BIOL 251: Chapter 15 - translation
1/47
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
48 Terms
What were the two different types of fungal experiments/mutations, and which is which?
beadle and tatum (experiment)
Srb and Horowitz (mutations experiment)
what were the Beadle and Tatum experiments
grew mutated spores in complete medium
grew spores in minimal medium
why were spores grown in different types of media?
grew spores in different types of media to identify specific molecules missing
What did it mean if the spores in the minimal medium were unable to grow?
there is a mutation in pathway to biological molecule
what was the Srb and Horowitz mutation experiment
spores grown in media w/ precursors to biological molecules
allowed for determination of which step was inhibited by mutation
what did the Srb and Horowitz experiment conclude
each gene controls biosynthetic pathway
what was the one gene, one enzyme hypothesis?
genes function by encoding enzymes, and each gene encodes separate enzyme
You are studying a biochemical pathway and isolate mutants I, II, and III. Mutant I can grow if you supplement the medium with Z. Mutant II can grow if you supplement the medium with X, Y, or Z. Mutant III can grow if you supplement the medium with X and Z, but not if you supplement the medium with Y. What is the order of X, Y, and Z in this biochemical pathway?
what are enzymes
Proteins that catalyze biochemical reactions by lowering activation energy, thus increasing reaction rates.
are amino acids enzymes?
yes
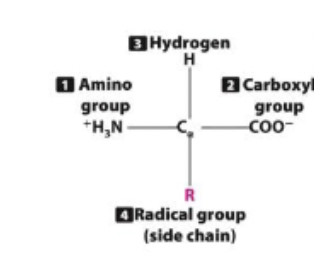
what is the structure and function of amino acids?
proteins are polymers of amino acids
all amino acids have central carbon w/: amino group, carboxyl group, R group
what are polymers
Long chains of repeating molecular units, such as amino acids in proteins.
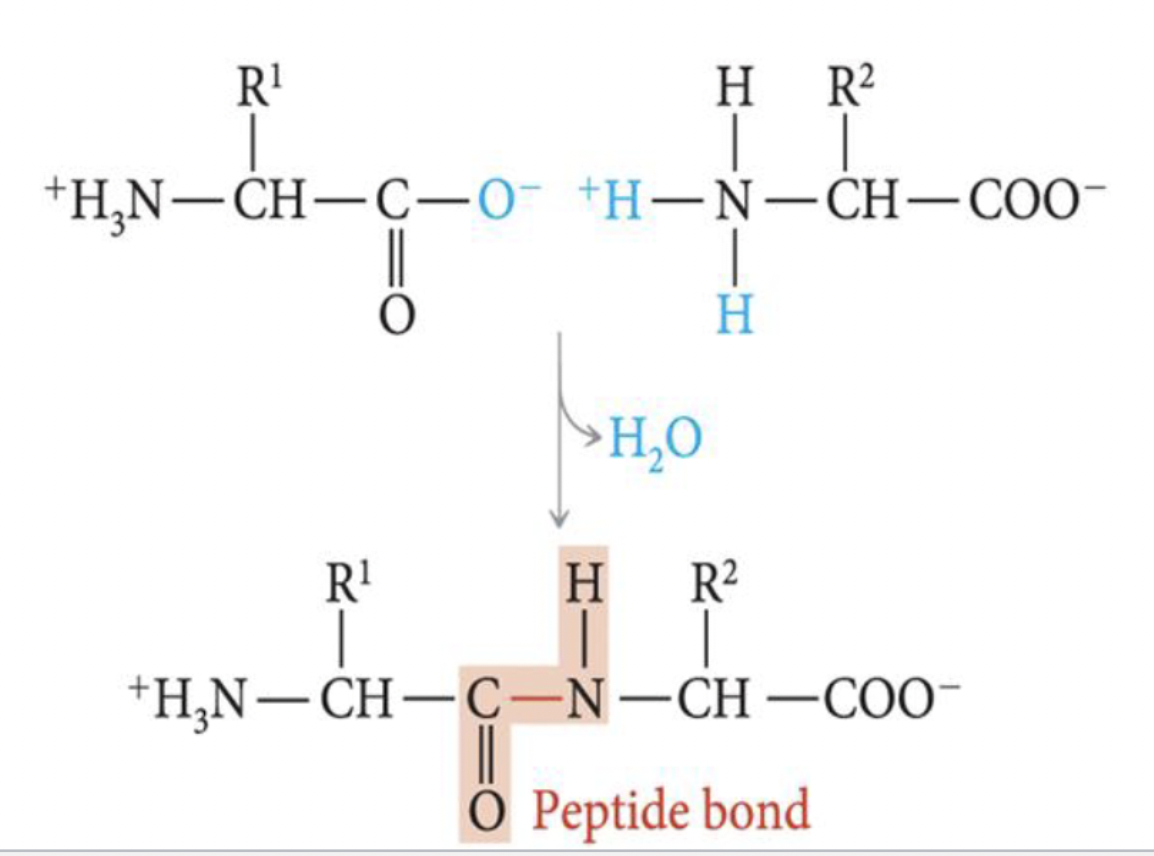
what is the structure of proteins?
formed by peptide bonds b/w amino acids
at least 50 amino acids connected to form chain
protein structures (primary, secondary, tert, quat)
primary: sequence of amino acids (series is based on codons)
secondary: folds and twists due to interactions b/w neighboring amino acids
tertiary: additional folds from amino amino acid interactions (3D structure)
quaternary: 2 or more tertiary structures (polypeptide chains) associate (multiple RNAs coming tg)
what is the domain in proteins?
group of amino acids that form discrete functional unit within protein
what did the Nirenberg and Matthaei experiments do and how?
calculate probability bases of codons
3C:2A
ex: CCA probability: 3/5×3/5×2/5=18/125
What was the procedure for the Nirenberg and Matthaei experiments?
1.) created homopolymer
2.) added homopolymer to tubes w/ individual amino acids
3.) protein only formed in 1 tube indicating amino acid coded for homopolymer
what were the Nirenberg and Leder experiments doing and what were the steps?
ribosome binding
mRNA that coded for known amino acids made
codon added to ribosomes, tRNA w/ amino acid
ribosome binds tRNA to mRNA
mixture filtered
what is the outcome of ribosome binding
tRNAs paired w/ ribosome-bound mRNA stuck to filter
unbound tRNAs pass through filter
what are the stop codons, how many are there, and what are their function?
UAA, UAG, UGA
3
terminate translation, do not code for amino acids
which are the sense codons and what do they do?
every one except for stop codons
encode amino acids
what are synonymous codons?
different codons that specify same amino acid
what are tRNA codons?
each type attaches to a single type of amino acid
some amino acids carried by iso-accepting tRNAs
three-nucleotide sequences within a tRNA molecule
what are iso-accepting tRNAs
accept same amino acid but have different anticodon sequnce
how does binding tRNA and mRNA codons occur?
5’ end of codon forms hydrogen bond w/ third 3’ base of anticodon
middle bases of codon and anticodon bind
What is wobbling when tRNA and mRNA codons bond?
weak pairing of third bases on codon and anticodon
especially when bases do not form Watson-Crick pair
what is the reading frame when the code is non-overlapping?
way that nucleotide sequence is read in groups of three nucleotides during translation
(overlap codons when reading the sequence until you reach then no more overlapping them when reading the sequence in groups of 3 (example in NB))
what is the initiation codon and which one is it for eukaryotes/bacteria cells?
1st codon of mRNA to specify amino acid
eukaryotes cells: Met (AUG)
bacteria cells: fMEt (AUG)
ALWAYS start w/ methayanine
There are three types of RNA-RNA interactions of protein synthesis, what are the mRNA-mRNA interactions?
ribosome attaches to 5’ end of mRNA
There are three types of RNA-RNA interactions of protein synthesis, what are the mRNA-tRNA interactions?
tRNA brings amino acid based on codon (ex: anticodon for AUG is UAC.)
There are three types of RNA-RNA interactions of protein synthesis, what are the tRNA-tRNA interactions?
ribosome hold tRNA in place so peptide bond can form
what are the 4 stages of protein synthesis(translation), and which are different/same for eukaryotes/bacteria?
tRNA charging (same for both)
initiation (different for each)
elongation (same for both)
termination (different for each)
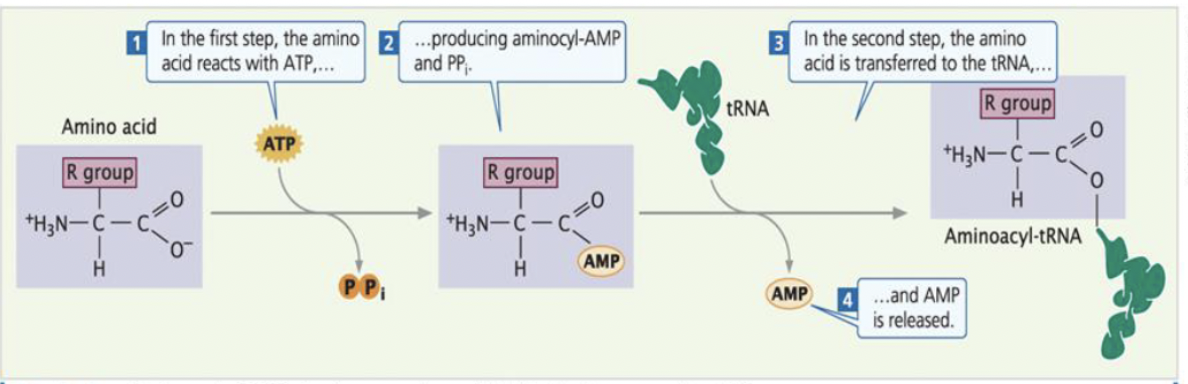
what are the steps for charging tRNA?
aminoacyl-tRNA synthetase per amino acid
recognition of amino acid based on different sizes, charges,a nd R groups of amino acids
amino acid + tRNA + ATP » aminoacyl-tRNA + AMP +PPI
LOOK AT SLIDE
what are the 4 steps to initiation in bacteria?
ribosome has 30S (small) subunit and 50S subunit
mRNA binds to 30S and initiation factors prevent 50S from binding
ribonuclease degrades mRNA except regions covered by ribosome (includes start codon)
Shine-Dalgarno sequence pair w/ complementary nucleotides in 16S rRNA to create initiation complex
what is the 30S complex, and what are the 5 requirements to create the 30S complex?
initiation complex that binds to mRNA during translation. The five requirements include:
mRNA
small ribosomal unit (16S)
Initiation factors (IFs)
fMet-tRNA
1 molecule of GTP.
what is needed to create the 70S complex?
GTP hydrolyzed
initiation factors dissociate from complex
large subunit binds
is the shrine-dalgarno seqeunce in both eukaryotes and bacteria?
no, only in bacteria
what are the 7 steps to initiation in eukaryotes
CBC binds to 5’ cap, which aids in export of mRNA from nucleus to cytoplasm
5’ cap recognized by initiation complex of: small ribosomal subunit, initiation factors, and initiator tRNA w/ amino acid
pioneer translation checks for errors in mRNA
CBC replaced by initiation factor that promotes translation
poly-A tail proteins interact w/ 5’cap proteins to enhance binding of mRNA w/ ribosome
Kozak sequence surrounds start codon (protector)
initiation factors relecase to allow large subunit to join
what does APE stand for in elongation?
aminoacyl site, peptidyl site, exit site
what are the 4 steps to elongations?
P site occupied by fmet tRNA
complex enters A site of ribosome
formation of peptide bond b/w amino acids that are attached to tRNAs in P and A sites
translocation: moving ribosome down mRNA
how does termination occur in bacteria?
protein release factors bind to A site
RF1 and Rf2 bind to stop codons
RF3 binds to GTP
promotes cleavage of tRNA in P site and releases polypeptide chain
mRNA and ribosome dissociate
how does termination occur in eukaryotes?
only 2 RF proteins:
RF1 for stop codons
RF3 for GTP
what is the difference b/w termination in bacteria and eukaryotes?
prokaryotes extra RF factor (RF2), while eukaryotes do not
polyribosomes
multiple ribosomes on transcript
what is the mRNA surveillance of polyribosomes?
non-sense mediated mRNA decay
errors in splicing or mutations lead to nonfunctional proteins
rapid eliminations of mRNA containing premature termination codons
what is ribosome stalling in polyribosome?
no termination codon present due to mutation or transcription error
how does the stalled ribosome get removed?
transfer-messenger RNA (tmRNA) removes it and adds amino acids tag to polypeptide so cell degrades incomplete protein
what are post-translational modifications in eukaryotes?
ubitiquin added to protein to signal degradation
removal of signal sequence
chaperone proteins aid in folding and preventing damage
trimmed and modified if subunit of larger protein
carbohydrate activation
phosphates, carboxyl groups, and methyl groups added to amino acids