Unit 8: DNA Replication and Gene Expression
1/41
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
42 Terms
The Role of DNA
DNA Replication
Transcription/Translation
The effects of mutations
How do we know that DNA is the hereditary molecule
The process by which cells use hereditary information stored in DNA is very elegant
DNA must be retained intact, yet copied to make new cells (DNA replication)
It must be turned into working copies in the form of mRNA (Transcription)
mRNA molecules must be read and decoded to form the enzymes/structural proteins of the cell (Translation)
System must have ability to deal with damage (DNA repair)
The Griffiths Experiment
Some genetic factor can be transformed between S and R bacteria
Took heat killed infectious bacteria that were lysing and mixed it with live noninfectious bacteria and inserted it into the mouse. The mouse then died
Avery, MacLeod, McCarthy Experiment
Attempted to determine if it was DNA, RNA, or proteins that were transferrable from the Griffith’s experiment
Used mixtures obtained from S strains (infectious) but digested away the other components (proteases for proteins, RNAase for RNA, DNAase for DNA)
Only the mixture with DNA left could still transform R cells into S cells indicating that was the molecule responsible
Hershey-Chase Experiment
Used Radioactive labeling of either proteins or DNA in bacteriophages
They let the labeled phages infect bacterial cells, then determined where the tag ended up for each set up
Only the labeled phage DNA went into the bacterial cells further proving that DNA was the hereditary molecule in these experiments
DNA Replication is Semiconservative
Each time the dsDNA is copied, each copy carries one strand of the original molecule and newly made strand
Structure of DNA
Each nucleotide building block of DNA consists of a five carbon sugar (2-deoxy/ribose), a phosphate group attached to carbon 5 of the sugar, a nitrogenous base attached to carbon 1
Complementary
Phosphodiester covalent bonds form the sugar/phosphate backbone of each strand
Higher GC content more stable the DNA, organisms with low GC content have mechanisms to deal with stability
How DNA is packed
Bacteria: A single circular chromosome, has plasmids, histone like proteins but not homologous
Archaea: A single circular chromosome packaged around histone proteins, has plasmids
Eukarya: Multiple linear chromosomes packaged around histone proteins, rarely plasmids
Histones
The wrapping of dsDNA
Helps compact the very large chromosome structures of eukaryotic cells
Bacterial Exception
D. radiodurans may possess 4 to 10 copies of its genome
The stacked genome copies may help give this bacterium its strong radiation-damage-resistance trait
If one of the copies is mutated the others will take over
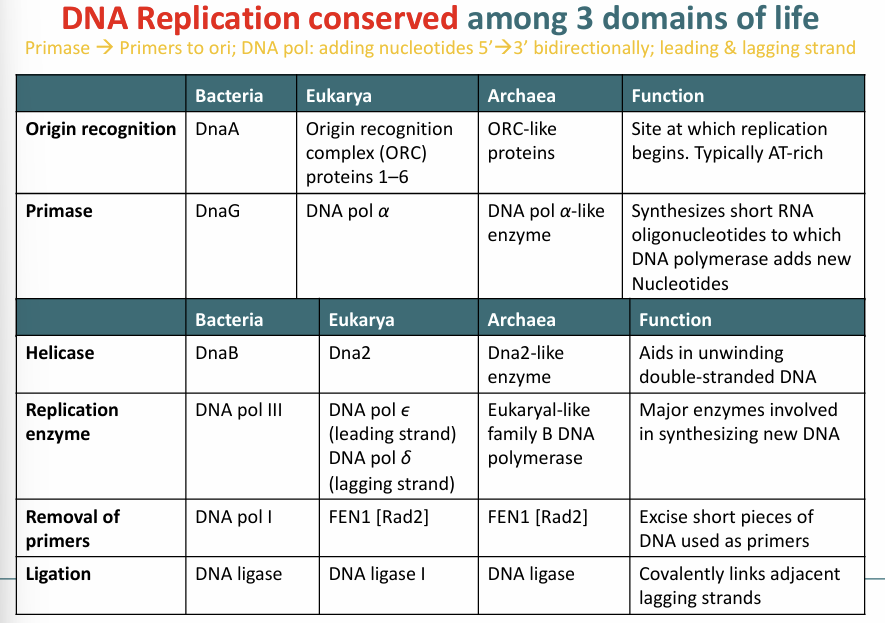
Bacteria DNA Replication: Initiation
DNAa protein binds to 9-bp repeats of oriC and subsequently w/13-bp repeat this separates the strands (particular part where DNA replication starts)
DNAb (helicase) is recruited with DNAc (helicase loader) for further unwinding
DNAg (primase) is recruited to lay initial complementary RNA primers needed for DNA polymerase to work (ESSENTIAL STEP FOR DNA REPLICATION)
Single stranded DNA binding proteins are recruited to help keep the DNA unwound
This all forms the replication fork/bubble
Eukaryotic DNA Replication: Initiation
Multiple origins of replication on each chromosome
Needs multiple enzymes to initiate and create the replication fork
Chromosomes are much larger this causes multiple origins of replication (every 10-100,000 bp)
ARS: Autonomously replicating sequence forms binding site for initiator ORC
ORC recruits additional proteins to unwind DNA
Archaea are very similar to Eukaryotes
Bacteria DNA Replication: Elongation
Occurs bidirectionally (helicases continue unwinding DNA)
Production of new DNA strands complementary to old strands catalyzed by DNA polymerase
Primase RNA polymerase synthesizes short single stranded primer to start the strand (This must be removed later)
Once replication fork forms DNA polymerase III adds nucleotides to the 3’ OH group of initial RNA primers (5’→3’) using DNA as a template
A continuous leading strand and a discontinuous lagging strand both synthesized at the same time
One molecule of double stranded DNA leads to 2 daughter dsDNA
Okazaki Fragments
Formed on the lagging stand
Strand can only be synthesized 5’ to 3’ so opposite strand must synthesize in fragments
DNA pol I removes the RNA primers by endonuclease activity and fills gaps with new nucleotides
Fragments are later joined together by DNA ligase (5’ phosphate and 3’ OH sugar backbone)
Eukaryotes DNA Replication: Elongation
Replication is highly conserved
It is largely the same in bacteria and Eukarya
Bacteria DNA Replication: Termination
Termination of DNA replication mediated by ter sites on opposite side of oriC (DNA polymerase makes its way bidirectional to ter sites)
Tus proteins bind to tern sites and stops the forward process of one of the replication forks
Two halves of newly synthesized DNA are joined when other replication for arrives causing two identical circular dsDNA molecules
Topoisomerase II forms transient double strand break then disentanglement (then pushed towards middle by parM)
DNA Replication Overview
DNA structure clearly determines the mechanism of replication
Allows the production of two new molecules identical to original
Transcription
The conversion of dsDNA into ssRNA
Has specific initiation, elongation, and termination processes
Gene
A segment of DNA that gets transcribed into +ssRNA
Associated DNA regions necessary for transcription
Differences between RNA and DNA
Ribose in RNA, Deoxyribose in DNA
Uracil in RNA, Thymine in DNA
The RNAs
Different forms of RNA and they each serve a different purpose
mRNA: 500-10,000 nucleotides, coding molecule translated into proteins (template for translation)
tRNAs: 75-100 nucleotides, Involved in translation charged with amino acids (not translated but participate in translation)
rRNA: 1500-1900 (small unit) 2900-4700 (large)
miRNA: less than 100 nucleotides functions to regulate gene expression (not translated but participate in translation)
Transcription Initiation Bacteria
Must identify the beginning and end of the segment
Starts at a promoter where DNA dependent RNA polymerase binds to promotor and separates the DNA and synthesizes complementary strand
RNA Polymerase reads the template DNA 5’ to 3’ but does not need a primer
RNA is the same sequence but there is U not T
RNA Polymerase Bacteria
Consists of 5 different polypeptide chains, alpha, beta, beta’, omega, and sigma
Sigma factor bound to RNA polymerase core enzyme and directs the combined holoenzyme to a promoter (this is very important in order for RNA polymerase to bind and the disassociation causes the DNA to unwind)
The better the binding the better the transcription and the more mRNA produced
How does Transcription begin Bacteria
At a consensus sequence at positions -10 (Pribow-Box or TATA box) and -35 in bacteria
Differences in the sequence dictates the affinity of the sigma factor
Eukarya Transcription: Initiation and elongation
Resembles bacteria
However…
Eukarya possesses 3 different RNA polymerases (I, II, III)
RNA pol II transcribes all protein encoding genes (similar to bacteria)
RNA pol I: rRNA genes
RNA pol III: tRNA and miRNA
There are individual transcription factor proteins that must associate with the promoter regions first
RNA pol is recruited to the transcription factor/DNA complex
Binding initiates the unwinding of DNA and the start of transcription
Archaea Transcription: Initiation and elongation
Not well understood
Possess only one DNA dependent RNA polymerase like bacteria but it looks more like eukaryotic RNA polymerase II
RNA pol does not bind to DNA directly
Transcription factors are homologous to eukaryotes and it directs RNA pol to promotor regions like Eukarya
Identifying DNA Binding Proteins
Can use EMSA (Electrophoretic Mobility Shift Assay)
Many DNA binding proteins like histones, DNAa, Orc, Sigma factors
Band shift indicates DNA protein interactions
Smaller migrates further, larger migrates shorter
How can this be used: You can test if protein binds with DNA after incubation, this would show if there is binding or not
Bacteria Transcription Termination
Rho dependent, a rho factor follows RNA pol and pops it off the DNA when it reaches a termination sequence
Rho independent, genes and GC rich repeat followed by string of adenines forms an RNA hairpin loop structure that causes RNA polymerase to dissociate
Eukarya Transcription Termination
More complex
RNA pol I resembles rho dependent
RNA pol III resembles rho independent
RNA pol II: cleaved by specific endonucleases
Post Translational Modifications
Eukaryotic pre-mRNA (product of RNA polymerase II) modified after transcription in 3 ways
Modified nucleotide (7-methylguanosine) added to 5’ end transcript and 5’ cap added
Poly A tail added by polyA polymerase
Introns (non-protein coding regions in eukaryal pre-mRNA) spliced out and exons are joined
5’ cap and poly A protect against degradation (RNA need to be protected because transcription and translation are separate)
Translation
Information contained in the mRNA molecule is decoded to form proteins
Extremely energy intensive so it is tightly controlled
Ribosomes
Interact with three nucleotides (codons) on mRNA and tRNAs which are charged with amino acids
Each nucleotide triplet matches complementary anticodon on each tRNA, amino acid is determined by the anticodon sequence
Ribosomes read the mRNA sequence 5’ to 3’ and recruits the tRNA
Peptide bonds are then formed between amino acids that are delivered
Structure of tRNA
Cloverleaf structure
Undergo intra molecular base pairing
High degree of hairpin structure which allows for stability
Anticodon must remain unpaired to match mRNA
Highly specific aminoacyl-tRNA-synthetase enzymes achieve charge this charge must be before translation
One tRNA for each amino acid (20 different tRNAs) so there are 20 different genes for tRNA
The Genetic Code
4 bases made in mRNA creates 64 codons
61 codons, 1 Start codon (AUG), 3 Stop codons (UAA, UAG, UGA)
Identical in all organisms
The codon on the mRNA does not need to be exact to match the anticodon
This is known as a wobble (3rd base is not essential)
How does Ribosome know where to start
fMET-tRNA (initiator tRNA) binds to AUG after the Shine-Dalgarno sequence mediated by initiation factor 2 and modified to N-formyl-methionine
First AUG after Shine-Dalgarno box will be the start
Bacteria Translation: Initiation
Initiation depends on interaction between small ribosome subunit (16s) and the Shine-Dalgarno sequence on mRNA
This aligns all the machinery to start
Bacteria Translation: Elongation
Charged tRNA with complementary anticodon enters aminoacyl A site of ribosome
Peptide bonds form in peptide P site
Empty tRNA exits ribosome from exit site (uncharged)
When ribosome reaches a stop codon, release factors cause complex to come apart, releasing new protein for folding and modification
Bacteria and Shine Dalgarno sequences
mRNA is polycistronic (monocistronic in eukaryotes)
Which means it can code for more than one protein
Contain the genetic information for more than one gene however, every single gene has its own Shine-Dalgarno sequence this leads to faster translation as a ribosome can bind to each one
Role of Nuclear Membrane
Because of the lack of a nuclear membrane in bacteria transcription and translation are coupled
Also the lack of introns and PTMs of mRNA make coupling easier
Mutations
All of the following only refer to a mutation in a coding region
Silent: Usually the 3rd AA results in no change
Missense: A change in a codon that results in coding for a different AA
Nonsense: A change that forms a stop codon where is should not be
Frameshifts: insertions or deletions can change how ribosome reads and can drastically alter
Inversion: DNA becomes inverted at the same location
Translocation: DNA segment breaks off and reattaches at different chromosomes
Mutations can be good or bad
If a mutation was in a non-coding region (depending on what it is) would affect the protein
Mutations From Envionment
Chemicals (nitrous acid) can induce mutations by removing amino groups
Ultraviolet light can cause higher than expected mutation rate in DNA by forming thymine dimers
Ionizing Radiation can cause double strand to break in phosphate sugar backbone
Repair of Mutations
DNA pol can proof read
Mismatch repair systems
Photolyase cleaves covalently linked T-T (thymine dimer)
Alkyl transferase can remove methyl groups adding to bases by DNA modifying agents
DNA glycosylases recognize damaged bases and initiates excisions