MCB104 - Lecture 12 - Gene Regulation
1/63
Earn XP
Description and Tags
NO EPIGENETICS
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
64 Terms
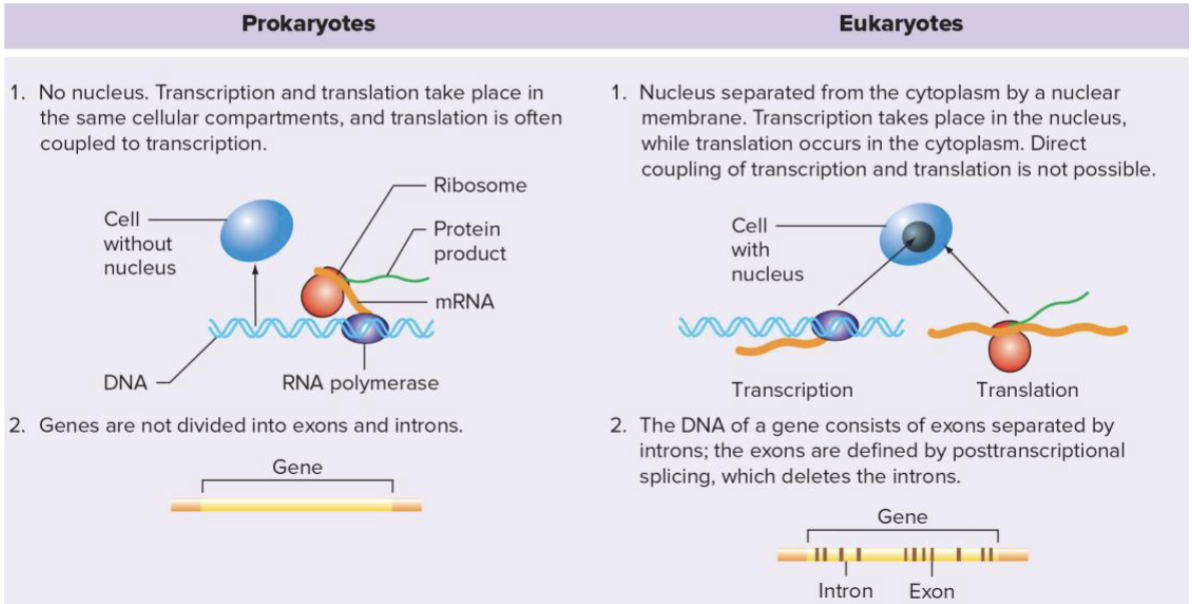
What is the difference in prokaryotic vs. eukaryotic gene regulation?
What are enhancers and why do they matter?
Unlike prokaryotes, eukaryotes have chromatin
Chromatin must be cleared from promoters for transcription
Done by various chromatin remodeling proteins
These often bind to enhancers
DNA sequences away from the promoter that stimulate transcription
Can be very distant, range is 100bp-1Mbp, but typically 1-100 kbp
Can be up or downstream of the promoter
What is Translation Initiation?
Bacterial mRNAs are often multicistronic
Have multiple ribosome binding sites (RBSs) which means multiple polypeptides
Small ribosomes subunit initiates by binding the RBS
In Eukaryotes (typically) only one start site
Small ribosome subunit binds 5’ cap first and scans to the start

What is mRNA processing?
Eukaryotes require splicing of primary transcript due to pressure of introns
Prokaryotes typically no introns
Eukaryotes, but not prokaryotes, add methylated cap (5’ end) and poly-A tail (3’ end)
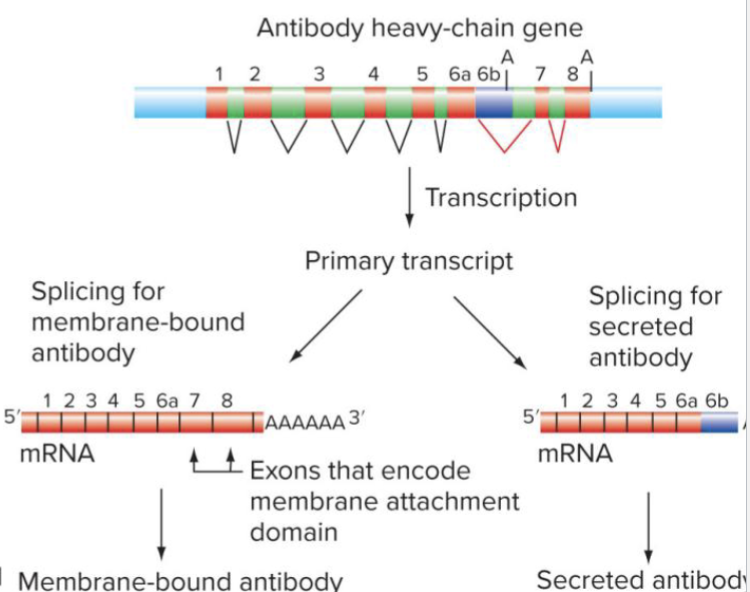
What is alternative splicing?
Alternative splicing produces different mRNAs from the same primary transcript
EXON 6…
If the sequence required to stick in membrane (start/stop transfer sequence)
If in Exon 7 or 8
For VerA: 6a gets cut to 7 (membrane bound anitbody)
For VerB: If regulatory protein it blocks splicing to occur and the A is the PolyA sequence and instead, 7 & 8 gone
Now you have all the stuff that would have been here is in the antibody but it’s no longer attached to membrane and can be secreted
What is the 5’ methylated cap?
Capping enzyme adds a methylated “backward” G to the 1st nucleotide of a primary transcript
What is the transcription product?
Primary transcript is the single strand RNA result of transcription
In prokaryotes, primary transcript is mRNA
In eukaryotes, primary transcript is processed to make an mRNA
5’ methylated cap
3’ poly-A tail, 100-200 A bases
Introns removed by RNA splicing
Where does regulation of gene expression occur?
EVERY
Regulation can occur at every step of the expression process
Chromatin structure
Transcription initation
Recruitment/blocking of RNA polymerase
Co and post transcriptional: mRNA processing
Splicing, base modifications, etc
Pre-translational: mRNA stability and avaliability
Translation initation
Post translational modifications
Phosphorylation, glycosylation, etc
What is regulatory elements?
DNA sequences that are NOT transcribed but play a role in regulating other nucleotides sequences
Promoters, enhancers, silencers, etc
CIs regulatory element: a regulatory element on the same chromosome as the target
What are regulatory genes?
Genes whose products interact with other sequences and alter their transcription or translation
Trans acting proteins: A regulatory protein whose target can be on a different chromosome
What is Constitutively expression?
Continuously expressed under normal conditions
What is positive control?
Stimulate gene expression
What is negative control?
Inhibit gene expression
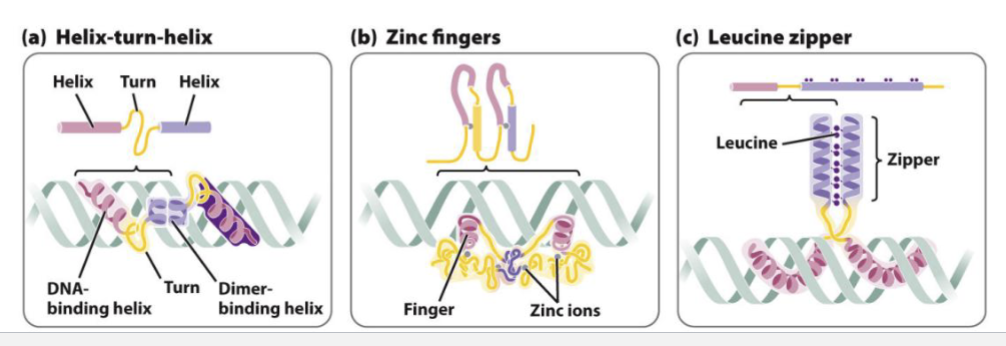
What are DNA binding proteins?
~60-90 amino acids, responsible for binding to DNA, typically via hydrogen bonds
Motif: a simple structure that occurs frequently
DNA binding motifs typically interact with the major groove
Distinctive types of DNA-binding proteins based on the motif
3 examples:
What are operons?
Bacterial gene expression often uses operons
Opersons consist of a promotor, regulatory sequences (operator) and structural genes
Often controlled by regulatory genes that are not part of the operon
What is inducible operons?
Transcription is usually off and needs to be turned on
Negative inducible operons have an inducer inactivate a repressor
Typically catabolic pathways
What are repressible operons:
Transcription is normally on and needs to be turned off
Typically anabolic pathways
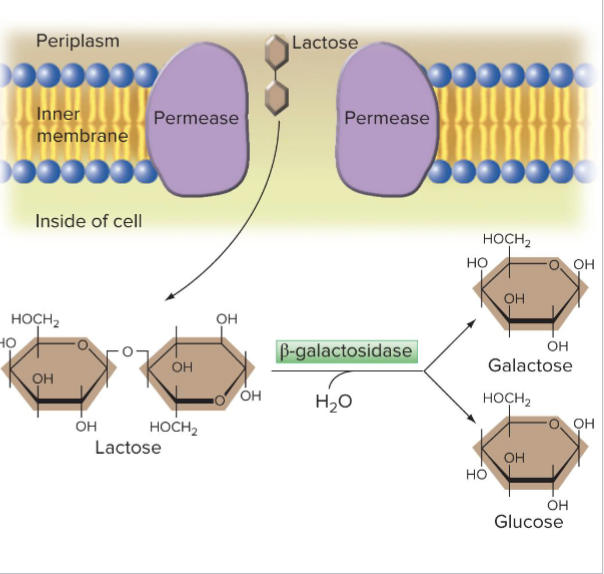
What is the E.coli Lactose Metabolism
Permease symporter transports lactose into the cell
Lactose is hydrolyzed by beta-galactosidase
Often called just B-gal
Products are glucose and galactose
How is lactose used as a gene regulation model?
The lac genes are not essential for survival
If both glucose and lactose are present, E.coli cells will use glucose first
Simple assays for lac expression ONPG or X-gal as substrates for B-gal produce colored products
Lactose induces a 1000-fold increase in B-gal activity
Many lac- mutants have been identified
DESCRIBE THE LAC OPERSON COMPONENTS AND PROTEINS
Lac Operon has several components:
3 Structural Genes:
LacZ: B-galactosidase
LacY: Permease
LacA: Acetyltransferase
Promotor (lacP): RNA Polymerase binding site
Operator (lacO): Binding site for lac repressor
CRP binding site: BInding site for CRP
There are 2 regulatory proteins:
lacl: encodes lac repressor protein
CRP: cAMP receptor protein
Also called CAP: Catabolite Activated Protein
How does Lac operon repression work?
The lac operon is normally inactive
The lac repressor protein binds to LacO
The Operator is located overlaps with the RNA polymerase binding site and covers the transcription start site
How does the lac repressor work?
lac repressor is a tetramer, with each subunit containing a DNA-binding HTH motif
lac operson has 3 operators (O1, O2, O3) each of which contains 2 recognition sequences for lac repressor
O1 has the strongest binding affinity for lac repressor
Maximum repression occurs when all 4 repressor subunits are bound (cooperatively)
2 repressor subunits bind to O1
2 repressor subunits bind to O2 or O3
How does induction of the lac operon work?
When inducer (lactose or IPTG) is present
Inducer binds the lac repressor
Allosteric inhibition of the repressor
Reversibly changes shape and cannot bind to the operator
RNA polymerase may now bind the promoer
How does glucose and the lac operon relate?
When both glucose and lactose are present, only glucose is used
When glucose is low, adenyl cyclase has higher activity
cAMP binds to CRP proteins
cAMP-CRP binds to CRP binding sites and recruits RNA polymerase
Without CRP bound, recruitment of RNA polymerase is very inefficient
Catabolite repression: the product of the catabolic pathway blocks the expression of genes involved
Describe lac mutant analysis
Mutants reveal roles of lac operon components
Table shows B-gal activity under different conditions
Partial diploidy via introduction of a plasmid can show cis vs. trans activity
For a table, the plasmid contains all components except lacZ
(lacZ minus plasmid that has lacI-, if I mutate the operator (LacO-) i will have constitutent actvity but if LacI- can operate in trans and can inhibit operon even if not on same DNA sequence)
LaclS is a gain of function that cannot be bound by inducer
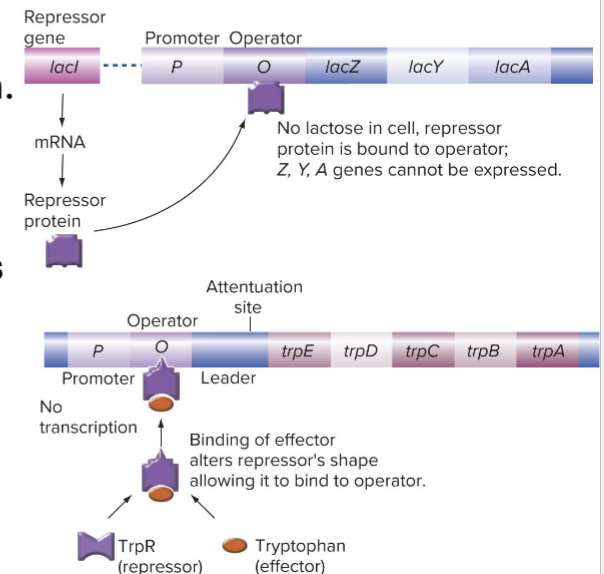
What is the Trp Operon?
Anabolic synthesis of tryptophan (Trp) is regulated by the trp operson
When tryptophan is present, it binds to the repressor
Trp bound repressor can bind to TrpO, blocking transcription
What is the Trp Attenuator
The trp RNA leader can fold into 2 different conformations
The 3 to 4 stem loop is a transcription terminator
The 2 to 3 stem-loop prevents formation of the 3 to 4 stem - loop so it is an antiterminator
A terminator may halt transcription or block the ribosome
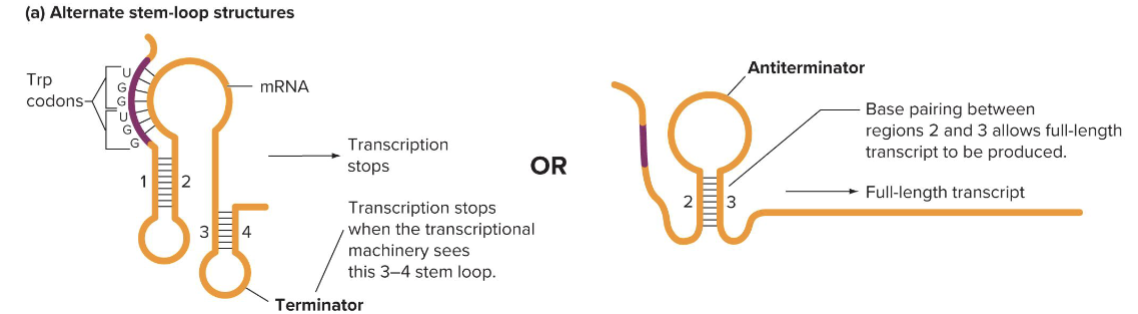
How is the attenuator regulated by avaliability of tRNATrp
RNA leader includes 2 trp codons
When tRNATrp is present rapid ribosome movement results in 3 to 4 stem loop formation, which is a terminator
When tRNATrp is low, the ribosome stalls, allowing the 2-3 loop to form, blocking the 3 to 4 loop
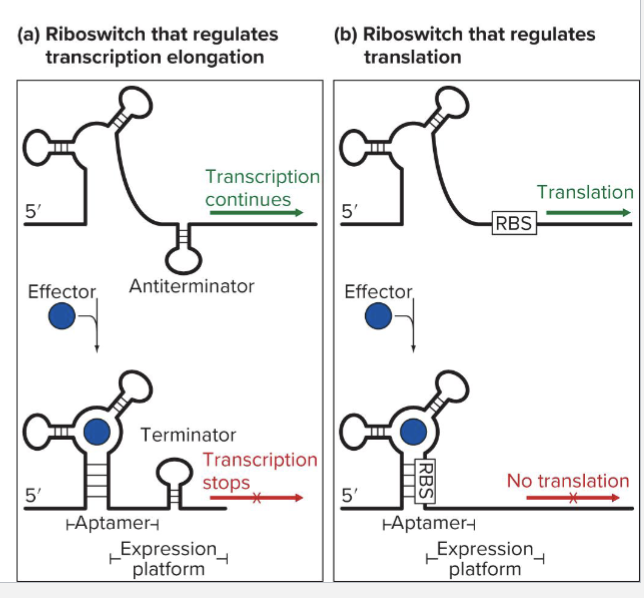
How does riboswitch work?
RNA leader with several conformations
Binds small molecule through a short sequence called an aptamer
Conformation of the “expression platform” changes if aptamer is bound or unbound
A riboswtich can regulate transcription or translation
How is gene regulated by small RNAs? (sRNA)
Usually inhibit translation by base-pairing with RBS
Can also activate translation by disrupting stem loops
Some lead to mRNA degradation
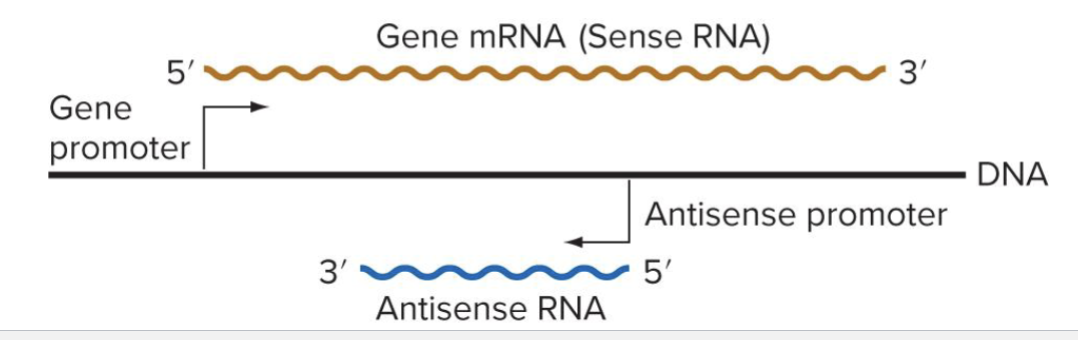
How is gene regulated by antisense RNA?
Produced by transcription of the strand of DNA opposite the template strand (antisense)
May inhibit translation by base pairing with the same strand
May lead to degradation of mRNA
May interfere with transcription of sense gene
What is Eukaryotic Gene Expression?
Many steps can be regulated to control the amount of active gene product
Transcription initiation
Transcript processing
Export from nucleus
Translation of mRNA
Protein localization
Protein modification
What is Eukaryotic Gene Regulation: Cis elements - PROMOTERS
Promotors: DNA sequence that is usually directly adjacent to the gene
Bind RNA polymerase
Often have TATA box (TATA (T/A) A(A/T)
Allow basal level of transcription
What is Eukaryotic Gene Regulation: Cis elements - ENHANCERS
DNA sequence that can be far away from gene
Augment or repress the basal level of transcription
May be located either 5’ or 3’ to the transcription start site
Still function when moved to different positions relative to promoter
What are the cis elements of eukaryotic gene expression?
Enhancers and Promoters
What are reporter genes?
Enhancers can be identified by making a conduct with
Putative enhancer
Minimal promoter
Reporter protein (GFP)
An organism with that sequence will express GFP when that enhancer is active
What are eukaryotic trans factors:
Transcription Factors
What is the eukaryotic trans factors - Transcription Factors
General term for any DNA binding protein that regulates transcription
Binds to promoters and enhancers
Recruit other proteins to influence transcription
3 types: Basal Factors, Activators, and Repressors
What are Basal Factors?
The core transcription machinery
Ordered pathway of assembly at promoter:
1) TATA-binding protein (TBP) binds to TATA box
2) TBP associated factors (TAFs) bind to TBP
3) RNA pol II binds to TAFs
What is the mediator complex?
Transcription of many eukaryotic genes requires the mediator complex
Mediator is a complex of more than 20 proteins
Doesn’t bind DNA directly, bridges between
The promoter
Activator or repressor proteins at
Describe Activators
Activators positively regulate transcription
2 main mechanisms:
Recruiting basal factors and RNA pol II to promoters
Often involves mediator for this
Recruiting coactivators to open chromatin structure
What is the activator structure?
Activator proteins have at least 2 functional domains
DNA binding domain: binds to specific enhancer
Activation domain: binds to other proteins (basal factors or coactivators)
Dimerization domain: some activators also have a domain that allow them to interact with other proteins
Homodimers pair with themselves
Heterodimers pair with another activators
Provides coincidence detection
What are Repressor Proteins?
The opposite of activators but same principles
2 main mechanisms:
Recruit co-repressors to prevent RNA pol II complex binding
Recruit co-repressors to close chromatin structure
How can the same transcription factor play different roles?
Transcription factors’ impacts on transcription are context dependent
The presence or absence of other transcription factors can cause them to act as either repressors or activators
Ex: Dorsal is an activator unless it is near dead ringer (dri) at which point they bind the corepressor Groucho
How does transcription factor regulation work?
The function of trans acting proteins changes by
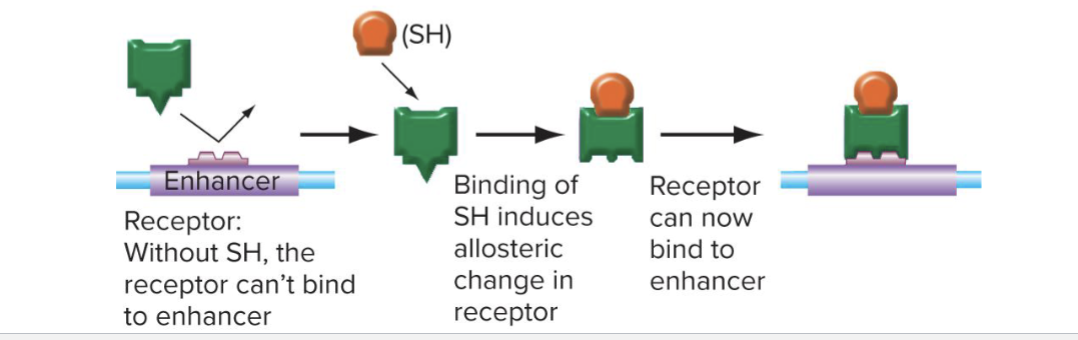
Allosteric interactions (ex: Ligand binding)
Modificaton of transcription factors (ex: phosphorylation)
Transcription factor cascades
How does fine-tuning gene expression work?
In humans, approximately 2000 genes encode transcriptional regulatory proteins
Each regulatory proteins can act on many genes
Each enhancer has binding sites with varying affinities for activators and repressors
Post-translational modifications and ligand binding can alter TF activity
What are indirect repressors?
An indirect repressor interferes with the function of an activator
Competition due to overlapping binding sites
Repressor binds to activation domain (quenching)
BInding to activator and keeping it in cytoplasm
Binding to activator and preventing homodimerization
What are insulators?
Insulators are sequences located between an enhancer and a promoter that block access to the promoter
Human insulators bind CTCF proteins to form loops called topologically associating domains (TADs)
Enhancers activate promoters located in the same loop
How do you detect TADs?
Chromatin Conformation Capture:
Cross-link proteins and DNA in close proximity within chromatin
DNA is fragmented and DNA ligase joins ends
High-throughput methods used to define TADs
Measures frequency with which 2 sequences were ligated together
How can you study transcription factors using reporters?
GFP reporters can be used to screen for mutations in transcription factors
Mutations in a gene encoding in an activator reducing expression of the reporter
Mutations in a gene encoding a repressor increase expression of the reporter
How can you study transcription factors using CHIP-Seq?
Co-immunoprecipitation: Use an antibody against a target (Bait) protein to isolate it and anything attached to it (Prey) for analysis
Chromatin immuniprecitation: Co-IP where the bait is a TF and the prey is the DNA
Steps:
Crosslink DNA
Fragment DNA
Bind with an antibody specific to the target protein
Purify complexes with antibody, target protein, and DNA fragments
Sequence DNA
How does posttranscriptional regulation work?
Posttranscriptional regulation can occur at many steps
At level of RNA:
Splicing, stability, and localization
At level of protein:
Synthesis, stabillity, modification, and localization
What is Splicing Regulation?
Fruitless (fru) in Drosophila is required for courtship
Both male and female flies produce same primary transcript
Females express Transformer (Tra) protein that remove an exon from the final transcript
Result is sex specific Fru proteins
How can translation initiation be regulated?
Control of translation often occurs at initiation
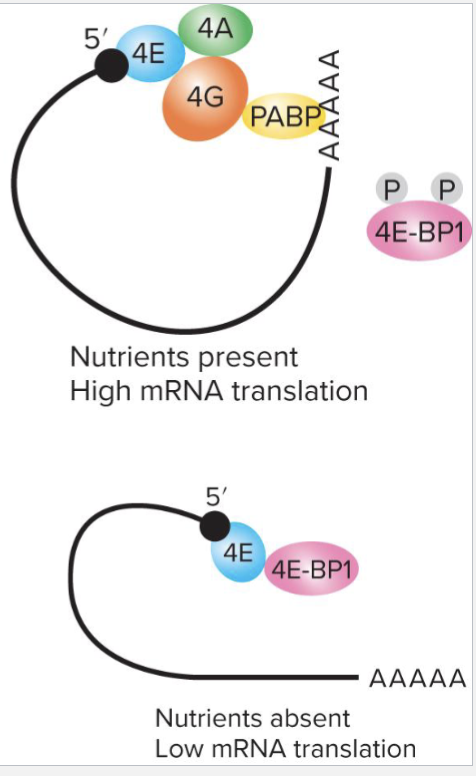
Small subunit of ribosome recognizes a complex structure around 5’ cap
eIFG protein binds to poly-A binding protein (PABP) at poly-A tail to circularize mRNA
Example Regulation:
4E-BP1 binds to initiation elF4E, blocks initiation
Presence of nutrients and growth factors in environment leads to phosphorylation of 4E-BP1, preventing association
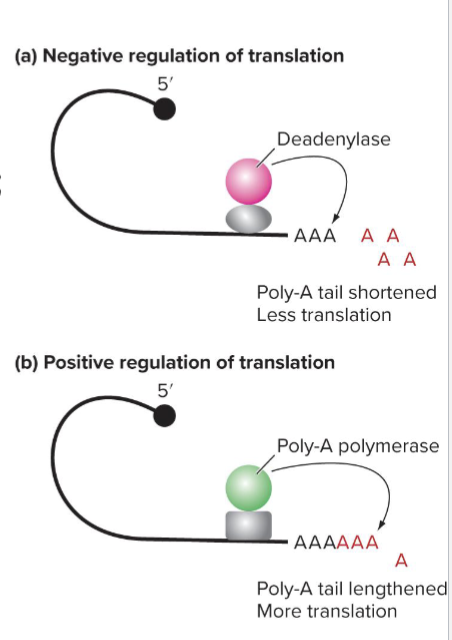
How does Poly-A tail length work?
Longer poly-A tails bind PABP more efficiently
Translation initation complex forms more efficiently
Once tail is removed, mRNA is targeted for degradation
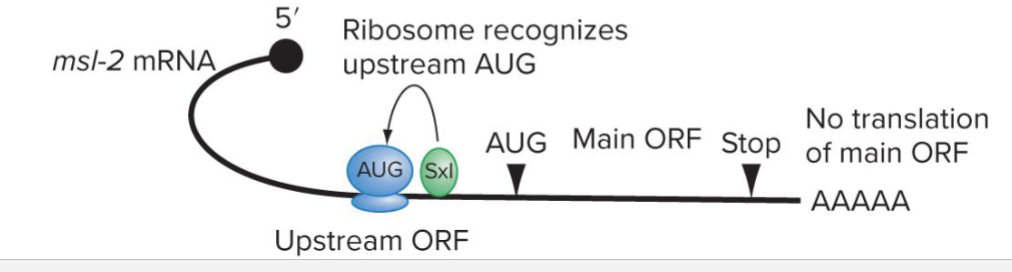
How does Translational Control by ORF work?
Upstream open reading frame can be translated instead of the main ORF
Regulators can bind the uORF to encourage or discourage its translation
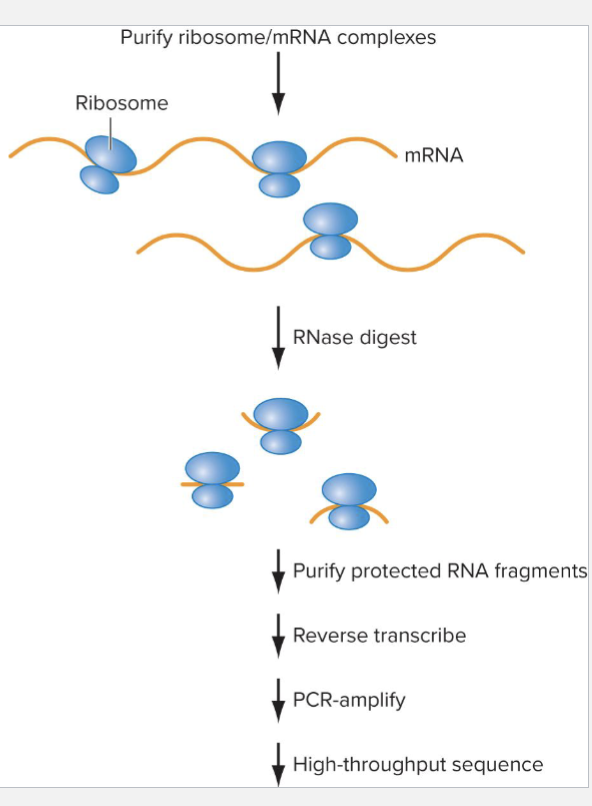
How does Ribosomal Profiling work?
mRNA level doesn’t always correlate with protein level
Ribosome profiling allows researchers to observe positions of ribosomes on mRNAS
Steps:
Purify (often crosslink) ribosome/mRNA complexes
Digest with RNAase to eliminate uncovered RNA
Sequence RNAs to determine what is being transcribed
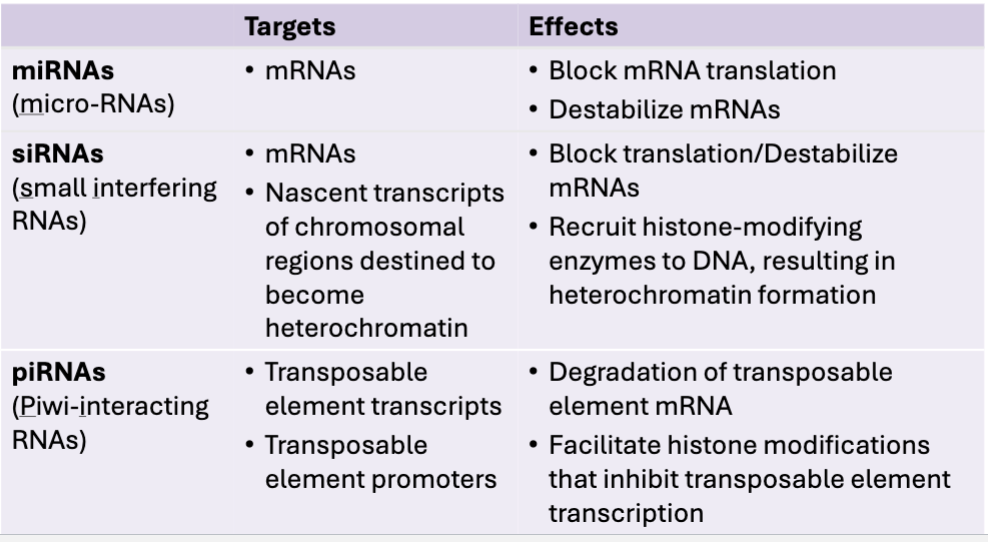
Think of miRNAs, siRNAs, piRNAS and their targets & effects
What are miRNAs?
Most miRNAs are transcribed by RNA polymerase II
The primary transcripts have double stranded stem loops
Drosha excises stem-loop from primary miRNA (pri-miRNA) to generate pre-miRNA
Dicer processes pre-miRNA to a 21-25nt long duplex miRNA
One strand is incorportated into miRNA-induced silencing complex (miRISC)
What is miRNA regulation?
miRNAs bind to complementary sequences of other RNAs
When complementarity is perfect, target mRNA is degraded
When complementarity is imperfect, translation of mRNA target is repressed
What are siRNAs?
siRNAs follow the same pathway as miRNAs except:
Source is different: exogenous dsRNAs or transcription of both strands of an endogenous genomic sequence
A few different components in RISC complex
Almost always a perfect match, so almost always degrades target mRNA
Some riRNAs bind to genomic sequence and recruit factors that convert it to heterochomatin
The siRNA response is called RNA interference (RNAi)
RNAi is a common genetic tool, permitting the knockdown of target genes without having to make mutants
Discovered in C. elegans
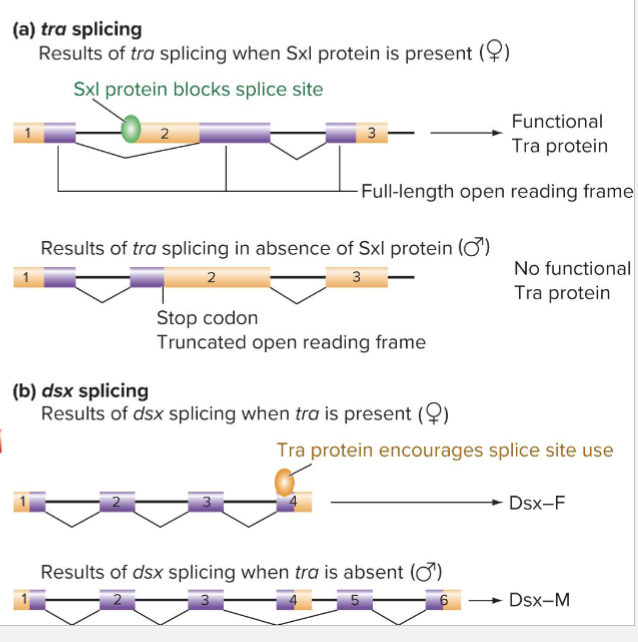
What is Drosophila Sex Determination?
Sex lethal (Sxl) gene encodes an RNA binding protein that controls the alternative splicing of RNA targets
Required for female-specific development
In early embryos, Sxl is transcribed only in females
XX cells express Sxl from establishment promoter (Pe)
Genes encoding activators for Pe are on the X chromosome
Dose dependent: 2 copies required to meet threshold
How does Sxl regulate alternative splicing?
After early embryogenesis, transcription of Sxl occurs from the Pm promoter
Transcripts in males have a stop codon in exon 3
In females, Sxl includes the exon 3, leading to protein production
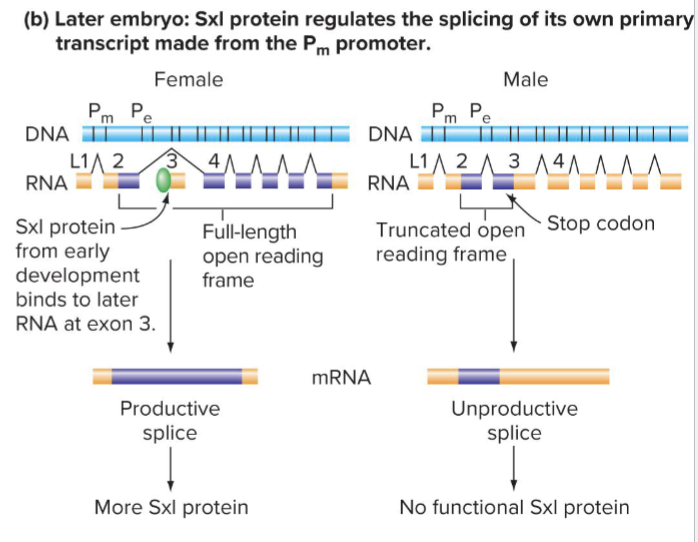
What does Sxl trigger?
Splicing “cascade”
Sxl regulates splicing of transformer (tra) mRNA
Tra protein regulates splicing of dsx mRNA (and many others)