: The folded shape of a protein is stabilized by bonds and interactions between its atoms. This stability is necessary to maintain the protein's structure and function under different conditions.
4. Regulation: Proteins can change shape in response to signals, allowing them to turn their functions on or off. This flexibility helps regulate processes in the cell.
5. Assembly: Some proteins need specific shapes to join with others and form larger structures or complexes. These complexes perform more complex tasks than individual proteins alone.
In summary, the shape of a protein is not just a structural feature—it's critical for how proteins perform their roles in cells and organisms.
factors that effect protiens
Certainly! Here's a simpler explanation of factors that affect proteins:
1. Temperature: Proteins have an optimal temperature range for function. High temperatures can unfold (denature) proteins, disrupting their structure and function.
2. pH: Proteins also have an optimal pH range. Changes in pH can alter protein shape and stability, affecting their ability to function properly.
3. Salt Concentration: Proteins are sensitive to salt levels. High salt concentrations can affect protein solubility and stability by shielding charges on the protein surface.
4. Solvents: Chemicals like organic solvents or detergents can disrupt protein structure by affecting hydrophobic interactions.
5. Co-factors: Some proteins require specific molecules (co-factors) to function. Changes in co-factor availability can impact protein activity.
6. Post-Translational Modifications: Proteins can be modified after they are made, which can affect their function, localization, or interactions.
7. Oxidation: Reactive oxygen species can damage proteins by oxidizing amino acids, altering their structure and function.
8. Mutation: Changes in the DNA sequence can lead to mutations in proteins, affecting their structure and function.
9. Protein Interactions: Proteins often work together in complexes. Changes in how proteins interact with each other can affect their overall function.
These factors play critical roles in determining how proteins behave and function within cells and organisms. Understanding them helps scientists study protein function in health and disease.
factors that effect enzymes
temperature
pH
inhibitors
the rate of an enzyme controlled reaction is affected by…?
concentration of reactants and of the enzyme
explain the induced fit model
Starting Point: Initially, the enzyme's shape isn't a perfect match for the substrate.
Binding: When the substrate approaches the enzyme, they interact through weak interactions such as hydrogen bonds, van der Waals forces, and hydrophobic interactions. When the substrate (like a key) fits into the enzyme's active site (like a lock), they interact and bind together.
Adjustment: The enzyme changes its shape slightly to better fit the substrate. This is called induced fit.
Catalysis: With the substrate snugly in place, the enzyme can now catalyze the reaction more efficiently.
Release: After the reaction, the products are released, and the enzyme returns to its original shape or adjusts again for another substrate.
In essence, the induced fit model explains how enzymes and substrates interact dynamically, with the enzyme adapting its shape to enhance the catalytic process.
differential gene expression
Describes the differences in gene expression patterns among different cell types, developmental stages, or environmental conditions. It explains how cells with the same DNA can have different functions and identities.
controlled gene expression is about the regulatory mechanisms within a single cell, while differential gene expression explains the diversity of gene activity across different cells or conditions.
Controlled Gene Expression:
It focuses on the precise regulation of gene activity to ensure proper cellular function. It involves mechanisms that ensure genes are expressed at the right levels, times, and places.
controlled gene expression is about the regulatory mechanisms within a single cell, while differential gene expression explains the diversity of gene activity across different cells or conditions.
what allows some genes to be expressed and some to be silenced?
Transcription factors.
For a gene to be activated and its protein expressed, transcription factors must bind to the gene to allow the attachment of RNA Polymerase. Without transcription factors, RNA Polymerase cannot bind and the gene is not transcribed and is effectively silenced. In multicellular organisms, specific transcription factors activate some genes and silence others, resulting in differentiated cell types with specialised functions.
What are introns?
they are non-coding sections of the DNA strand. they are weeded out by transcription factors if that specific gene dosent need them.
how do cells adapt to their envrioments via gene expression?
Cells adapt to their environments through changes in gene expression, which allow them to respond to signals like food availability, temperature, or threats.
1. Sensing Signals: Cells can detect changes in their environment, like low nutrients or stress.
2. Switching Genes On and Off: Depending on what the cell senses, it turns certain genes on or off.
3. Adapting: By changing which genes are active, cells can make proteins that help them survive, repair damage, or grow differently.
4. Examples: In response to a lack of a tyrosine from the envrionment, the cell can turn on a gene that produces tyrosine. if tyrosine becomes avalibe again that gene is switched off so as to not wste resources.
In summary, cells use gene expression changes to adjust to their surroundings, helping them stay healthy and functional in different conditions.
when does transcription and traslation happen?
All the time (in interphase) its is a constant process.
herterochromatin
Heterochromatin is a tightly packed form of DNA where the genes are inaccesable to transcription
euchromatin
Euchromatin is a lightly packed form of chromatin that is often (but not always) under active transcription. this version of chromatin is accessable to transcription.
what is chromatin remodeling
the addition or removal of chemical groups such as acetyl and methyl groups from histones.
what does adding acetyl groups to histones do? (acetylation)
makes chromatin less condensed and avalible to transcribe (euchromatin)
what does adding methyl groups to histones do? (Histone methylation)
makes chromatin more condensed and inaccesable to transcription (herterochromatin)
what is DNA Methylation
cells can switch genes on and off by removing methyl groups from cytosine nucleotides. it is facilitated by DMNTs, a group of enzymes called methyltransferases.
what happens to genes that are highly methylated?
they are switched off and silenced as RNA Polymerases cannot bind and transcribe DNA effectively.
what happens when methyl groups are removed from cytosine nucleotides
it allows transcription factors and RNA Polymerase to bind and transcribe genes that were previously silenced by methylation.
is DNA methylation inherited?
yes, genes stay methylated through cell divisions as DMNTs methylate daughter DNA molecues after DNA replication, ensuring methylation patterns are passed onto daughter cells.
what is epigenetic control
the control via chromatin modification and DNA methylation over what cells do, overceeding the sequence of genes.
what are proto-oncogenes?
they are genes that regulate cell division in eukaryotes. they code for growth factors, receptors and enzymes that promote cell division and tumour suppressor genes.
what are tumor suppresor genes
Tumor suppressor genes are a class of genes that help regulate cell growth, division, and apoptosis (programmed cell death) to prevent the formation of tumors. They act as the "brakes" of the cell cycle, ensuring that cells do not proliferate uncontrollably.
how do epiggenetic factors cause cancer and dieases?
Epigenetic factors can cause cancer and other diseases by changing how genes are turned on or off without altering the DNA sequence. These changes can disrupt normal cell functions, leading to uncontrolled growth or other issues. Here’s how it works in simpler terms:
epigenetic factors can:
Impact on Cancer
Turning Off Tumor Suppressor Genes: Genes that usually keep cell growth in check get turned off, leading to uncontrolled cell growth.
Turning On Oncogenes: Genes that promote cell division can be turned on too much, also leading to cancer.
Spreading Cancer: Changes in gene activity can help cancer cells move and invade other parts of the body.
Drug Resistance: Cancer cells can become resistant to treatments due to changes in gene activity.
Impact on Other Diseases
Neurodegenerative Diseases: Changes in gene activity can affect brain cells, contributing to diseases like Alzheimer's.
Heart Diseases: Epigenetic changes can impact genes involved in heart function and lead to conditions like heart attacks.
Autoimmune Diseases: These changes can affect immune cells and contribute to diseases where the immune system attacks the body, like lupus.
Metabolic Disorders: Changes in genes related to metabolism can lead to obesity and diabetes.
what are mutations?
changes in nucleotide sequences
what is a point mutation
What is a Point Mutation?
- Single Nucleotide Change: A point mutation affects just one nucleotide (the building blocks of DNA) in the DNA sequence.
Types of Point Mutations:
1. Substitution: One nucleotide is replaced by another. For example, an adenine (A) might be replaced by a guanine (G).
- Silent Mutation: Doesn't change the amino acid sequence of a protein.
- Missense Mutation: Changes one amino acid in the protein.
- Nonsense Mutation: Creates a stop codon, shortening the protein.
what are frameshift mutations?
Frameshift mutations are genetic mutations caused by insertions or deletions of nucleotides in the DNA sequence that change the reading frame of the genetic code. Here’s a simple explanation:
What are Frameshift Mutations?
Insertion: Adding one or more nucleotides into the DNA sequence.
Deletion: Removing one or more nucleotides from the DNA sequence.
what are all the different types of mutations
Substitution: One nucleotide is replaced by another. For example, an adenine (A) might be replaced by a guanine (G).
Insertion: Adding one or more nucleotides into the DNA sequence.
Deletion: Removing one or more nucleotides from the DNA sequence.
what is a mutagen?
A mutagen is any agent that can cause changes or mutations in the DNA sequence of an organism. These changes can be harmful, beneficial, or neutral, but they alter the genetic material.
list mutagens
Types of Mutagens:
Physical Mutagens:
Radiation: Such as X-rays, gamma rays, and ultraviolet (UV) light. These can break DNA strands or cause incorrect bonding between nucleotides.
Chemical Mutagens:
Base Analogues: Chemicals that resemble DNA bases and can be incorporated into DNA during replication, causing incorrect base pairing.
Alkylating Agents: Chemicals that add alkyl groups to DNA bases, leading to incorrect base pairing or strand breaks.
Intercalating Agents: Chemicals that insert themselves between DNA bases, causing frame shifts during DNA replication.
Biological Mutagens:
Viruses: Some viruses can insert their genetic material into the host DNA, disrupting normal gene function.
Bacteria: Certain bacterial infections can cause chronic inflammation, increasing the rate of mutation.
what are the differences of mutations in germ and somatic cells?
Germ Cell Mutations: Passed on to offspring, affecting future generations. every cell in the organisum carries the mutation.
Somatic Cell Mutations: Affect only the individual in which they occur, contributing to diseases like cancer. the mutation is localised in one area.
how do mutation cause cancer?
Oncogenes Activation: Some mutations can turn normal genes (proto-oncogenes) into oncogenes. Oncogenes promote cell division and growth excessively when mutated, leading to uncontrolled cell proliferation.
Example: A mutation can cause an oncogene to be stuck in the "on" position, continuously signaling cells to divide.
Proto-oncogenes excess: proto-oncogenes are made too many times resulting in multiple proto-oncogenes proteins promoting cell division
Tumor Suppressor Genes Inactivation: Other mutations can inactivate tumor suppressor genes, which normally prevent cells from growing and dividing uncontrollably.
what is PCR
Polymerase Chain Reaction (PCR) is a laboratory technique used to amplify (make many copies of) a specific segment of DNA. It is widely used in various biological and medical applications.
What are the steps of PCR
Steps of PCR
1. Denaturation:
- Purpose: Heating the reaction mixture to around 94-98°C separates the double-stranded DNA template into two single strands.
- How It Works: The high temperature breaks the hydrogen bonds between complementary base pairs, turning the double-stranded DNA into single strands.
2. Annealing:
- Purpose: Cooling the reaction mixture to a temperature typically between 50-65°C allows specific primers to bind (anneal) to the complementary sequences on the single-stranded DNA template.
- How It Works: Primers are short DNA sequences designed to be complementary to the sequences flanking the target DNA region to be amplified.
3. Extension (Elongation):
- Purpose: Raising the temperature to around 72°C activates the DNA polymerase enzyme, called taq polymerase, which synthesizes a new DNA strand complementary to the template strand.
- How It Works: taq polymerase adds nucleotides to the primer in a 5' to 3' direction, extending the primer sequence by using the single-stranded DNA template.
4. Repeat Cycles:
what are the components of PCR and their purpose?
DNA Template: Contains the target DNA sequence to be amplified.
Primers: Bind to the specific sites on the DNA template to provide a starting point for DNA synthesis.
DNA Polymerase: Synthesizes new DNA strands by adding nucleotides to the primers.
Nucleotides (dNTPs): Serve as the building blocks for new DNA strand synthesis.
Buffer Solution: Maintains the optimal conditions for DNA polymerase activity.
MgCl₂: Acts as a cofactor necessary for DNA polymerase function.
what is the difference between electrophoresis and gel electrophoresis
Electrophoresis: A general technique for separating charged particles in various mediums under an electric field.
Gel Electrophoresis: A specific type of electrophoresis using a gel medium for the separation and analysis of DNA, RNA, and proteins based on size and charge.
what are the materials needed for gel electrophoresis
Gel Matrix: Agarose (for DNA/RNA) or polyacrylamide (for proteins).
Electrophoresis Buffer: Maintains pH and ionic strength (e.g., TAE or TBE for DNA, SDS-PAGE buffer for proteins).
Sample: DNA, RNA, or protein.
Loading Dye: Helps to visualize the sample when loading and tracking its progress during electrophoresis.
Molecular Weight Marker/ Ladder: Standard reference to estimate the size of separated molecules.
Electric Power Supply: Provides the electric field.
Staining Solution: Visualizes separated molecules (e.g., ethidium bromide or SYBR Green for DNA, Coomassie Blue or silver stain for proteins).
what is the process of gel electrophoresis?
Preparation: Gel and samples are prepared, and the gel is set up in the tank.
Loading: Samples and molecular weight markers are loaded into the wells.
Electrophoresis: An electric field is applied, causing the molecules to migrate through the gel.
Staining: The gel is stained to visualize the separated bands.
Analysis: Bands are compared to a molecular weight marker to estimate sizes.
what is the purpose of gel electrophoresis?
Separation of Molecules:
DNA/RNA: Gel electrophoresis separates DNA or RNA fragments based on their size. Smaller fragments move faster and further through the gel than larger ones.
Proteins: Proteins are separated based on their size and charge. The gel matrix acts as a sieve, allowing smaller proteins to move more quickly.
Analysis of Genetic Material:
DNA Fingerprinting: Used in forensic science to compare DNA samples from crime scenes with those of suspects.
Genetic Testing: Identifies genetic mutations, diagnoses genetic disorders, and screens for hereditary diseases.
Restriction Fragment Length Polymorphism (RFLP): Analyzes DNA fragments cut by restriction enzymes to study genetic variation.
and more
what size of dna fragments go further in gel electrophoresis?
the shorter the fragment is the further it will go
how do you read a DNA sequencing gel (chain termination method (sanger method))
Understand the Layout:
The gel will typically have four lanes, each corresponding to one of the four nucleotides: A (adenine), T (thymine), C (cytosine), and G (guanine).
Each band in a lane represents a DNA fragment that has terminated at the respective nucleotide.
Visualize the Bands:
The gel is stained and viewed under appropriate conditions to see the bands. Common stains include ethidium bromide or fluorescent dyes.
Bands appear as dark lines on the gel.
Identify the Starting Point:
DNA fragments at the bottom of the gel are the smallest and represent the nucleotides closest to the primer (the start of the sequencing reaction).
Fragments further up the gel are larger and represent nucleotides further from the primer.
Read the Sequence from Bottom to Top:
Start at the bottom of the gel and read upwards, moving from the smallest to the largest fragments.
For each band, note the corresponding lane (A, T, C, or G). This indicates which nucleotide is at that position in the sequence.
what is the chain termination method (sanger method)
Sanger sequencing, also known as the chain termination method, is a technique used to determine the sequence of nucleotides in a DNA strand. It involves synthesizing the DNA strand in the presence of special terminator nucleotides (ddNTPs) that halt the DNA synthesis process when incorporated. These terminated fragments are then separated by size using gel electrophoresis, and the sequence is deduced by analyzing the order of terminated fragments from shortest to longest. This method has been pivotal in genetic research, diagnostics, and understanding genetic variations.
Read this
G A T C
| | | |-------| (C)
| | |-------| | (T)
| |-------| | | (A)
|-------| | | | (G)
| | | |-------| (C)
| | |-------| | (T)
|-------| | | | (G)
| | | |-------| (C)
| | |-------| | (T)
what is the sequence?
The sequence is: C-T-G-C-T-G-A-T-C
what is the whole-genome shotgun approach
The whole-genome shotgun sequencing approach is a method used to sequence entire genomes by breaking DNA into small fragments, sequencing them, and then assembling the genome computationally.
what is the technique used to create electropherograms
electrophoresis
what are short tandem repeats (STRs)
DNA sequences consisting of 2-6 base pairs repeated multiple times in a row.
what does 6,9, mean in a STR dna profile
6 repeats on the maternal chromosome, 9 on the fraternal
maternal always goes first
what does 12 mean in a STR dna profile
they have 12 repeats on both the maternal and fraternal sides. this can also be expressed as 12,12,
what is the difference when gel electrophoresis is used for DNA sequencing and str analysis
Key Differences:
Goal:
DNA Sequencing: To determine the precise order of nucleotides in a DNA segment.
STR Analysis: To compare the lengths of specific DNA regions containing short tandem repeats.
Fragment Generation:
DNA Sequencing: Uses chain-terminating nucleotides to create fragments of every possible length from the primer to the end of the sequence.
STR Analysis: Uses PCR to amplify specific repeat regions, resulting in fragments that vary in length based on the number of repeats.
Reading the Gel:
DNA Sequencing: Reads the sequence of nucleotides based on the position of the bands (each band represents a terminated fragment at a specific nucleotide).
STR Analysis: Measures the length of amplified fragments to determine the number of repeats in each STR region.
how can STR DNA profiles be presented
data table or a gel or electropherogram
gel is used when gel electrophoresis is used
electropherogram is used when capilary gel electrophoresis is used
what is capilary gel electrophoresis
Capillary gel electrophoresis (CGE) is an advanced analytical technique used to separate and analyze DNA, RNA, and proteins. It utilizes thin capillary tubes filled with a gel matrix, through which an electric field is applied to move molecules based on their size and charge. This method provides high resolution, faster analysis, and greater automation compared to traditional gel electrophoresis. CGE requires only small sample sizes and produces precise quantitative data, making it ideal for applications in DNA sequencing, forensic analysis, genetic studies, and protein analysis. The results are detected and visualized as an electropherogram, allowing for accurate identification and quantification of the separated molecules.
what are single nucleotide polymorphism (SNP)
SNPs are single base pair changes in the DNA sequence that are common in the population. They can occur in both coding and non-coding regions of the genome and can have neutral, harmful, or beneficial effects. SNPs are valuable in genetic research, medical research, personalized medicine, and forensics due to their role in genetic variation and disease susceptibility.
how do you read SNPs
Example of Reading an SNP:
Reference Sequence: ATCGGA
Sample Sequence: ATCAGA
SNP Identification: The fourth nucleotide (G) in the reference sequence is replaced with an A in the sample sequence. This indicates the presence of an SNP at this position.
what is genetic engineering
Genetic engineering is the direct manipulation of an organism's genes using biotechnology. It involves modifying the genetic material of an organism to achieve desired traits or outcomes.
the basis of it is that dna is the same in all living things so dna from one living thing can be transcribed and translated in another
what is dna cloning
DNA cloning is the process of creating multiple identical copies of a specific segment of DNA. This technique is fundamental in molecular biology for studying genes, producing proteins, and manipulating genetic material.
what is recombinant dna
Recombinant DNA (rDNA) refers to artificially created DNA sequences that combine genetic material from different sources, often different species, into a single molecule.
what is gene cloning
where a recombinant cell is replicated many times in which the split dna is carried on
what is a vector
In molecular biology, a vector refers to a DNA molecule used as a vehicle to transfer foreign genetic material into a target cell, typically for the purpose of gene cloning, gene expression, or genetic modification. Vectors are crucial tools in genetic engineering and biotechnology.
what are the types of vectors
Plasmids: Small, circular DNA molecules found in bacteria that replicate independently of the bacterial chromosome.
Viral Vectors: Derived from viruses modified to carry foreign DNA and deliver it into host cells. Examples include retroviruses and adenoviruses.
Bacterial Artificial Chromosomes (BACs): Large DNA constructs used for cloning large DNA fragments, including entire genes or gene clusters.
what is a resrtiction enzyme
A restriction enzyme, also known as a restriction endonuclease, is a type of enzyme that cuts DNA molecules at specific nucleotide sequences, known as recognition sites or restriction sites.
what is a restriction site
A restriction site, also known as a recognition site, is a specific sequence of nucleotides in a DNA molecule that is recognized and cut by a restriction enzyme.
what is a feature of restriction sites that invole the 5’3 and 3’5 strands
Restriction sites are often palindromic, meaning they read the same backward and forward on complementary DNA strands. For example, if a restriction enzyme recognizes the sequence 5'-GAATTC-3', its complementary sequence on the opposite strand is also 5'-GAATTC-3'.
what are sticky ends
Sticky ends are single-stranded overhangs that result from the action of certain restriction enzymes when they cut DNA molecules at specific sequences known as restriction sites.
what are blunt ends
Blunt ends refer to DNA fragments that have been cut by restriction enzymes in such a way that both strands of the DNA molecule are cleaved at the same position, resulting in no overhanging single-stranded regions.
what is a probe and what does it do
In molecular biology and genetics, a probe refers to a small, single-stranded DNA or RNA molecule that is used to detect complementary sequences in a sample of DNA or RNA.
Probes are designed to bind specifically to a complementary sequence of nucleotides in a target DNA or RNA molecule. They are used to identify or "probe" for the presence of specific sequences of interest in a biological sample.
what is electroporation
Electroporation is a laboratory technique used to introduce DNA, RNA, or other molecules into cells by applying a brief electric pulse to create temporary pores in the cell membrane.
Membrane Permeabilization: The electric field disrupts the lipid bilayer of the cell membrane, creating temporary pores or channels.
what is microinjection
Microinjection is a technique used in molecular biology and genetics to introduce materials such as DNA, RNA, proteins, or other substances directly into individual cells using a fine needle or pipette.
Procedure:
Needle/Pipette: A very fine needle or glass pipette is used to physically inject materials into the cytoplasm or nucleus of a single cell.
Micromanipulator: The needle or pipette is controlled with precision using a micromanipulator, which allows for accurate targeting of the cell and injection site.
what is tranformation
Transformation in molecular biology refers to the process by which foreign DNA is introduced into a host organism, such as bacteria or yeast, resulting in the uptake and incorporation of this DNA into the host's genome.
what is a virus vector
A virus vector, also known as a viral vector, is a virus that has been genetically modified to deliver foreign genetic material (such as DNA or RNA) into a host cell's genome
what is a phage (bacteriophage)
A bacteriophage, often referred to simply as a phage, is a virus that infects and replicates within bacteria.
process of crispr
CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats) is a revolutionary gene-editing technology that allows scientists to make precise changes to DNA within cells. Here’s a simplified explanation of the CRISPR process:
CRISPR Process:
Identifying the Target Sequence:
CRISPR RNA (crRNA): A short RNA sequence that is designed to be complementary to the target DNA sequence where editing is desired.
TracrRNA (Trans-activating crRNA): A trans-activating RNA that forms a complex with crRNA to guide the Cas enzyme to the target DNA.
Cas Protein:
CRISPR-associated Protein (Cas): A protein, such as Cas9, that acts as a molecular scissors capable of cutting DNA at specific locations guided by the crRNA.
Cas9: One of the most commonly used Cas proteins in CRISPR technology, derived from bacteria like Streptococcus pyogenes.
Guide RNA (gRNA):
crRNA and tracrRNA Fusion: In many applications, crRNA and tracrRNA are fused together to form a single guide RNA (gRNA), simplifying the delivery of the targeting sequence.
DNA Cleavage:
Binding: The gRNA guides the Cas protein to the complementary target sequence on the DNA.
Cutting: Cas protein introduces a double-stranded break (DSB) at the specific site, disrupting the DNA sequence.
Repair Mechanisms:
Non-homologous End Joining (NHEJ): Cells may repair the DNA break by ligating the ends together, often resulting in small insertions or deletions (indels) that can disrupt gene function.
Homology-directed Repair (HDR): Researchers can provide a DNA template along with CRISPR components, allowing precise edits to be made by using the cell’s HDR machinery.