L10: Post-transcriptional regulation
1/26
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
27 Terms
What is post-transcriptional regulation? (review/ key concepts)
Regulation of gene expression at the levels of RNA and translation, including RNA stability, degradation, riboswitches, attenuation, and translational control.
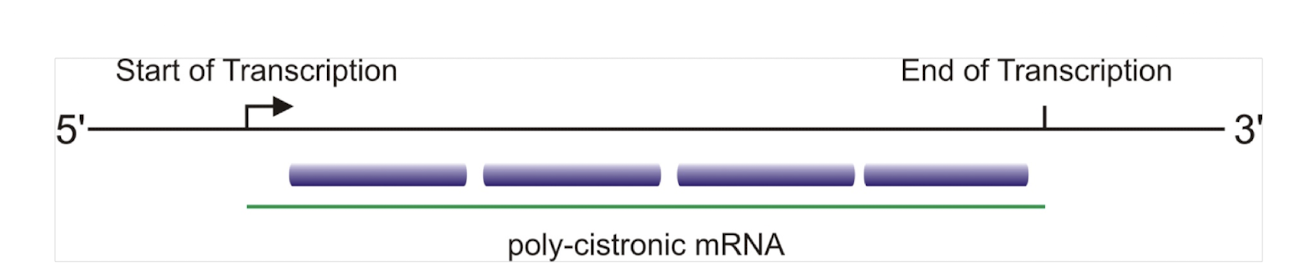
How can expression levels differ within a polycistronic operon?
Through differences in mRNA stability, degradation rates, translational efficiency, Shine–Dalgarno sequence strength, and structural features such as hairpins.
Why is RNA difficult to work with experimentally?
mRNA is highly unstable, especially bacterial mRNA with no poly-A tail
How does RT-PCR help measure RNA and an advantage + limitation?
Reverse transcriptase converts RNA to cDNA (complementary DNA), which can be amplified by PCR/quantitative PCR to compare amounts of RNA.
Advantage: sensitive quantification of amount of RNA present.
Limitation: does not provide transcript size.
What technique measures both size and amount of mRNA?
Northern blotting, involves measuring mRNA which is highly unstable and then radiolabelled DNA probes are use to hybridise RNA on a membrane:
Amplifies the signal and allows us to read the amount and size of transcript directly
what are the key steps of northern blotting
1. Extraction of RNA: Isolate total RNA from a sample.
Electrophoresis: Separate RNA fragments by size using gel electrophoresis (e.g., agarose gel).
Transfer (Blotting): Transfer the separated RNA from the gel to a solid support membrane (e.g., nylon membrane).
Hybridization: Incubate the membrane with a labeled probe (DNA or RNA, typically radiolabeled) that is complementary to the target RNA sequence.
Washing: Remove unbound probes.
Detection: Detect the hybridized probe (e.g., autoradiography) to visualize the target RNA bands.
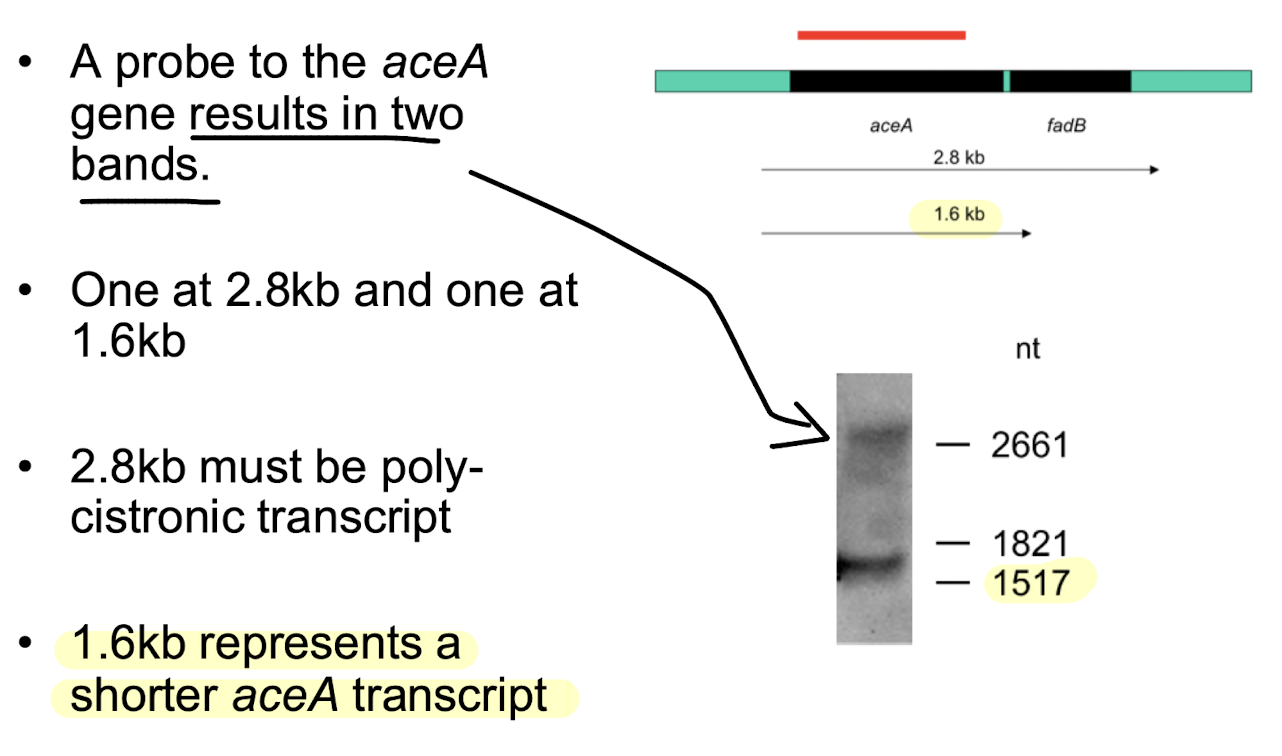
what is aceA?
AceA is a gene that encodes for an enzyme involved in the metabolism of acetate in bacteria, specifically as part of the glyoxylate cycle.
What did the aceA Northern blot reveal and why?
Two transcripts: 2.8 kb (polycistronic full transcript) and 1.6 kb (shorter transcript).
Caused by RNA degradation producing stable partial fragments
Fast degradation i.e. thanks to short half-life of mRNA and RNAse activity can allow a rapid change in message
What are ribonucleases and why are they important?
Enzymes that degrade RNA from 5′→3′ or 3′→5′.
They prevent mRNA accumulation and allow rapid gene expression changes such as responses to environmental changes.
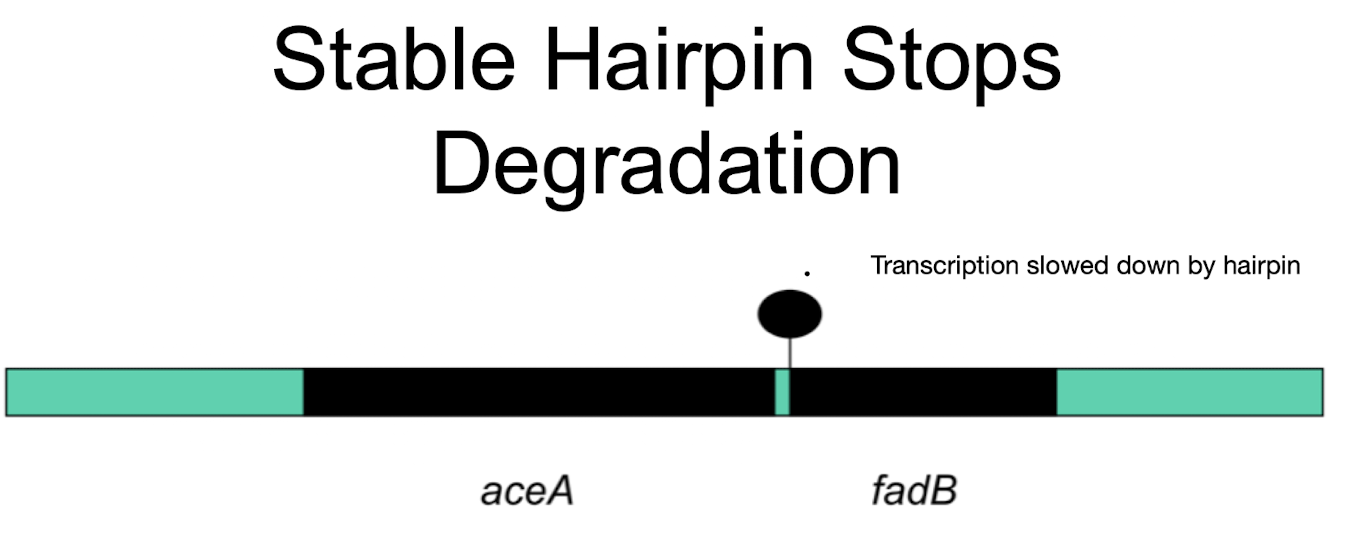
How can RNA structure affect stability?
Hairpin loops can protect downstream RNA regions from degradation, increasing stability and translation
How does a hairpin affect aceA and fadB expression?
A stable hairpin protects aceA, yielding more aceA mRNA than fadB even though they are in the same operon
In Rhodobacter capsulatus, why are pufBA transcripts more abundant than pufLMX?
pufBA mRNA has a longer half-life (~20 min) vs. pufLMX (~3 min) due to more stable secondary structures.
This increased stability allows for greater accumulation of pufBA transcripts, enhancing their expression relative to pufLMX.

Why must puf gene expression be tightly regulated?
The photosystem is oxygen-sensitive, so expression is required only under anaerobic light conditions
How does oxygen regulate pufBA transcript levels?
A specific endonuclease degrades pufBA when oxygen is present, decreasing half-life from 20 min to 2 min
This regulated degradation reduces the stability of pufBA mRNA, leading to lower transcript levels in aerobic conditions.
How does Shine–Dalgarno sequence variation regulate translation?
The Shine–Dalgarno sequence varies in its complementarity to the 16S rRNA, affecting ribosome binding affinity.
This variation influences the efficiency of translation initiation, regulating protein synthesis.
What are translational repressors?
Proteins that bind mRNA to block 30S ribosomal subunit access to the RBS, reducing translation.
Common in ribosomal protein gene regulation and can be involved in various stress responses in cells.
What is the role of the 5′ untranslated region (5′ UTR)?
Contains regulatory structures like aptamers and expression platforms that influence transcription termination or translation initiation.
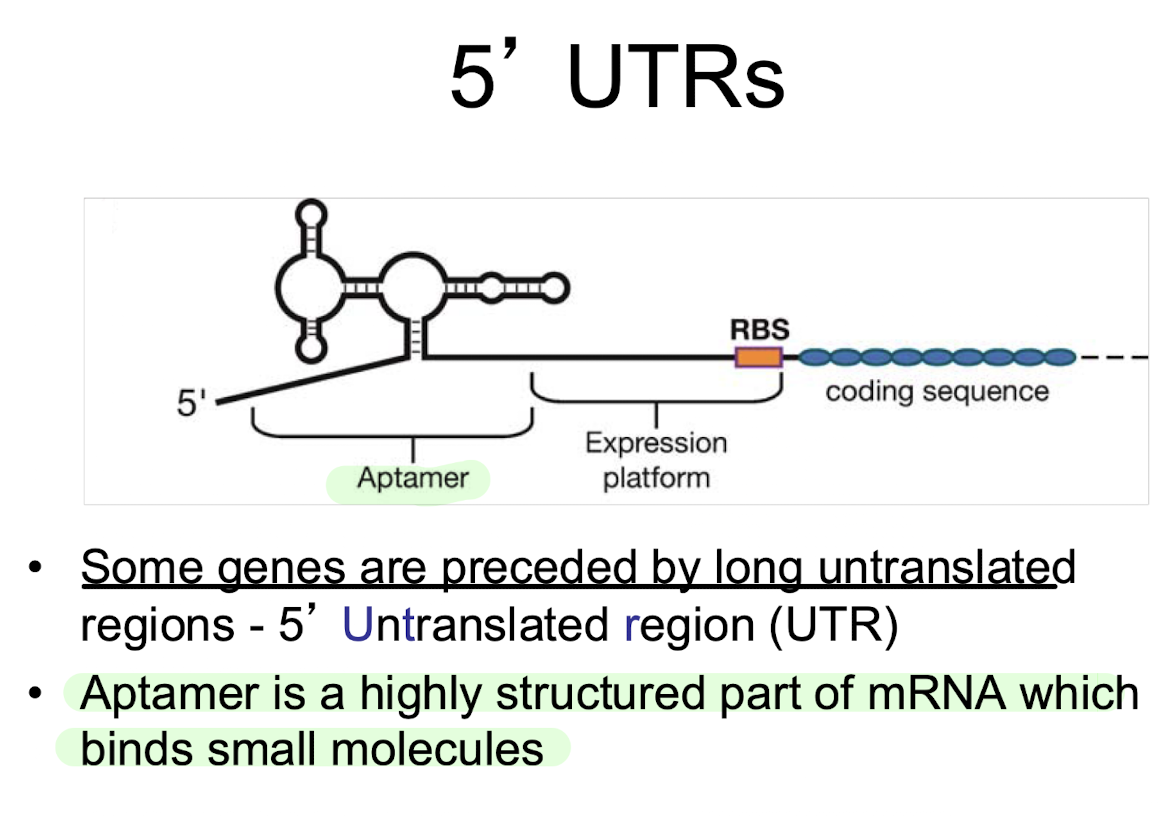
aptamer
highly structured part of mRNA which binds small molecules
expression platform
contains the part of the mRNA that controls the RBS (ribosome binding site)
• The expression platform can be converted into an “ON” or “OFF” position depending on secondary structure.
• These are referred to as Riboswitches
What is a riboswitch?
A cis-acting RNA structure where binding of a small molecule to an aptamer induces a conformational change controlling premature transcription termination
How does the Vitamin B12 riboswitch regulate transcription?
Low B12: Anti-terminator forms → sequence binds to left side of terminator sequence → terminator doesn’t form → transcription continues of biosynthesis genes
High B12: Aptamer binds B12 → disrupted anti-terminator binding → intrinsic terminator hairpin forms → transcription stops
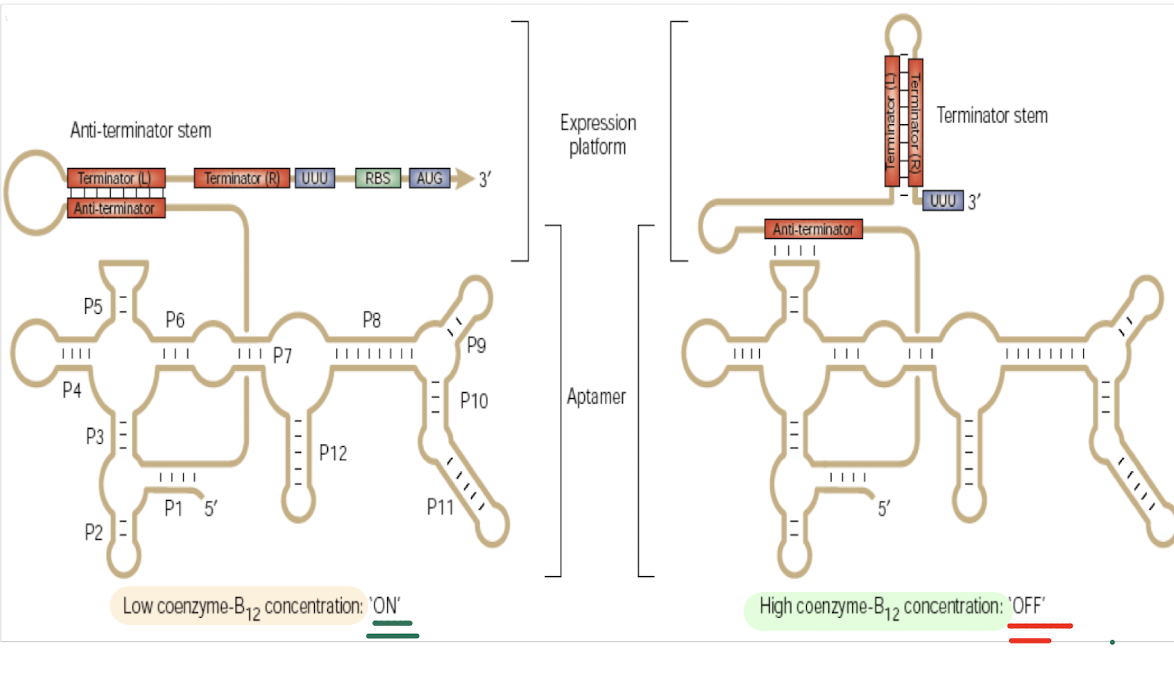
How does the Vitamin B12 riboswitch regulate translation?
Low B12: anti RBS RBS exposed → translation initiates.
High B12: Anti-RBS strand binds the RBS → translation inhibited.
What is attenuation?
A regulatory mechanism where transcription termination depends on translation activity, possible because transcription and translation occur simultaneously in bacteria
Allows bacteria to fine-tune gene expression in response to changing conditions.
Trp operon attenuation mechanism:
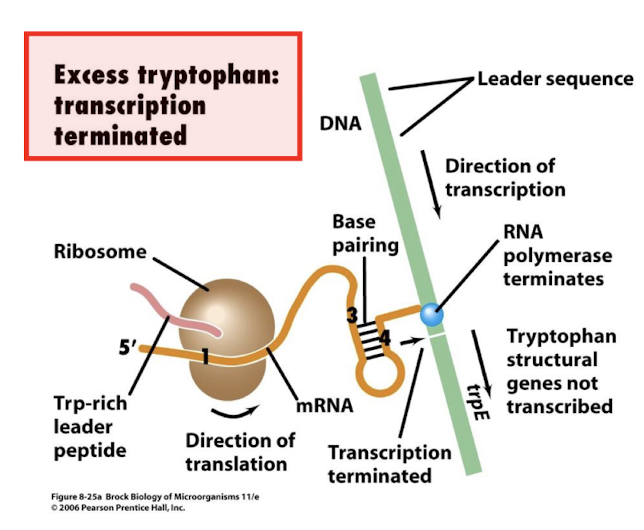
Trp operon has a Trp rich leader peptide
• Under normal circumstances a hairpin forms which terminates transcription (similar to normal termination)
• In the absence of Trp then the ribosome stalls at the leader peptide
• Now a different hairpin forms which is not a terminator
• Thus transcription termination is attenuated
How does attenuation work in the Trp operon?
High Trp: Ribosome quickly translates leader peptide → terminator hairpin forms → transcription stops.
Low Trp: Ribosome stalls → alternative hairpin forms (non-terminator) → transcription continues.

What are small inhibitory RNAs?
Short non-coding RNAs that bind complementary sequences on mRNAs to block the ribosome binding site, inhibiting translation
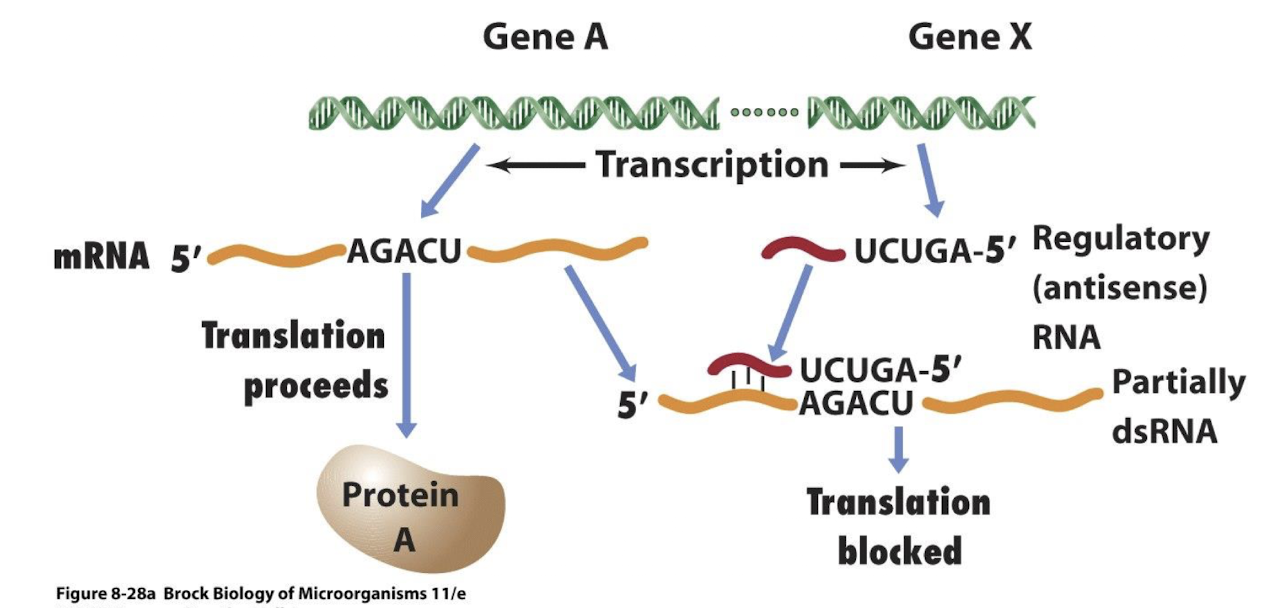
Why are sRNAs significant?
Their discovery revealed that post-transcriptional regulation is far more extensive than previously thought. Often dependent on global regulators like Hfq