Unit 7
1/30
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
31 Terms
Explain the different pahses of mitosis:
Interphase (23hrs)
G1 phase (11hrs): carries out its function+biomolecule synthesis
S Phase (8hrs): cells duplicate DNA and synthesize histones
G2 phase (4hrs): preparation for mitosis
Mitosis (1hr)
Whta is the special propertyof stem cells that keeps them from aging?
Divide without losing their telomeres, length is kept intact because tey posses telomerase (keeps them from shortening)
Also divide thru assymetric division, dividing thru mitosis to produce 2 different cells: a mature differentiated cell (called progenitor cell, will finish matuing into enterocyte which remain in G0 and don’t divide anymore) and another stem cell
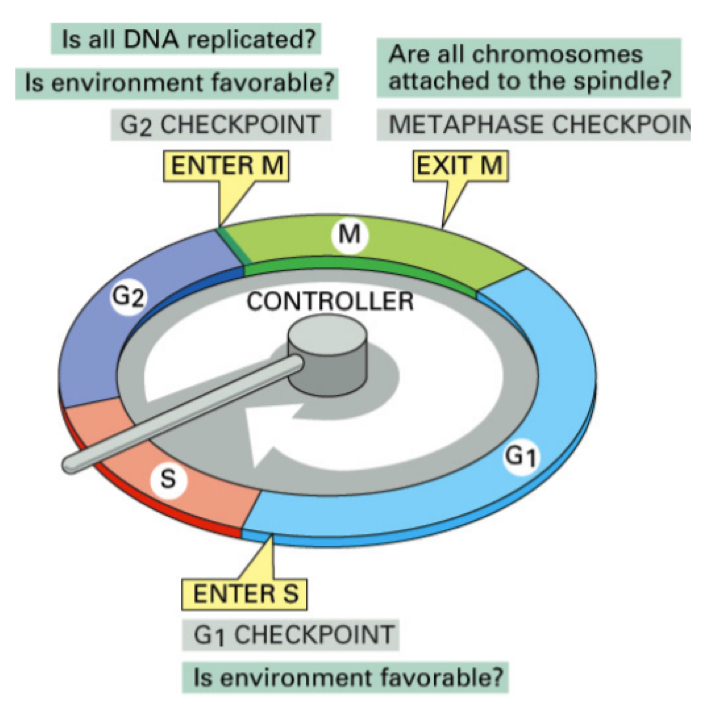
Whta are the cell cycle chekcpoints?
At end of G1: (((main chekcpoint)))
Cell decides whether to duplciate DNA or not, if outer conditions are desirbale for replication they do so and enter teh S phase
At end of G2:
Cell checks if DNA has been correctly replicated
During mitosis:
Cell checks if chromosomes are atached to spindles and correctly aligned
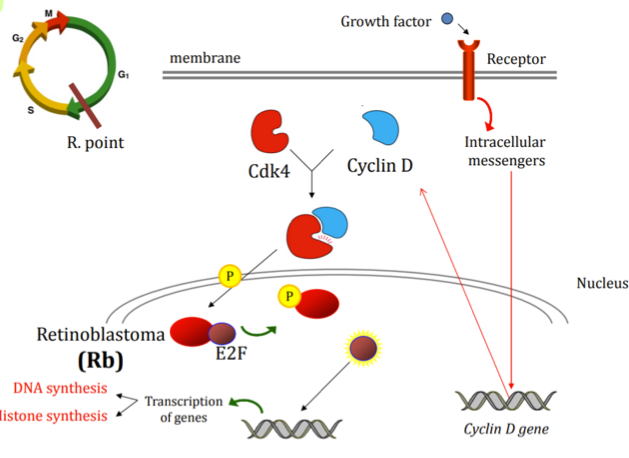
What specifically occurs at G1 checkpoint?
Receptor at membrane for nutrients, growht factors and hormones to check external conditions, signal transmitted intracellularly
Intracellular responsee is to stimulate transcription of cyclin D
Cyclin D binds to Cdk4 to form a complex that translocates to the nucleus
Cdk4-Cyclin D phosphrylates Rb which causes the separation of Rb and E2F.
E2F (now active) stimulates the transcription of different genes to begin cell proliferation
What can go wrong at the G1 checkpoint?
Inactivating mutations in Rb: Rb loses fnction, E2F always active- causes cancer, uncontrollled cell proliferation
Abnormally increase levels of cyclins: cells constantly replicate and create tumors.
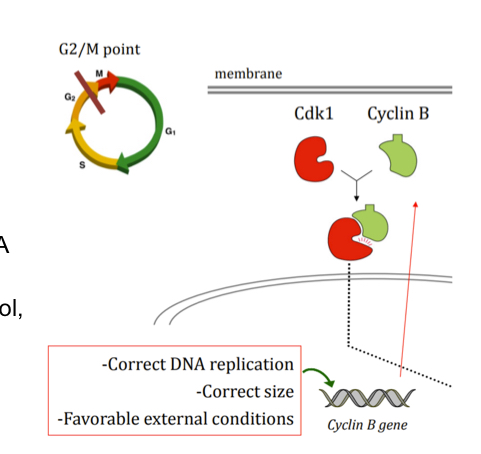
Whats pecifically occurs at G2 chekcpoint?
cyclin B gene transcribed and translated, binds to cdk1
Cdk1-cyclin B complex acts by phosphorylating different proteins:
In the nuclear lamina, phosphorylating it components (lamins) so they disintegrate ad chromosomes can condensate and spindle fibers can form/ attach
How can the cell cycle be inactivated?
inhibition of intracellular signalling
Cyclin degradation
Production of cdk-cyclin inhibitors
Dephosphorylation of Rb so that E2F can be rebind and remain inactive
When these inactivation mechanisms are impaired, tumorous cells appear
Keep this in mind:

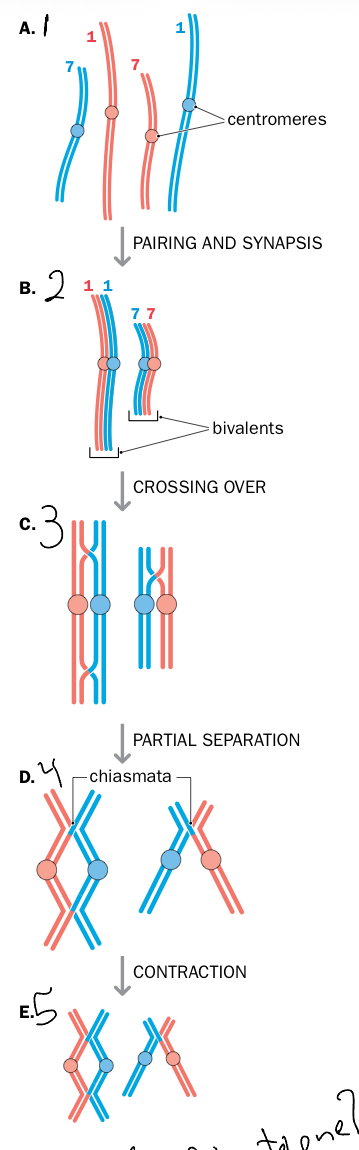
Explain the subphases of prophase in he first meiotic division.
Leptotene: chromatine starts to condensateand fom chromosomes
Zygotene (union): chromosomal pairing occurs resulting in synaptonemal complex (SC), held togetehr by proteins, have bivalence
Pachytene (thick): chromosomes fully visile, crossing over occurs, the chiasma forming at hotspot (sites of preffered recombination)
Diplotene: chromosomes separate+decondense, but remain bound by chiasmata
Diakinesis: chiasmatadisapear, nucleolus and nucleus dissapear, spindle fibers begin to form
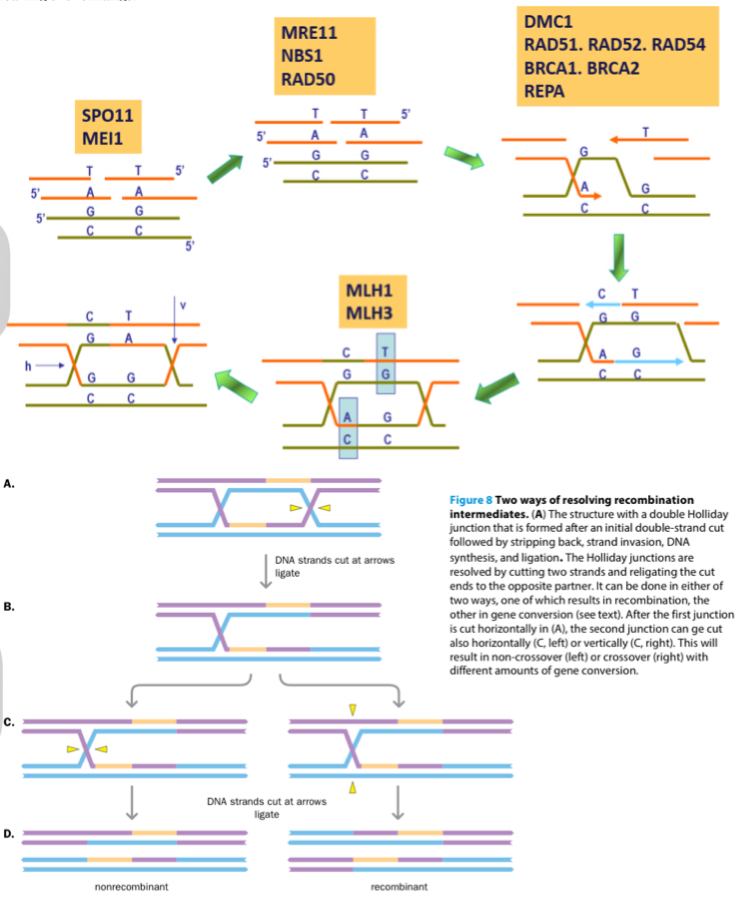
Explain recombination at a molecular level.
In pachytene of prophase I
Endonuclease create a double-strand break (DSB) at specific chormatid sites
A multi-protein exonuclease complex (MNR) digests the ends of the break to leave 3’ overhangs
Overhang searches and invades chromatid of homologous structure to create a holliday structure
The 3’ are extended by polymerase can result in one of the following
Crossover Gene conversion (recombinant chomatid in which sister chromatid gene was copied and attached to opposite)
Crossover w/out gene conversion (recombinant chromatid in which sise chromatid only exchanged starnds, did not copy DNA onto opposite strand)
Non-crossover gene conversion (non recombinant in which DNA was copied)
Non-crossover w/out gene conversion (non recombinant and no DNA replication)
Branch migration occurs followe by the resolution of holliday junctions by the cutting og two strands and religating the cut ends to the opposite partner
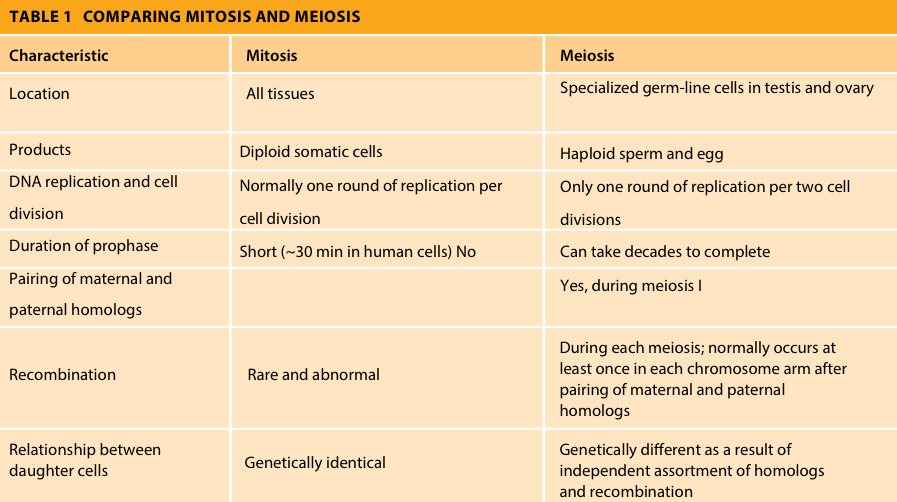
Meiosis/mitosis differences:
Why is metaphase I significant?
Bivalents are randomly oriented and assorted creating genetic variability
At what phase during prophase I one are female oocytes stuck in until puberty?
Diplotene phase
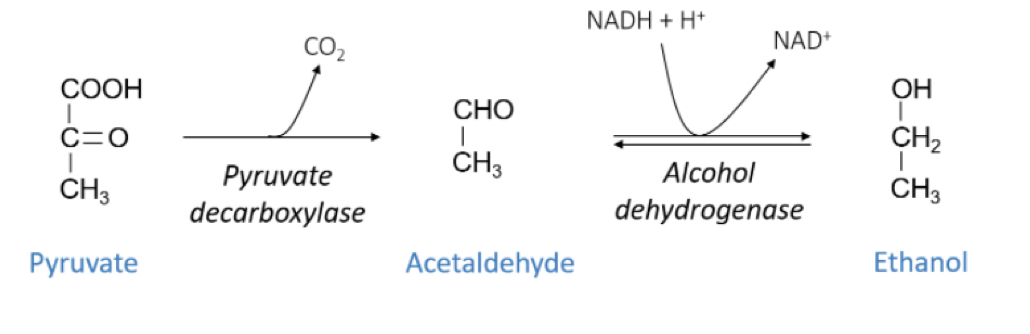
How is ethano produced from pyruvate?
Pyruvate decarboxylated into acealdehyde by pyruvate decarboxylase and alcohol dehydrogenase reduces it to Ethanol
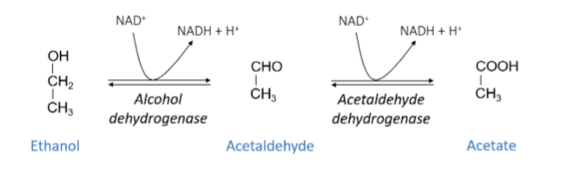
Describe Ethanol metabolismo in humans
Ethanol oidised to accetaldehyde by alcohol dehydrogenase and then acetaldehyde is oxidised again by acetaldehyde dehydrogenae to produce acetate
Acetaldehyde is toxic and belived to extert toxic action aginst the liver - responsible for hangover symptons
Acetate has no toxic effects
What happnes to individuals with a commons allelic variant of ALDH2?
Since the acetaldehyde oxidation in the humna liver t catalyzed by ALDH2, people with this variant have a decreased capaciy for acetaldehyde metabolism.
How is alcoholism treated?
With ALDH inhibitors (like dsulfiram) to help abstain from alcool intake— the inhibitors make you more drunk more quickly (from acetaldehyde accumulation), so you stop drinking.
What can prolonged alcohol abuse do to the body?
dangerous byproducts (acetaldehyde) created and accumulate
Chronic liver diseases such as fatty liver, hepatitis or cirrhosis can arise
It could inhibt fatty acid oidation, stimulate triacylglycerol synthesis, ketoacidosis, lactic acidosis and hypoglycemia.
Can also lead to vitamin deficiency (alcoholics tend to eat less), negatively impacts lining of gastointestinal tract+compromises absorption of nutrients
What is Wernicke-Korsakoff’s encephalopathy?
Lack of thiamine (vitamin B1) most common manifestation of this deficiency in alcoholics.
In what direction does DNA synthesis occur?
5’ to 3’
What is Ori in DNA replication?
The origin site, where replication begins, ofrming rpelication forks and move biderectionally along the DNA
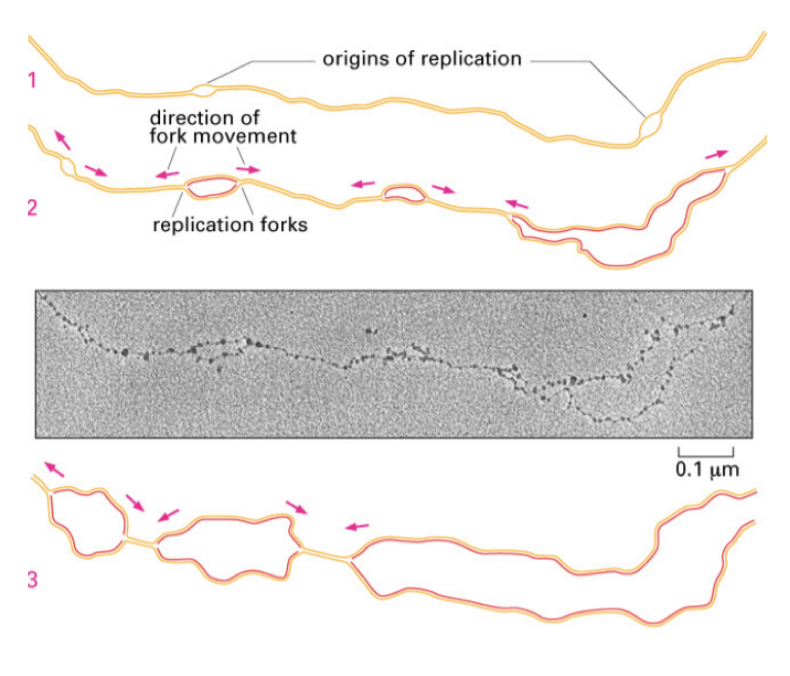
Explain the origin of rpelication in E Coli.
DnaA protein binds to binding site on DNA strand
The DnaA allows teh recruitment of helicase DnaB which breaks the hydrogen bonds in the DNA and unzips it.
The single starnd of DNA is then bound to SSB (single stranded DNA binding proteins, which stabilize the strand.
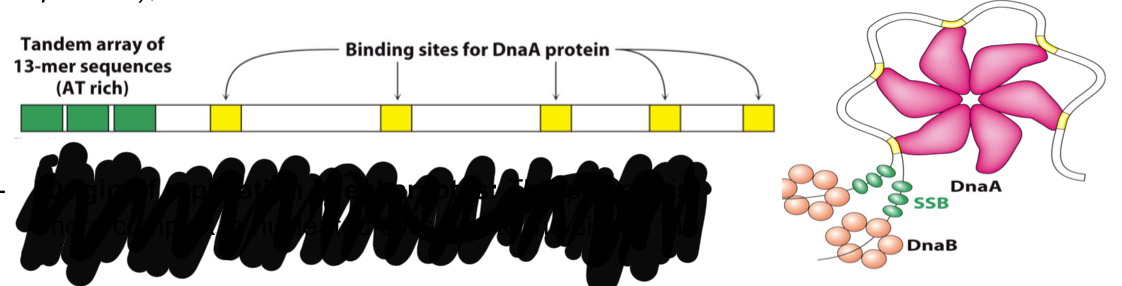
Describe the origin of DNA replication in eukaryotes.
Multiple origins of replication, no specific DNA sequences associated to these origins ( hoever, they ae usually rich in T and A bases (because they form double bonds and not triple which makes them easier to denature)
Give examples of eukaryotic origin of replication complexes (ORCs):
hexamer complex (similar to that of bacterial DnaA)
Helicase cmoplex formed by 6 subuntis of the Mcm2-7 protein
An SSB known as replication protein A
Polymerases: DNA-pol-alpha and DNA-pol-gamma
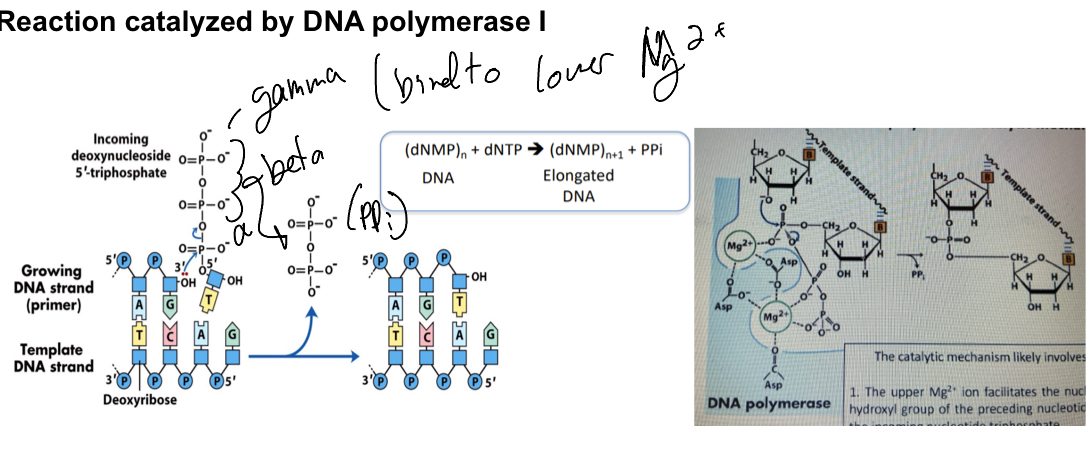
Explain the DNA polymerase I catalytic mechanism
DNA polymerase one accounts for 90% of DNA piolymease activity in E coli and has exonuclease proofreading. It tpically dissociates after 3-20 bases
Upper Mg2+ ion facilitates the nucleophlic attack of the 3-hydroxyl group of the preceding nucleotide on the alpha phosphate of the incoming nucleotide triphosphate
Lower Mg2+ helps remove the pyrophostphate (the beta and gamma phophate grroups)— hence linking this nucleotide to the one before it.
How is proofreading done on DNA?
Since for every 10^4 correct nucleotides added one is incorect,exonuclease removes the mispaired nucleotide and the poymerase bgins again.
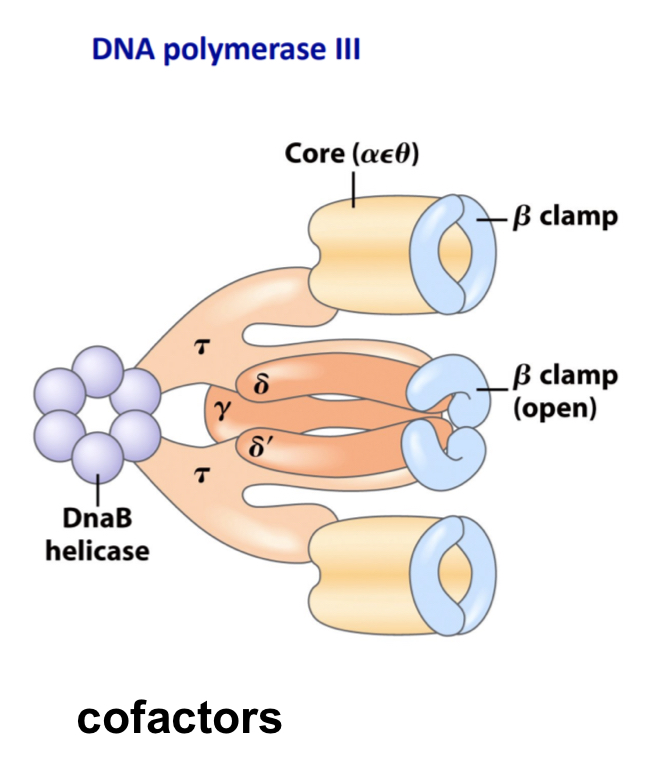
Describe the DNA polymerase III mechanism
10 types of subunits
polymerisation occurs in alpha subunits
Proofreading in epsilon subunit
High processivity provided by beta subunits as they associate in pairs to act as clamps preventing DNA dissoscation from DNA strands
Has 2 catalytic cores
What are Topoisomerases i and II?
Cope with the supercoiling of the DNA strands
Describe DNA replication mechanism in E coli
Initiation
proteins bind to DNA and open up the double helix
Prep of DNA for complementary pairing takes place
At least 9 protein cooperate in this step
Helicase denaturing
SSB (single stranded DNA binding proteins) prevent renaturalisation
Primer synthesized by primase
Elongation
2 catalytic cores ( made up of 2 beta subunits) carry out replication
Dna unwound by helicase, topoisomerases relieve the strand from supercoling
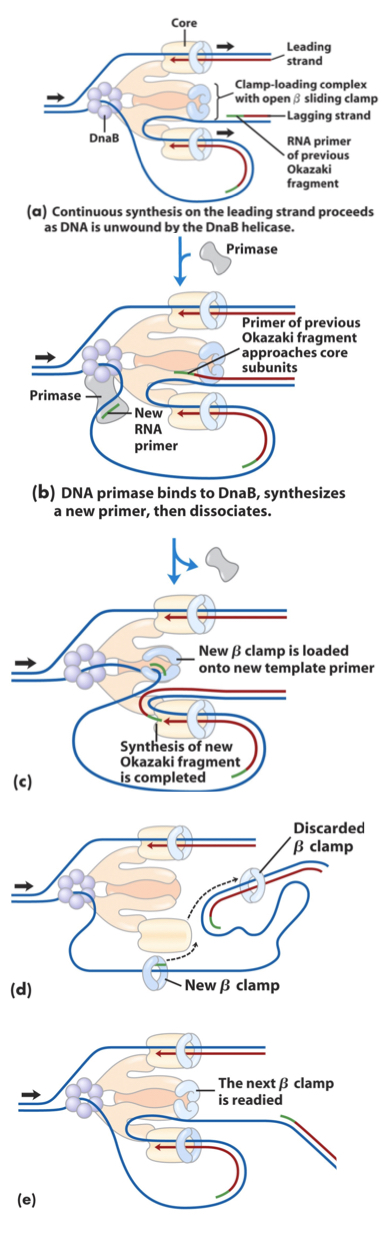
For okazaki segments: new primer added new beta clamp being added to primer spot and then moves along starnd synthesizing DNA
Then DNA polymerase I removes primer to replace it with DNA thru its exonuclease and polymerase activity…
the remaing nick is sealed byDNA ligase
Termination
replication forks encounter eachother where Ter sequences find each other functioning as binding sites for protein called Tus (terminus utilization substance).
Describe the mechanism of DNA replication in Eukaryotes
Same as E. Coli.. but…
Many more ORCs
polymerase alpha lacks 3’ to 5’ proofreading (exonuclease activity) but synthesizes RNA primers
Polymerase delta is equivalent of polymerase II in eukaryotes. PCNA subunit=beta clamp. Mainly performing elongation, used to detect tumors (appears in highly proliferating cells)
Polymerase epsilon eliminates primers from okazaki fragments
Polymerase gamma synthesizes mitochondrial DNA
