Dunn M&C - A bunch of terms for the final.
1/1026
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
1027 Terms
kinase
transfers a phosphate from ATP to something else.
use of phosphatases
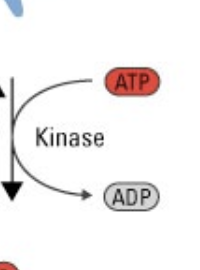
phosphorylation
a reversible post-translational modification that regulates protein function
causes conformational change in proteins that either activate (top) or inactivate (bottom) the protein’s function
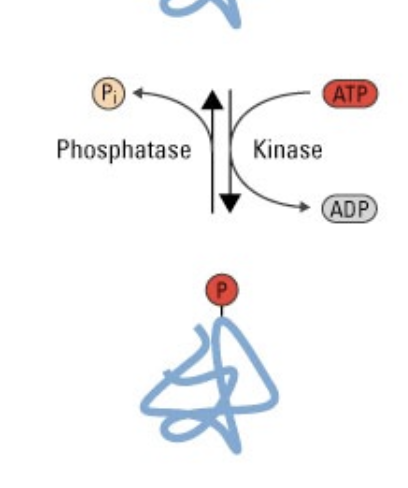
phosphatase
an enzyme that removes phosphate groups from molecules.
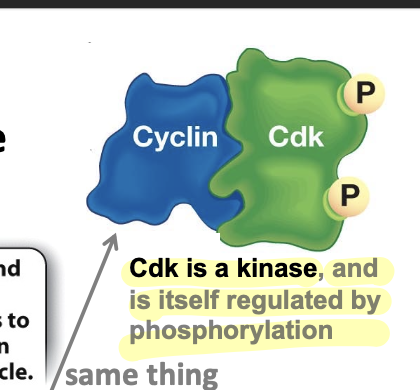
cyclin-Cdk complex
regulates the cell cycles and drives each phase of the cycle
cyclin concentrations changes, whereas the Cdk component is present at constant levels
when their job is done, cyclin is degraded (so cell cycle can only move in the forward direction)
contains heterodimers of cyclin and cDK
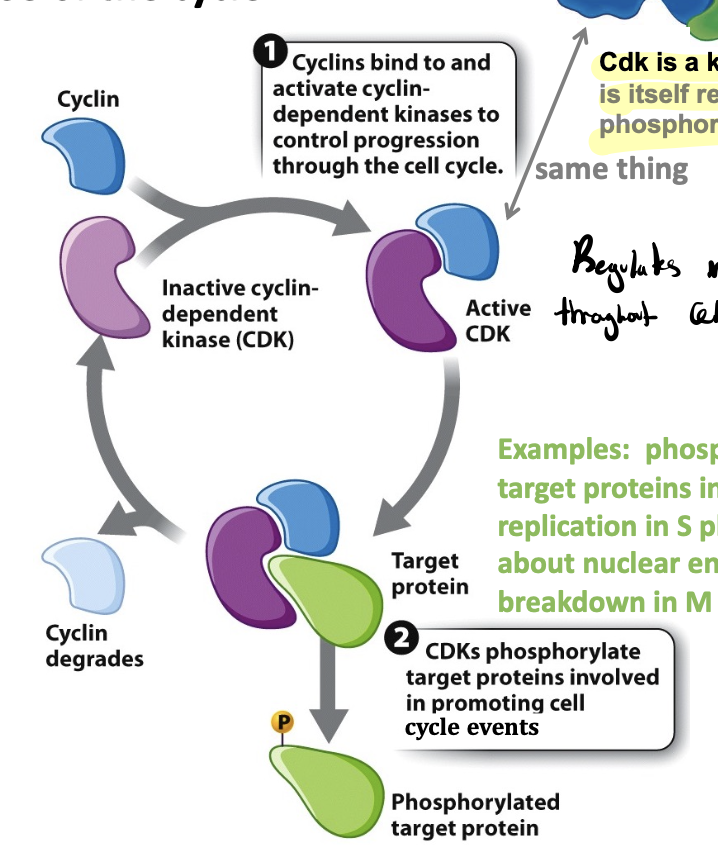
heterodimers
a complex formed by the association of two different protein subunits
e.g, cyclin-Cdk is one, cyclin and Cdk
Cdk
a kinase itself
intercellular signals
communication between cells
usually small soluble molecules that may be protein/nonprotein
intracellular signals
communication inside cells
usually small soluble molecules that may be protein/nonprotein
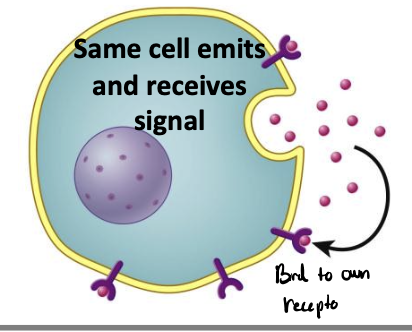
autocrine signaling
same cells emits and receives signal
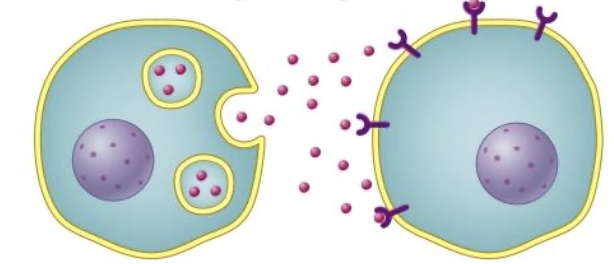
paracrine signaling
signal diffuses only microns away
many signals are peptide “growth factors”
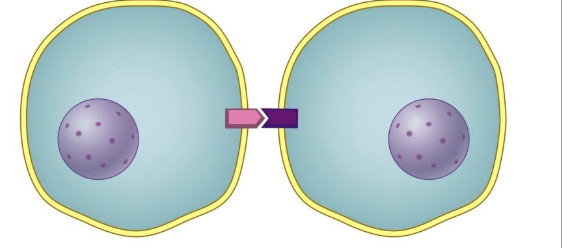
juxtacrine signaling
a.k.a contact-dependent signaling, the cells must be immediate neighbors because the signaling molecule is attached to the signaling cell
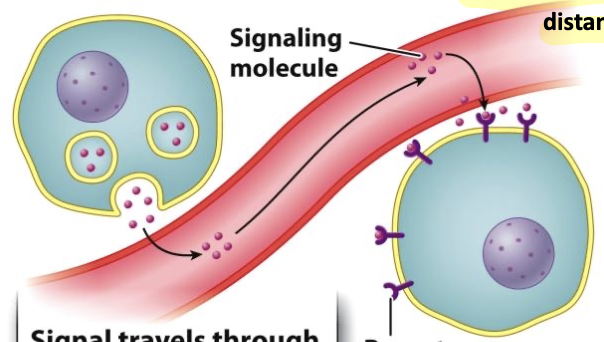
endocrine signaling
signals that diffuse over long distances
e.g, circulatory system through blood stream
receptors
signals are received by cells through these protein complexes
not all cells respond to all signals
a cell must express the receptor for that signal
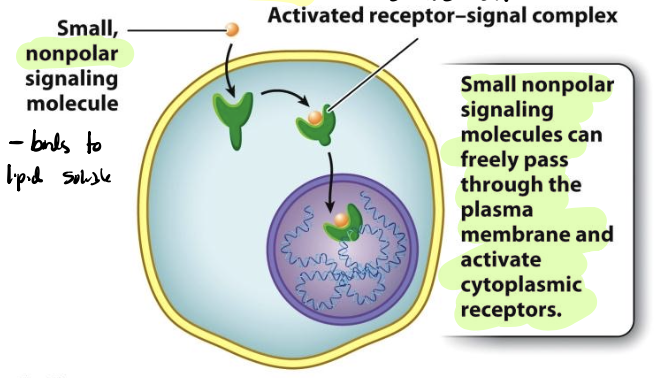
intracellular receptors
receptors residing inside the cell
small, nonpolar signaling molecule passes through the PM and activate cytoplasmic receptors
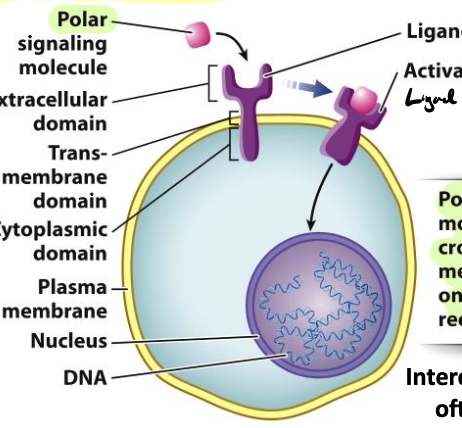
cell-surface receptor
receptor embedded in the plasma membrane
polar signaling molecules cannot pass the PM and rely on cell-surface receptor$
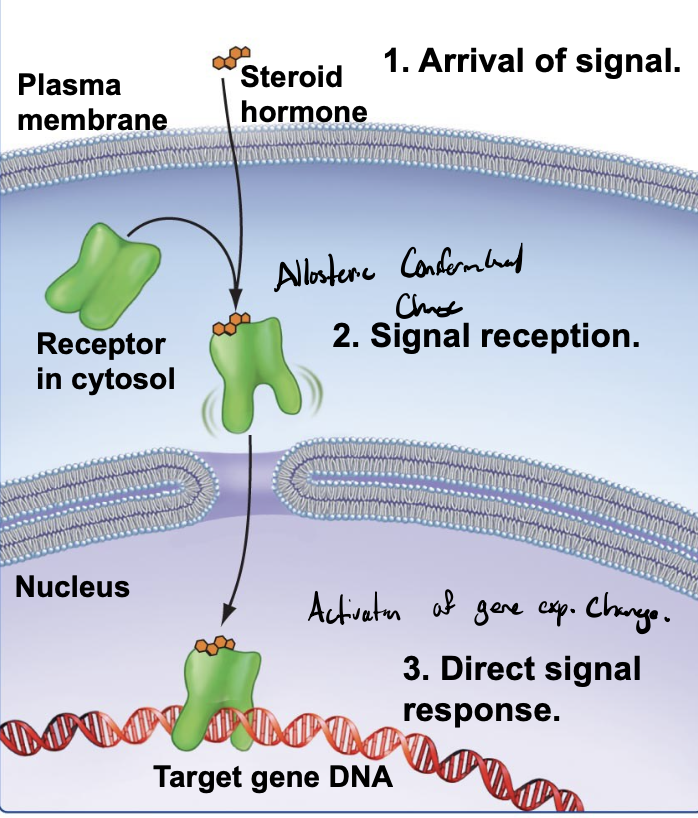
lipid-soluble signals
diffuse directly across the membrane and bind to intracellular receptors
signal directly by inducing a conformational change in an intracellular receptor, bringing a change in cell behavior
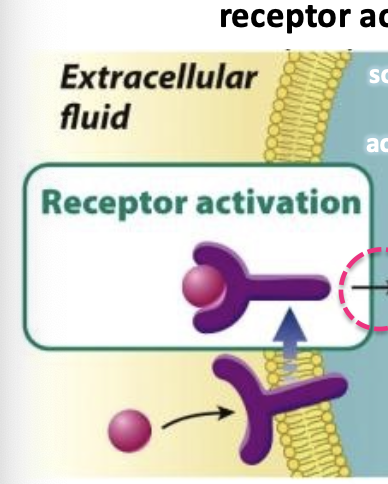
lipid insoluble signals
binds to cell surface receptors that link the external environment to the cell interior
hydrophilic
wont pass through PM
signal
change in gene expression or other cell behavior
signal-transducing receptors
on the surface of cells, these are what intercellular signals usually bind to
signal binds to receptor which is activated by a conformational chage
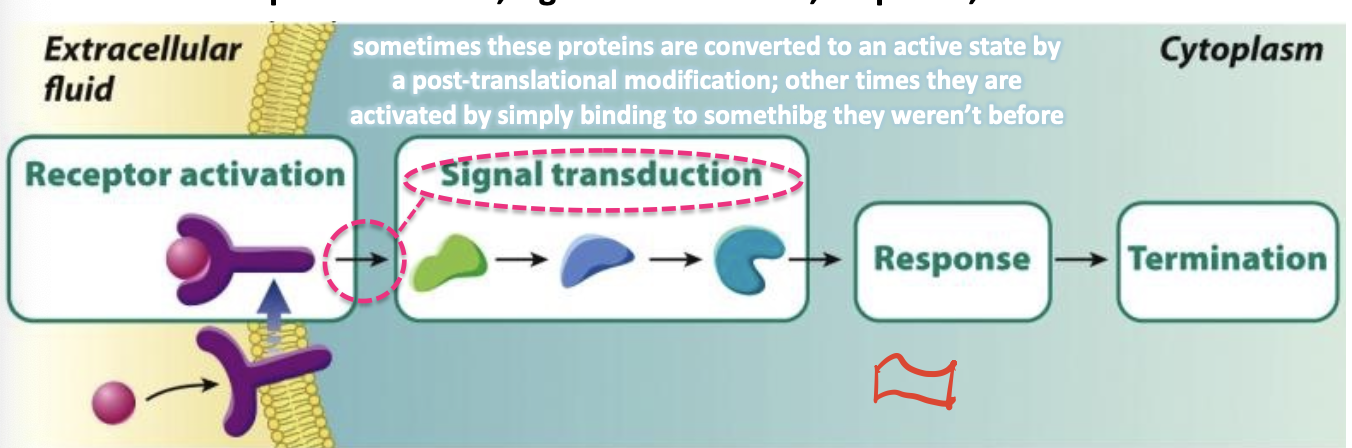
signal transduction
the conversion of the extracellular signal to an intracellular signal
kinase receptors
a second large class of receptors that are used for cell communication
receptor tyrosine kinase: epidermal growth factor
receptor tyrosine kinases (RTKs)
cell surface receptors that undergo ligand-induced dimerization
control a more limited set of cell behaviors — they usually promote cell proliferation, survival, and tissue growth
activated by ligand-induced dimerization
ligand
extracellular signal
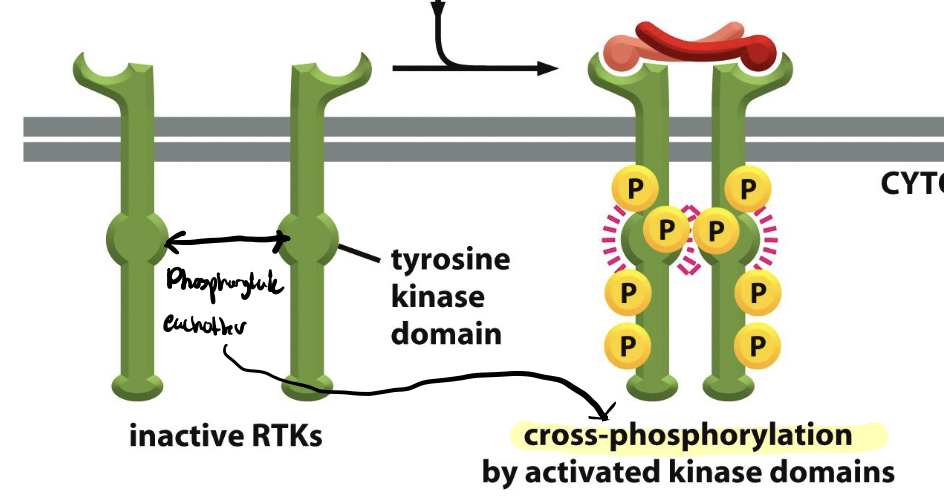
cross-phosphorylation
process where one protein kinase phosphorylates another protein kinase, eachother
two tyrosine domains
normal rtk activation
dimerization
process where two similar or identical molecules (monomers) combine to form a larger unit called a dimer
e.g, kinase domains in RTK activation
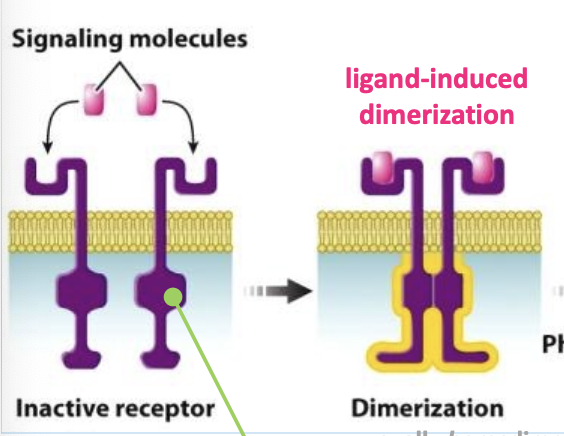
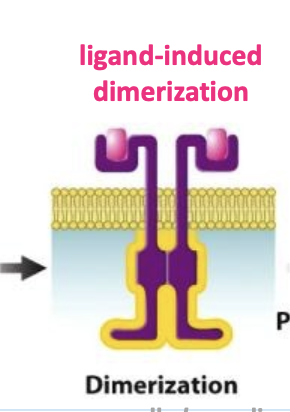
ligand-induced dimerization
process where a ligand triggers the formation of a dimer (two identical or similar molecules joined together) by binding to a receptor protein
kinase domain
a portion of the protein that can remove a phosphate from ATP and transfer it to something else
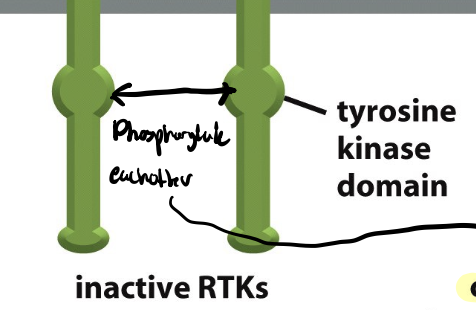
trans-phosphorylation
each member of the receptor pair attaches phosphate groups to the other member
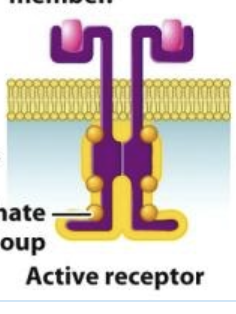
recruitment and activation of cytoplasmic signaling proteins
the attached phosphate groups provide binding sites for intracellular signaling proteins
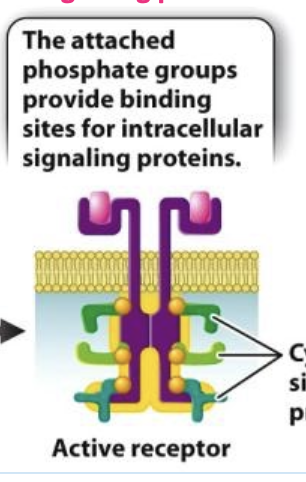
tyrosine side chains
Have a hydroxyl group to which phosphate can be covalently attached;
These phosphotyrosines create new binding sites for cytoplasmic signaling proteins, recruiting them to the membrane
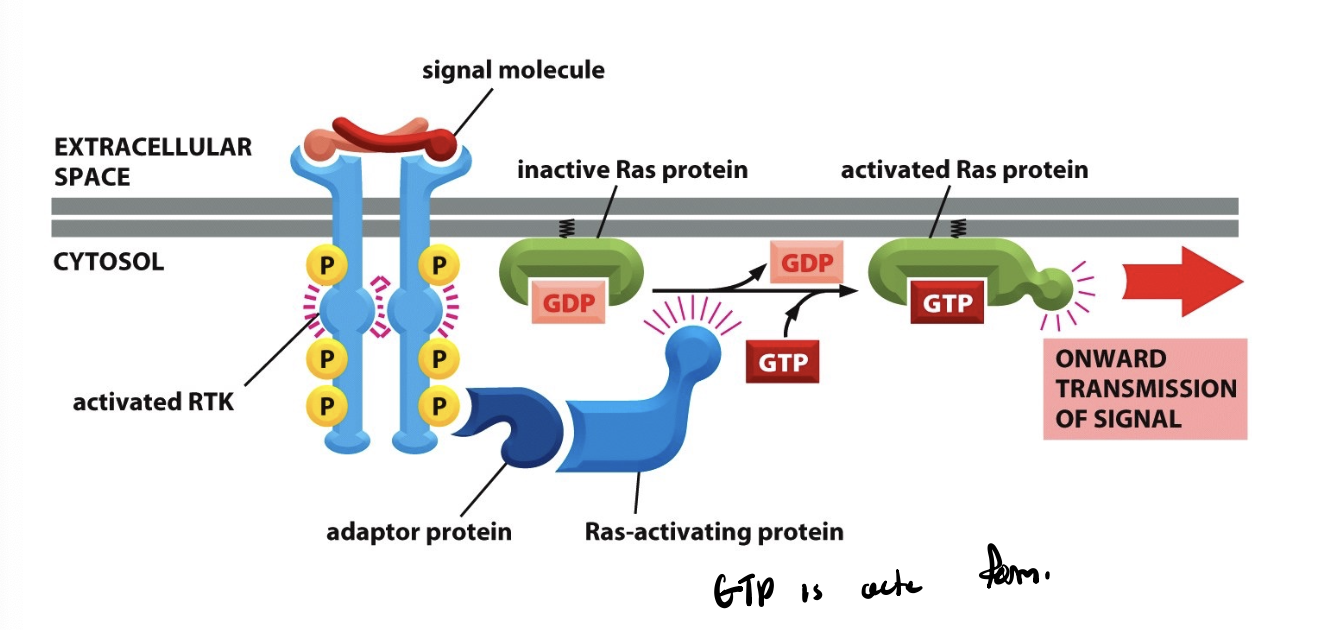
Ras protein
family of small GTPases that play a crucial role in activating the MAP kinase signaling pathway.
RTKs generate intracellular response through Ras
GTPase
family of enzymes that regulate various cellular processes by hydrolyzing guanosine triphosphate (GTP) to guanosine diphosphate (GDP
Ras GTPase
formation of this signaling complexes Ras to exchange its GDP for GTP, rendering it active
kinase cascade
leads to the phosphorylation of target proteins which brings about a change in cell behavior. each kinase phosphorylates and activates the next
activated by activated RAS
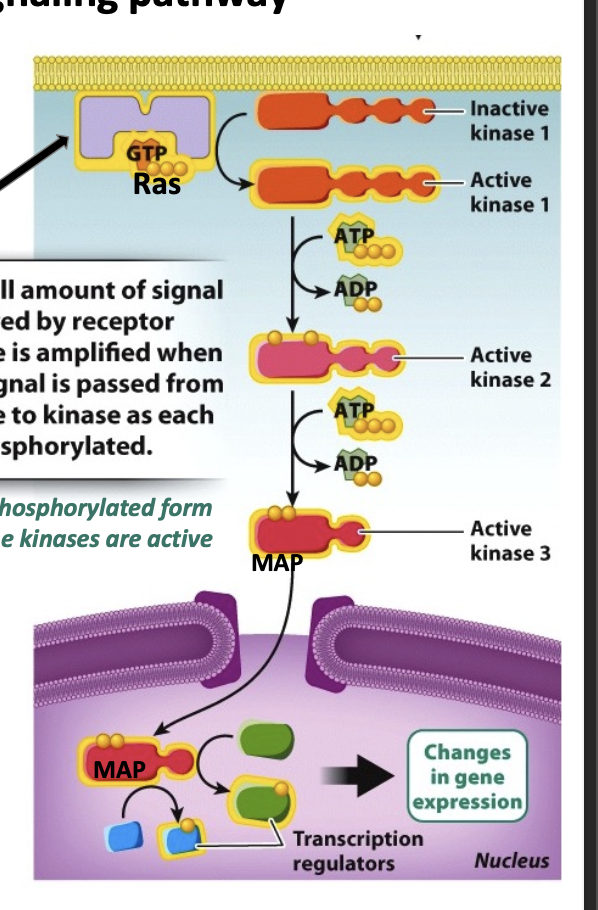
Ras-GTP
activates the kinase/phosphorylation cascade
active
Ras-GDP
inactive
GTP exchange
binding of cytoplasmic signaling proteins leads to stimulation. this exchange leads to a conformational change in the G-protein, activating it and initiating downstream signaling cascade
occurs when a G protein (=GTPase) releases its bound GDP and binds instead to a GTP
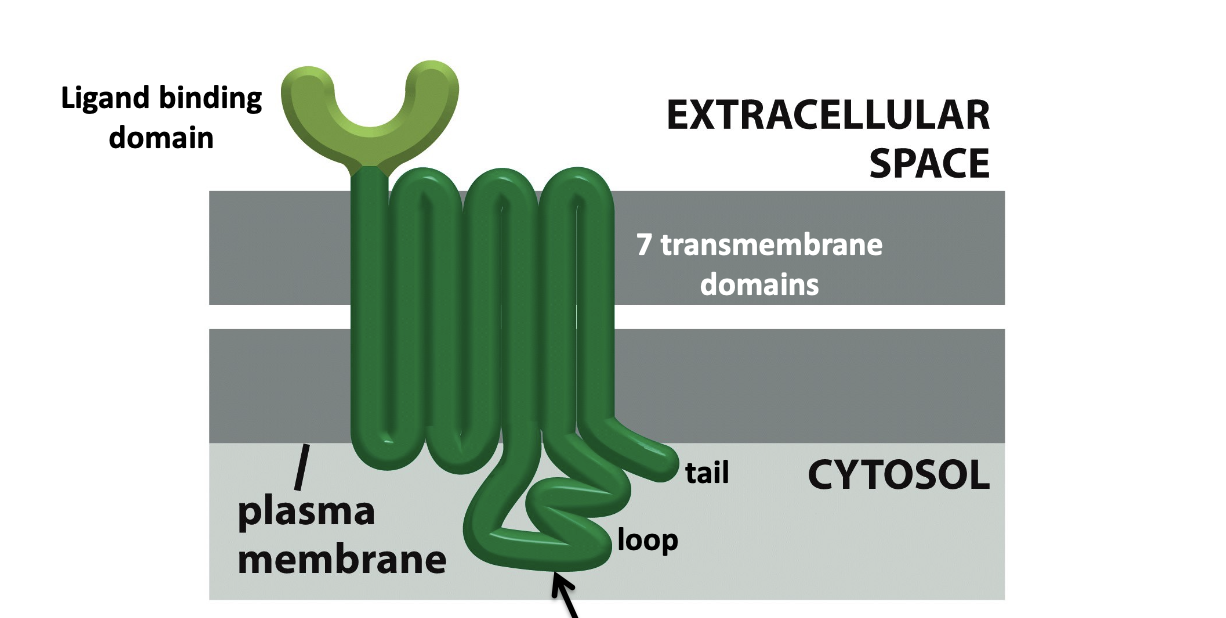
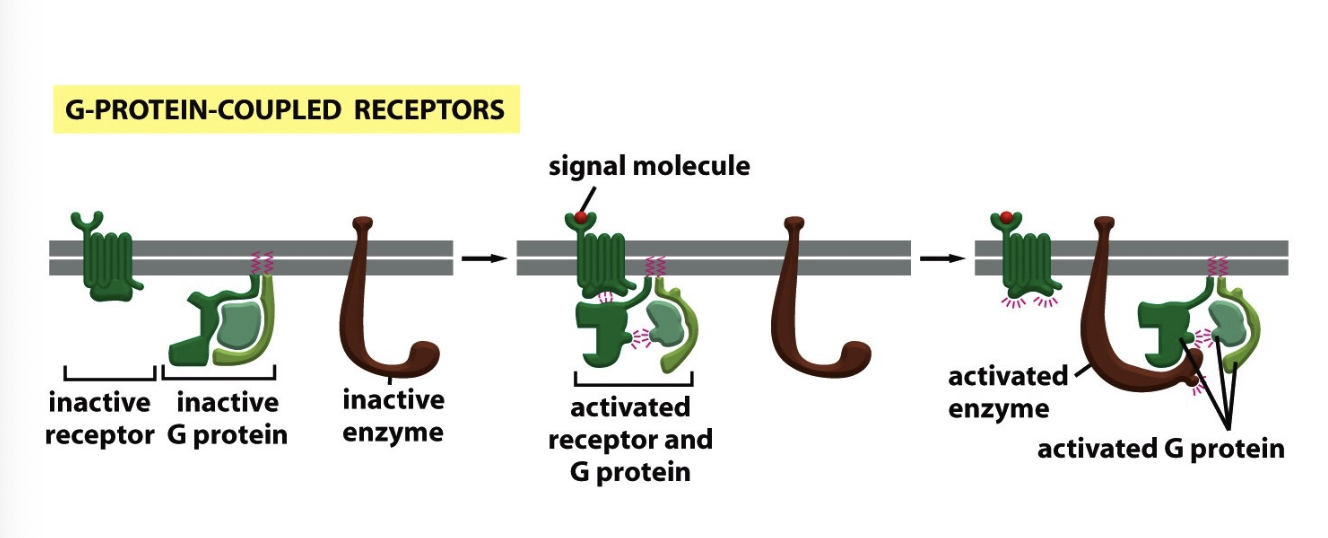
G protein-coupled receptors (GPCRs)
huge class of signaling receptors that share a common structure and mechanism of signal transduction
G protein binds to the cytosolic tail or loop
induces g protein to replace GDP with GTP (GTP exchange)
active version often change level of second messenger
G proteins
activated when a ligand binds to a G protein-coupled receptor (GPCR), triggering a conformational change in the receptor that in turn causes the G protein to release GDP and bind GTP
associated with membrane by lipid anchors
peripheral membrane proteins
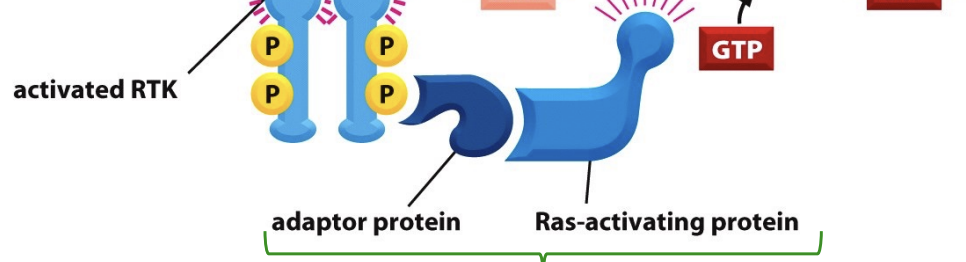
cytoplasmic signaling proteins
adaptor protein and ras activating proteins
second messenger
small intracellular signaling molecules that transmit signals from the cell surface to the inside of the cell, mediating the effects of first messengers
temporarily increases or decreases in response to an extracellular signal.
its small, diffuses rapidly and helps to relay the signal to other signaling proteins inside the cell
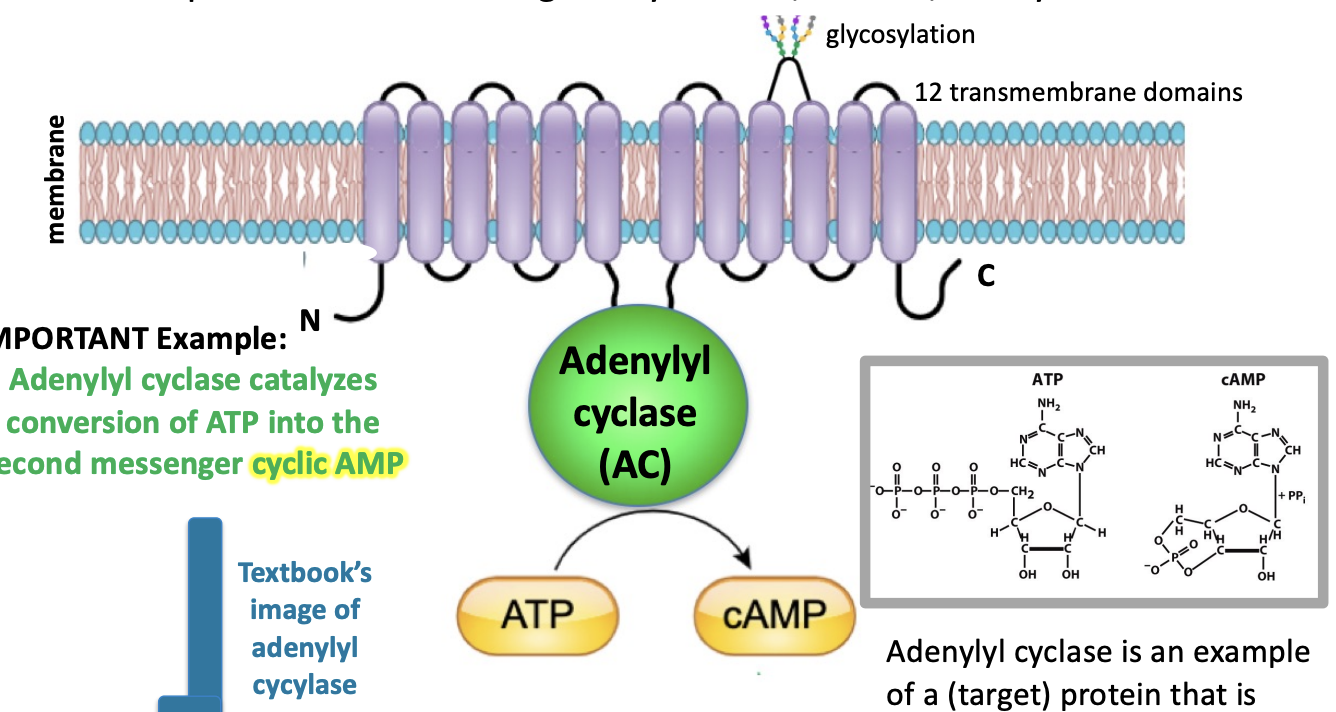
adenylyl cyclase
catalyzes conversion of ATP into second messenger cyclic AMP (cAMP)
GPCR amplification
occurs when one molecule in a relay leads to multiple active molecules
signal amplification
allows a cell to be (sometimes exquisitely) sensitive to incredibly small concentrations of signaling molecules
occurs when one ligand induces many copies of an intracellular signal. this can be accomplished by the activation of an enzyme
signal transduction
the conversion of the extracellular signal to an intracellular signal
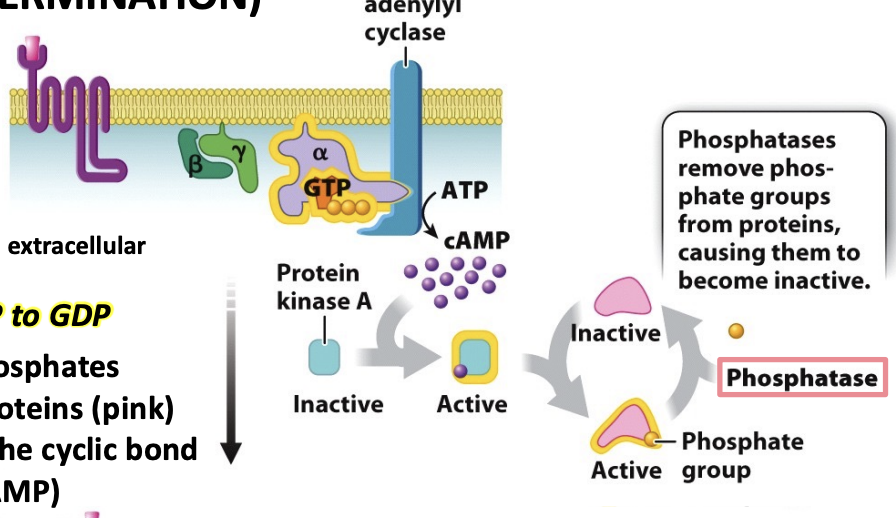
signal termination
signal goes away
G protein hydrolyzes GTP to GDP
phosphatases remove phosphates from activated targer proteins (pink)
Phosphodiesterases cut the cyclic bond in cyclic AMP (to yield just AMP, which is inactive)
binding affinity
The amount of time a signaling molecule remains bound to its receptor depends on how tightly the receptor holds on to it
GPCR functions
changing the intracellular concentration of a second messenger or by altering the activity of the ion channels, which changes the membrane potential
membrane potential
charge differential across plasma membrane
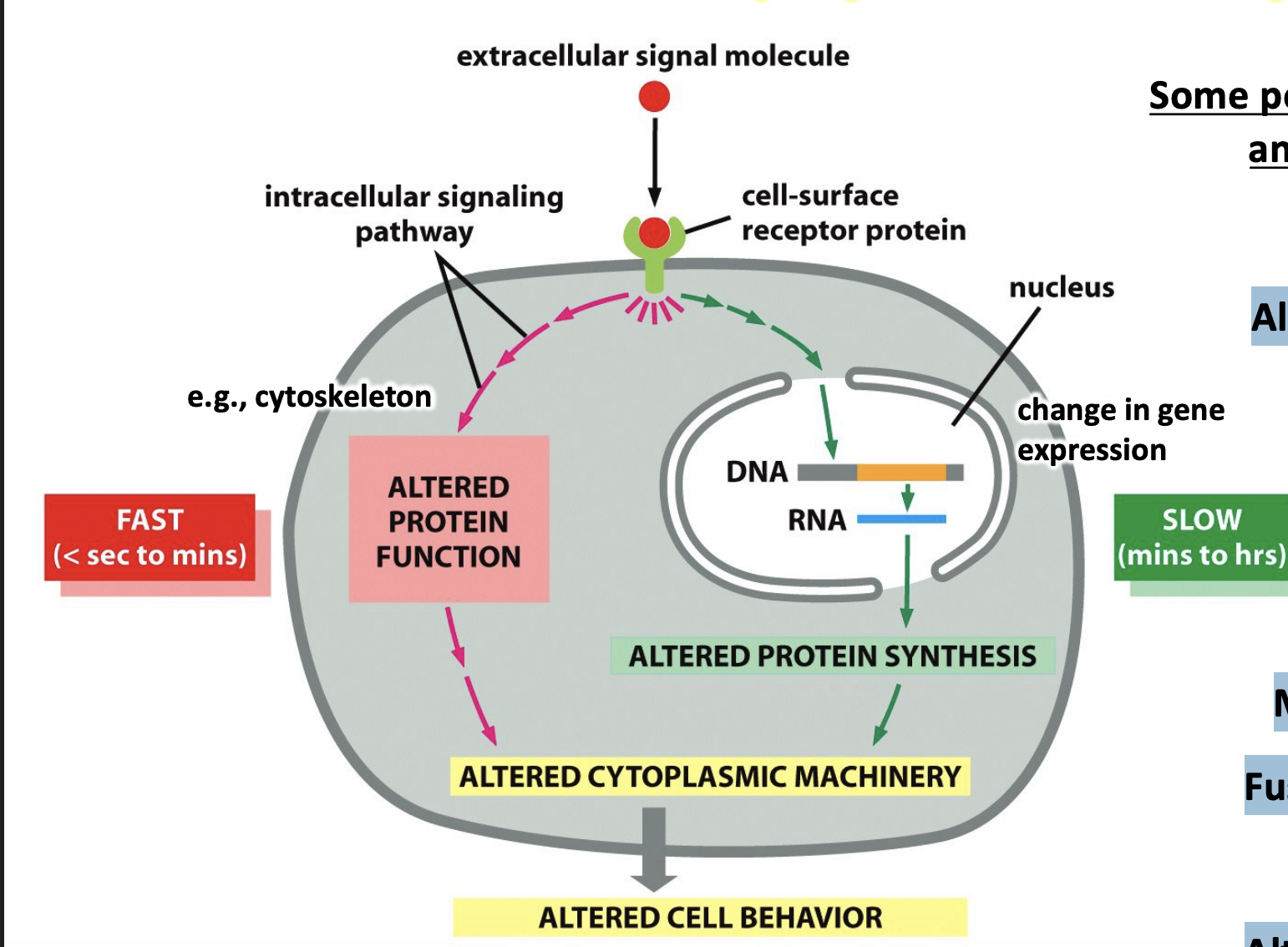
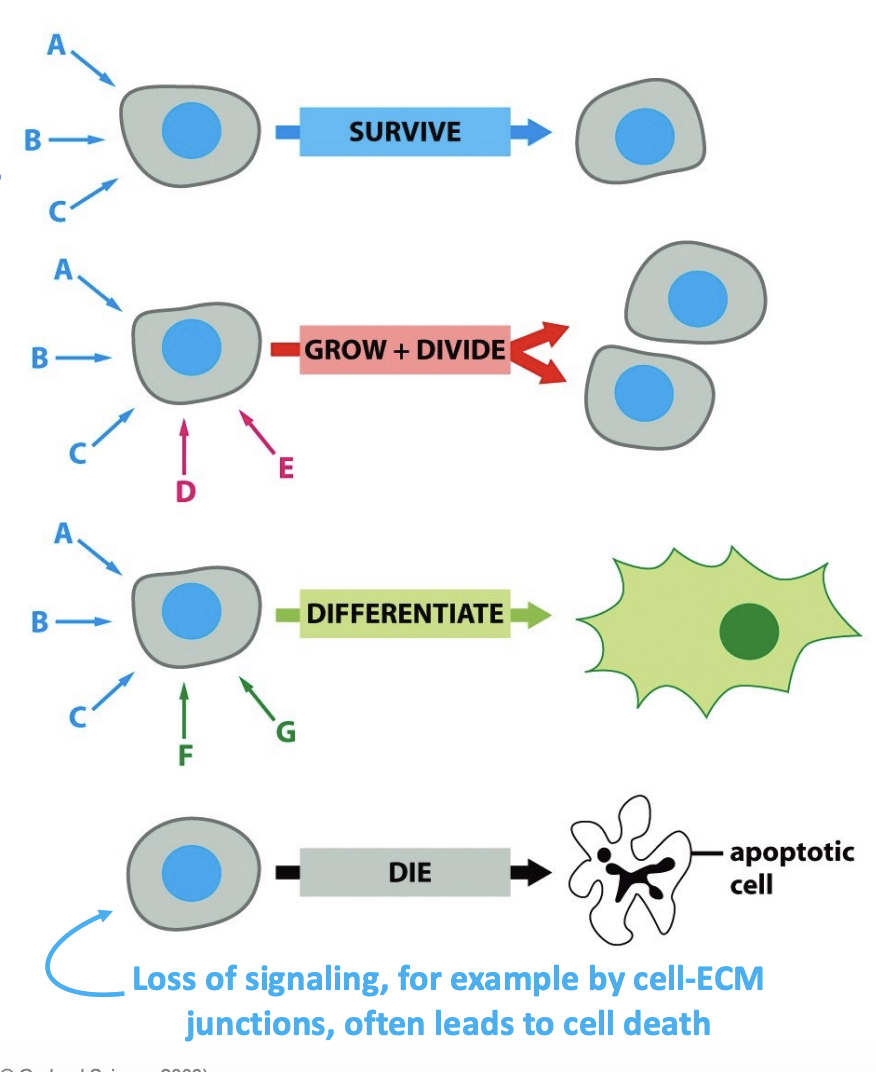
Cellular Response w/ speed & quality
Responses to a signal may vary according to the cell type because each cell only expresses a subset of receptors
e.g, Alter metabolism, Alter gene expression, Survive/die, Move/change shape, “Fire”, Proliferate (enter S phase), Alter ion permeability, Secrete something, Fuse with another cell
signal integration
information processing in the brain: multiple variables determine the outcome
cells usually integrate the signals they receive
combinatorial control and contributes to regulation of gene
expression
phosphodiesterases
cuts cyclic bond in cyclic AMP (cAMP) to yield just AMP, which stops the phosphorylation and activation of target proteins
protein functions
structural support
membrane channels (control the movement of ions and molecules across cell membranes)
act as enzymes
signaling (function as receptors, hormones, or intracellular messengers to regulate cellular activities)
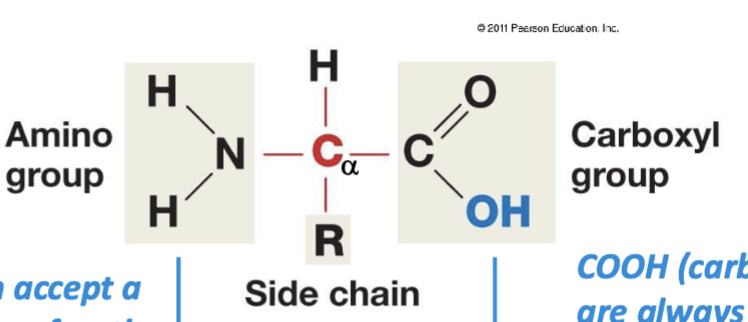
amino functional group
-NH2 — typically acts as a base, so it can collect a proton (H+). And when it collects a proton, it gains a full + sign.
carboxyl functional group
-COOH — typically acts as an acid, so it can donate a proton. And when it donates a proton, it gains a full - sign.
hydroxyl functional group
-OH; highly polar, so makes compounds more soluble through H-bonding with water; doesn’t dissociate or ionize in physiological conditions
phosphate functional group
several phosphate groups linked together, breaking these O-P bonds between them releases large amounts of energy
sulfhydryl functional group
when present in proteins, can form disulfide (S-S) bonds that contribute to protein structure
amino acid structure
N-C-C: amino group, a carbon (connected to H and R-Group/side chain), and carboxyl group. tetrahedral arrangement
side chains
also known as R-group, are the most important to the functional aspects of protein; gives amino acids its unique character
central carbon
also known as the alpha (α) carbon, is present in amino acids.
non-ionized form of amino acids
represented in solid state/neutral structure
ionized form of amino acids
represented in cells/aqueous state: N will often accept a proton as a fourth covalent bond, and carboxyl group are always willing to give up a proton at a physiological pH
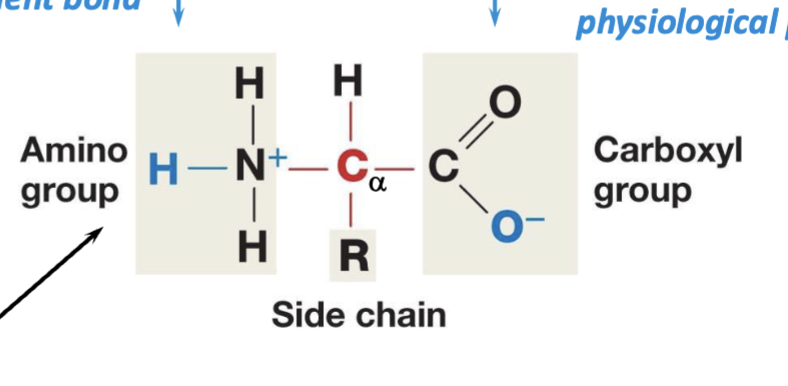
zwitterion
a molecule that has both a positive charge and a negative charge, but an overall neutral charge
amino acids types
there are 20 common amino acids; they are grouped based on how well they interact with water (philic or phobic), or whether they are basic or acid or polar uncharged.
Alanine
common amino acid; has a methyl group
Methionine
common amino acid; not too polar
Phenylalanine
common amino acid; bulky and hydrophobic with a carbon ring
glycine (gly, G)
a special amino acid; gives the amino acid more flexibility especially in the polypeptide backbone, the side chain is just a hydrogen group (H)
proline (pro, P)
a special amino acid; present in polypeptides and it can constrain how a polypeptide chain forms due to the restriction of rotation of the C-N backbone
cysteine (cys, C)
a special amino acid; the only amino acid with a sulfhydryl group, two of these can form disulfide bonds, forming cross bridges that connect different parts of the same protein or different proteins
amino acid w/ hydrophilic side chains
amino acids with polar side chains have a perm. partial charge. they are hydrophilic and tend to form hydrogen bonds w/ one another or water molecules. basic & acidic amino acids are strongly polar and hydrophilic. these basic (+) and acids (-) are charged and these charged groups can form ionic bonds with one another and other charged molecules.
process of creating peptide bonds between amino acids
the carboxyl group of one amino acid reacts with the amino group of another amino acid, and dehydration synthesis/condensation occurs (release of a water molecule). this is catalyzed in the large subunit of the ribosome. ** Practice drawing these
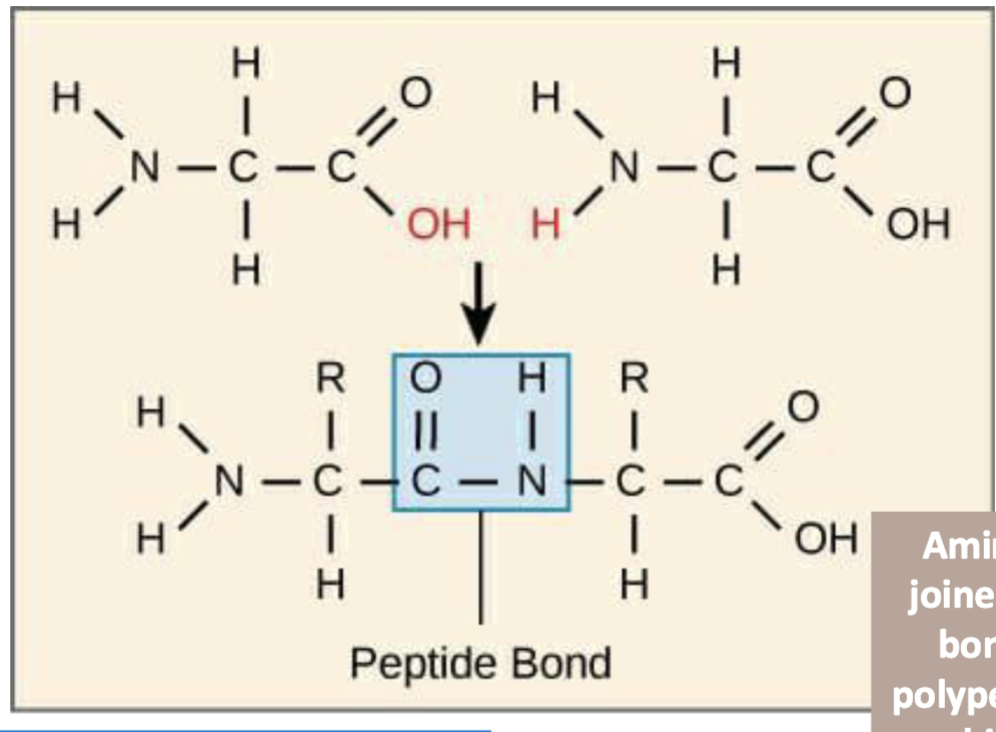
condensation reactions
build polymers from monomers; peptide bond formation
hydrolysis reactions
break polymers back into monomers; peptide bond cleavage
peptide bond cleavage
the process of breaking the covalent bonds that connect amino acids in a protein; catalyzed by proteases that require no energy
protease
a hydrolase enzyme; breaks polypeptides
primary structure of a protein
sequence of amino acids which determines the shape into which it will fold, which in turn determines how it will function
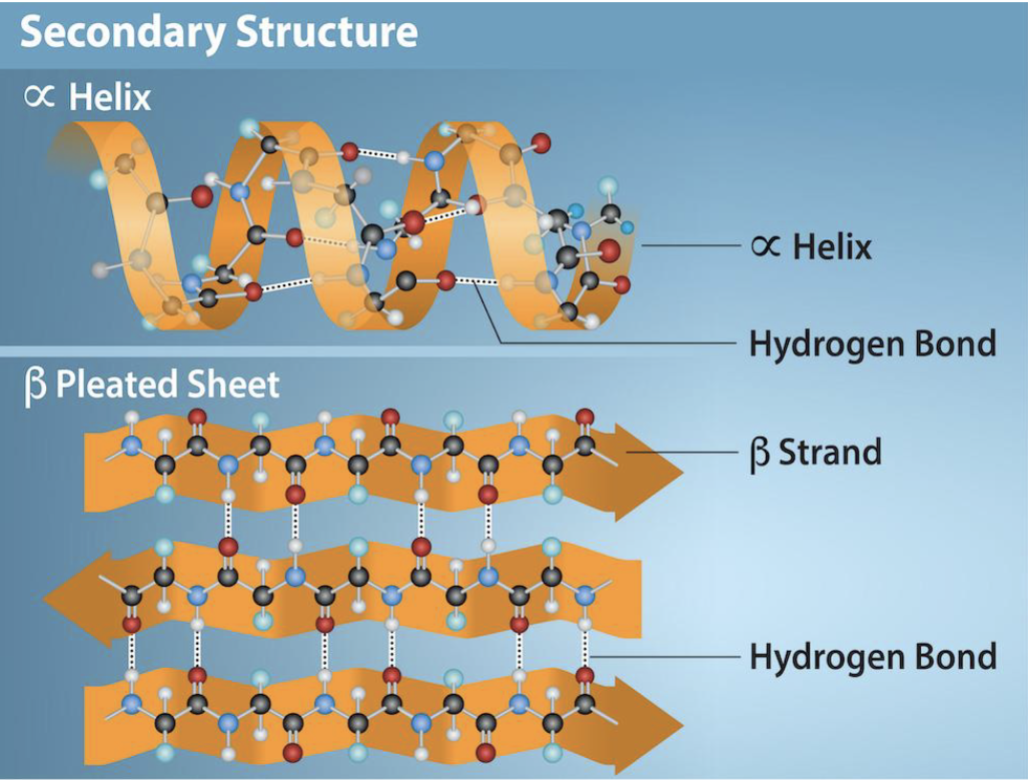
secondary structure of a protein
alpha helices and beta sheets, which result from hydrogen bonding interactions between backbone atoms; at least one (alpha/beta) is present in virtually all proteins. H-bonding is the backbone for secondary structures
tertiary structure of a protein
the three-dimensional shape, usually made of several secondary structure elements. this structure determines the proteins function.
quatenary structure
the interaction between more than one polypeptide chain; two or more tertiary structures come together to form the functional protein. these are held together by van der Waals forces, hydrogen bonds, disulfide bonds and ionic bonds. these still have N and C-termini for each polypeptide
amino (N-) terminus
corresponds to a protein’s first amino acid; proteins are always synthesized from amino- to carboxyl terminus
carboxyl (C-) terminus
corresponds to a protein’s last amino acid; proteins are always synthesized from amino- to carboxyl terminus
pentapeptides
several amino acids; have a backbone which will determine secondary structures (remains the same regardless of which amino acids are present_
alpha helices
A common secondary structure in proteins where the polypeptide chain coils into a right-handed spiral, stabilized by hydrogen bonds
beta sheets
A secondary structure in proteins where polypeptide strands run parallel or antiparallel and are held together by hydrogen bonds
denaturing of a protein
known also as unfolding, which can occur due to chemical treatment or high temperatures, leading to possible loss of function, insolubility and unstable. if optimal conditions are returned, the protein can often refold and regain function.
importance of non-covalent bonds
very important for biological functions, because they are easily broken and reformed, they allow for dynamic interactions
globular proteins
hydrophobic residues are buried in a hydrophobic core containing nonpolar side chains (in an aqueous environment), while the outside of the protein has polar side chains which can form hydrogen bonds to water.
homodimer
composed of two identical polypeptide chains
heterodimer
consists of two different polypeptide chains
annealing
when complementary single-stranded nucleic acids come together by base-pairing
native state
a functional protein; not denatured.
disulfide bonds
found in the secondary and tertiary structures of proteins. They can occur within a single polypeptide chain (intramolecular) or between two polypeptide chains (intermolecular
hydrogen bonds
primarily found in both the secondary and tertiary structures of a protein; in secondary structures, they occur between the backbone atoms of the polypeptide chain, forming structures like alpha helices and beta sheets, while in tertiary structures, they can form between the side chains of amino acids
Element
a distinct type of substance or matter. each element has atoms that contain a unique number of protons in nucleus.
Atom
The fundamental unit of an element, made of protons, neutrons, and electrons.
Ion
An atom or molecule that carries a full charge —positive if it contains more protons than electrons, and negative if it contains more electrons than protons.
Molecule
A structure made of two or more atoms held together by covalent bonds.
Covalent Bond
An attraction between two atoms based on shared electrons.