Biology: DNA, Cell Cycle, Mitosis, Meiosis
1/91
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
92 Terms
Friedrich Miescher’s scientific discovery (1868)
investigated the chemical composition of the nucleus and concluded that the nucleus contains material that is not a protein but another phosphorus containing material
used cells from the pus from hospital bandages
isolated an organic acid that was high in nitrogen and phosphorus (he called it nuclein) - importantly, it was NOT a protein
Frederick Griffith’s scientific discovery (early 1900s)
in trying to develop a vaccine, found that the nuclear material that Miescher discovered had the ability to transform non pathogenic bacteria into pathogenic bacteria (transforming principle)
injected mice with live and dead forms of strep pneumoniae to trigger antibody response
Rough (R) strain did not kill the mice
Smooth (S) strain killed the mice
something holding onto the trait for lethality was transferred
Oswald Avery, Colin MacLeod, & Maclyn McCarthy’s scientific discovery (1940)
concluded that the “transforming principle” was a nucleic acid
chemically separated deadly smooth strain bacterial cells by destroying different components
mixed with non-lethal rough strain and injected into mice
Erwin Chargaff’s scientific discovery
researched the nitrogenous bases in DNA by pooling and separating them, concluding with two “rules”
for all species, the percentages of A=T and G=C
between different species there can be some variation in A, T, G, C amounts
Wilkins and Rosalind Franklin’s scientific discovery (1940s-50s)
used x-ray crystallography to find that DNA was a long, thin molecule in a double helix shape, the rungs of the ladder were .34 nm, one twist was 10 rungs (3.4 nm) long, and width was consistent at 2 nm
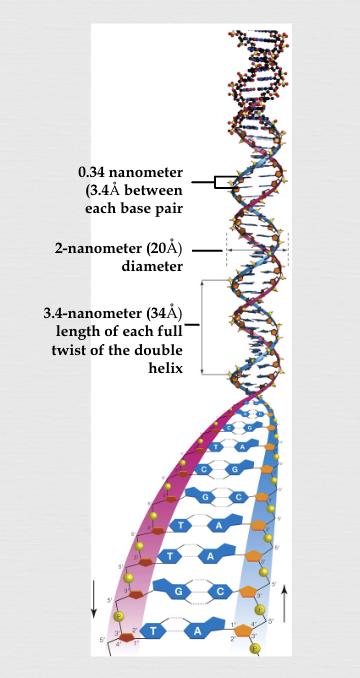
Watson and Crick’s scientific discovery (1953)
used Franklin’s x-ray results and other scientists’ discoveries as a basis to build a wood and metal model for the ground-breaking discovery of the “double-helix” structure for DNA (put all together into a one page Nature article):
consists of 2 strands of nucleotides (not 3 as Linus Pauling suggested)
sugars and phosphate make up the sides of the ladder
nitrogenous bases make up the rungs of the ladder (some had suggested they were on the outside)
nucleotide rung held together by hydrogen bonds (weak)
the bonds form when each strand runs in opposite directions (anti-parallel) of the other (an a-Ha discovery)
Structure of DNA
double helix
two strands twist around each other to form a double helix, a spiral staircase shape
sides run anti-parallel
backbone
dioxyribose sugar and phosphate groups form the backbone of each strand, running along the outside of the helix
bases
attached to each sugar molecule are one of the four nitrogenous base (A, T, G, C)
base pairing
the two strands are connected by hydrogen bonds between specific base pairs: A-T and G-C
complementary strands
because of base pairing, one strand of DNA can be used to determine the sequence of the other, making it possible to copy and replicate DNA
5 end has phosphate group
3 end has hydroxyl group
genetic information
the sequence of the bases encodes the genetic information, carrying instructions for making proteins and other biological molecules
chromatin
DNA in its un-condensed form; the complex of DNA and proteins, primarily histones, that makes up the genetic material of eukaryotic cells
exists in different forms within the nucleus
thin wispy, threadlike
in a non-dividing cell
usually too thin to be visible
DNA packaging
primary function is to package DNA into a compact form that fits within the nucleus
structure
basic unit is the nucleosome, where DNA is wrapped around histone proteins
dynamic nature
structure can change during the cell cycle, including condensation into chromosomes during mitosis
gene expression regulation
plays a role in regulating the gene expression by controlling access to DNA
cell cycle regulation
histone modifiers, which regulate chromatin structure, are also regulated by the cell cycle
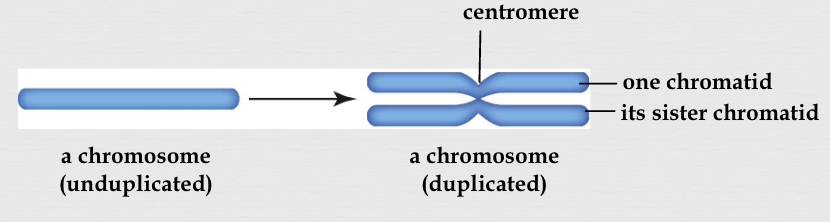
chromosome
DNA in a fully condensed form; a structure within a cell’s nucleus that carries DNA
made up of DNA tightly coiled around histone proteins
can be single stranded or double stranded (X shape)
DNA in preparation for cell division
DNA packaging
DNA is a very long molecule, so its tightly packed and organized into chromosomes to fit within the cell’s nucleus
structure
threadlike structures, visible under a microscope during cell division when DNA is most condensed
genes
contain genes, which are specific DNA sequences that code traits
human chromosomes
humans have 23 pairs of chromosomes, 46 total chromosomes
cell division
are essential for accurate DNA replication and distribution during cell division, ensuring that each daughter cell receives a complete set of genetic information
histone protein
basic proteins that play a crucial role in organizing DNA within the nucleus of eukaryotic cells and regulating gene expression
package DNA into structural units called nucleosomes
influence how tightly DNA is packed
chromatin wrap around to help 3m of DNA pack into a small nucleus (“spool of thread”)
DNA packaging
responsible for wrapping DNA around themselves, forming nucleosomes. which are then further organized into higher-order structures (ie. chromosomes, chromatin)
this process compacts DNA, allowing it to fit within the nucleus and enabling efficient cell division
gene expression regulation
packaging of DNA by histones can influence whether genes are accessible for transcription
histones can be modified to change accessibility of DNA, thereby influencing which genes are turned on or off
chromosome structure
provide structural support for chromosomes, ensuring proper organization and function during cell division
DNA protection
help protect DNA from damage by physical forces and chemical agents
nucleosome
the fundamental structural unit of chromatin, the DNA-protein complex that makes up eukaryotic chromosomes
consists of DNA wrapped around a core of histone proteins - this packaging allows for the efficient organization and compaction of DNA within the cell nucleus
components
made up of approx. 147 base pairs of DNA wrapped around a complex of 8 histone proteins
DNA wrapping
DNA is wrapped around the histone complex
structure
arranged in a beadlike structure along the DNA, with the DNA between nucleosomes called linker DNA
function
play a crucial role in DNA packaging and gene expression regulation
by organizing DNA into compact structures, nucleosomes help protect the genome from damage and regulate access to genetic information
dynamics
are dynamic structures that can be moved, assembled, and disassembled in a regulated fashion to allow for DNA replication and gene expression
sister chromatids
identical copies of a single chromosome that are formed during DNA replication and remain joined together by a centromere
essential for ensuring each daughter cell receives a complete set of chromosomes during cell division
formation
during the S phase of the cell cycle, DNA replication duplicates each chromosome, resulting in 2 identical copies
attachment
are attached to each other at the centromere, a constricted region of the chromosome
role in cell division
are crucial for accurate chromosome segregation during mitosis and meiosis
segregation
during anaphase, spindle fibers attach to the centromeres and pull the sister chromatids apart, ensuring each daughter cell receives one complete set of chromosomes
DNA repair & chromatin structure
play a role in DNA repair and maintain chromatin structure
centromere
a constricted region on a chromosome that serves as the attachment point for spindle fibers during cell division, ensuring accurate chromosome segregation
a specialized structure composed of both DNA and proteins
crucial for chromosome movement and control of gene expression
attachment site for spindle fibers
provide the anchor point for the microtubules of the spindle, which are responsible for pulling chromosomes apart during cell division
sister chromosome cohesion
holds sister chromosomes together until they are separated during anaphase
chromosome segregation
are essential for proper segregation of chromosomes into daughter cells
karyotype
a pictorial array of all the chromosomes in an organism’s cell during mitosis
arranged in pairs by length and centromere location and banding pattern
“map” of an individual’s chromosomes
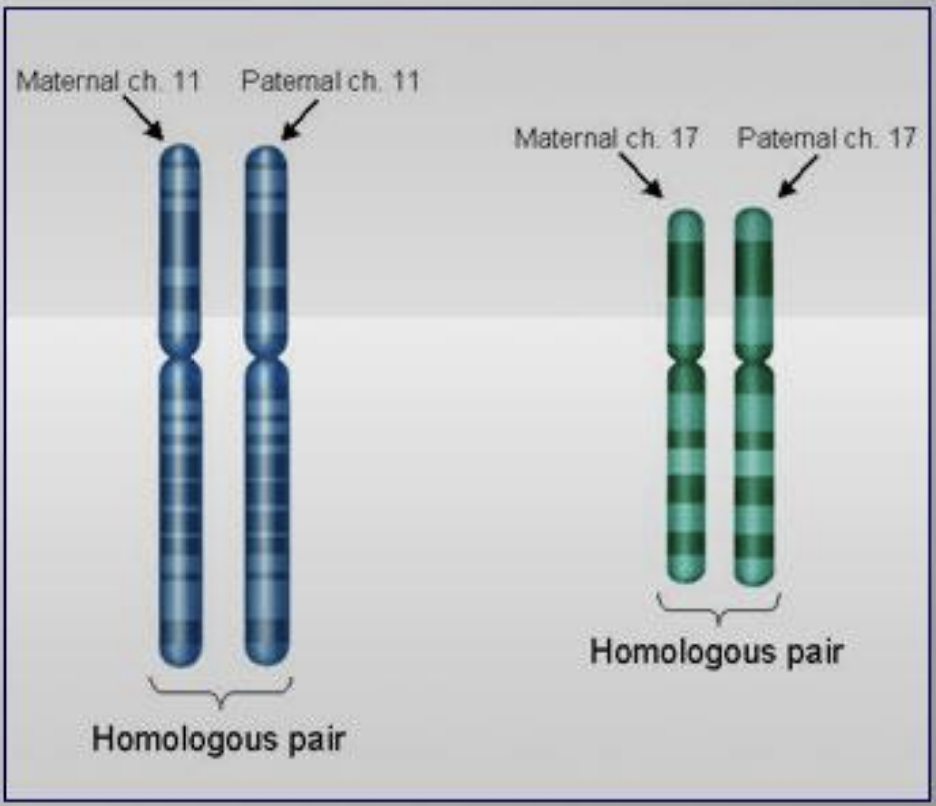
homologous chromosomes
matching pairs of chromosomes of approximately the same length, centromere position, and staining pattern that hold the same genes
in a diploid organism, where one chromosome is inherited from each parent
carry the same genes in the same order, but may have different versions of those genes (alleles)
chromosome number
the sum of all chromosomes in a cell of a given organism
autosome
chromosomes that are not involved in determining the sex of an organism
come in pairs - chromosome pairs #1-22
each carries numerous genes that control various traits and functions in an organism
sex chromosome
chromosomes that are responsible for determining the sex of an organism
one pair - chromosome pair #23
primarily determines biological gender
XX = female
XY = male
Okazaki fragments
short, newly synthesized DNA sequences on the lagging strand during DNA replication
formed because the lagging strand is synthesized discontinuously in a direction opposite to the replication fork
these fragments are later joined together by DNA ligase to form a continuous strand
lagging strand
during DNA replication, one DNA strand is synthesized continuously in the 5-3 direction, while the other strand (the lagging strand) is synthesized discontinuously
discontinuous synthesis
the lagging strand synthesis is discontinuous because DNA polymerase, the enzyme that builds new DNA can only move in the 5-3 direction, and the lagging strand runs in the opposite direction of the replication fork
formation
as the replication fork moves forward, DNA polymerase must detach and reattach at multiple points to synthesize the lagging strand in short fragments
RNA primers
each fragment is initiated by a short RNA primer, which is later replaced by DNA polymerase and the fragments joined together by DNA ligase
genome stability
proper processing of fragments is crucial for maintaining genome stability and preventing DNA damage
defects in this process can lead to DNA breaks and chromosome abnormalities
spindle fibers
array of microtubules that move chromosomes around the cells
crucial for cell division in eukaryotic cells
responsible for segregating chromosomes during mitosis and meiosis, ensuring each new cell receives the correct number of chromosomes
structure
formed by microtubules
function
chromosome movement
attach to chromosomes and use their motor proteins to pull them apart during cell division
cell division
help to push the cell poles apart, which is necessary for cytokinesis
formation
during prophase, spindle fibers start to form from centrosomes, which are located at the poles of the cell
centrosome (with centrioles)
organelle responsible for making microtubules (spindle fibers) that are used to move chromosomes around during cell division in animal cells
acts as the main microtubule-organizing center, regulating cell shape, polarity, and cilia formation
during cell division, centrosome duplicates and migrates to opposite poles of the cell, where it nucleates the mitotic spindle, ensuring chromosomes are evenly distributed to daughter cells
microtubule organization
primary site for microtubule nucleation, meaning it initiates the growth of microtubules
mitotic spindle formation
during cell division, the duplicated centrosomes move to opposite poles of the cell and act as the organizing centers for the mitotic spindle, a structure that separates chromosomes
asexual reproduction
form of reproduction in which offspring arise from one parent and inherit that parent’s genes only
offspring are genetically identical to single parent
sexual reproduction
form of reproduction in which genetically variable offspring are produced by the fusion of two parents
offspring have unique combination of genes from both parents
genetic variation
the differences in DNA sequences between individuals within a population or species
differences are essential for evolutionary change and are caused by various mechanisms
mechanisms:
mutations
gene flow
genetic recombination
importance
evolutionary change
adaptation
unique individuals
* crossing over (Prophase 1) and random alignment (Metaphase 1) ensure that no two reproductive cells are alike
interphase
longest phase of the cell cycle where a cell increases in mass, roughly doubles the number of components (organelles) in its cytoplasm, and duplicated its DNA
3 stages:
G1 (Gap 1)
S (Synthesis)
G2 (Gap 2)
G1 (Gap 1)
first interval where cellular contents, excluding the chromosomes are duplicated
cell growth and functioning (doing its job)
lasts 10-12 hours
S (Synthesis)
DNA replication in preparation for division
each of the 46 chromosomes is duplicated by the cell
lasts 4-6 hours
G2
second interval where cell makes proteins that will drive cell division
the cell “double checks” the duplicated chromosomes for error, making any needed repairs
lasts 4-6 hours
G0
resting phase of the cell cycle
cell cycle arrest
* not all cells are rapidly proliferating; the heart muscle, skeletal muscle, and nerve cells are “arrested” in G1 early in human development and never proceed past that point
proto-oncogene
genes that code for normal cell division that can turn into oncogenes - gene that transforms a normal cell into a cancer cell
checkpoints
the cell cycle has built-in checkpoints where specific proteins, products of checkpoint genes, monitor the structure of chromosomal DNA
two categories:
growth factors - stimulate cell division
tumor suppressors - control/slow/stop cell division
Spindle assembly checkpoint
check for chromosome attachment to spindle
G1 checkpoint
check for cell size, nutrients, growth factors, DNA damage
G2 checkpoint
check for cell size and DNA replication
* if a checkpoint gene mutates and its protein product no longer functions, the cell can lose control of the cell cycle (cell can become stuck in mitosis → tumors)
apoptosis
a natural, programmed form of cell death that is crucial for development and tissue maintenance in multicellular organisms
involves a tightly regulated process where the cell’s own enzymes break it down into small, membrane-bound fragments, which are then removed by other cells without causing inflammation
crucial role in various biological processes
controlling cell populations
eliminating damaged or infected cells
programmed cell death
is a genetically controlled process, meaning it follows a specific set of biochemical steps
morphological changes
during apoptosis, cells shrink, nucleus condenses, outer membrane forms small protrusions
cell fragmentation
cell breaks down into small, membrane-bound fragments called apoptotic bodies
phagocytosis
apoptotic bodies are engulfed and removed by neighboring cells
growth factor
a naturally occurring signaling molecule, often a protein, that stimulates cell growth, differentiation, survival, and sometimes tissue repair
accelerator - a mutation that keeps division happening
stimulate cellular processes
play a vital role in regulating cell growth and development by stimulating processes like:
proliferation - increase in cell number, often through mitosis
differentiation - cell becomes specialized to perform a specific function
survival - helping cells to resist apoptosis
tissue repair - wound healing and other repair processes
cell communication
act by binding to specific cell surface receptors, which trigger signals that affect gene expression and cell behavior
tumor suppressor
a gene that normally helps control cell growth and division, preventing cells from growing out of control and developing tumors
brakes - a mutation that slows down or stops division, maintaining cellular balance
normal function
encode proteins that regulate cell cycle, ensuring that cells divide at the right time and die when they should (apoptosis)
play a role in DNA repair, helping to correct errors that could lead to mutations
mechanism of action
work by inhibiting cell growth, promoting cell death, and ensuring proper cell-cycle checkpoints
loss of function & cancer
mutations in tumor suppressor genes can disrupt their function, leading to uncontrolled cell death, which is a hallmark of cancer
in many cancers, 1+ tumor suppressors are inactivated
tumor (neoplasm)
mass of cells dividing at an abnormally high rate that forms a mass or a lump
two types:
benign - slow growing tumor that stays localized in home tissue (ie. moles)
malignant - tumor cells that rapidly divide, metastasize, and invade other places in the body where they may start new tumors
metastasis
the process where cancer cells from a primary tumor spread to distant parts of the body, forming secondary tumors
cytokinesis
the process of cytoplasmic division during cell division, resulting in the formation of two daughter cells
follows mitosis or meiosis
ensures each new cell receives its share of organelles and other cytoplasmic components
cell division
is the second major phase of cell division, alongside mitosis or meiosis
Mitosis involves the division of the cell's nucleus, while cytokinesis separates the cytoplasm
cytoplasmic division
the cell’s cytoplasm, including organelles and other cellular components, is divided into two separate portions
daughter cells
result is two daughter cells, each containing a copy of the original cell's genetic material and its own set of cytoplasmic components
cleavage furrow
an invagination of the plasma membrane during cytokinesis
begins as a shallow groove and gradually deepens, eventually pinching the cell membrane in two to form two separate daughter cells
*animal cytokinesis
contractile ring
a ring-shaped structure of proteins that forms beneath the cell membrane during cell division, specifically during cytokinesis
constricts the cell, creating a cleavage furrow and physically separating the cell into two daughter cells
*animal cytokinesis
cell plate
a structure that forms to divide the cytoplasm and create a new cell wall between the two daughter cells
disc-like structure that builds a partition between the two new cells
*plant cytokinesis
haploid
a cell or organism containing only one complete set of chromosomes
has half the usual number of chromosomes as full complement
set represents one copy of each chromosome
does not possess any homologous pairs of chromosomes
ie. in humans, haploid cells are specifically the gametes (sperm and egg cells)
diploid
a cell or organism that has two complete sets of chromosomes
has two copies of each chromosome
in most sexually reproducing organisms, this second set of chromosomes is inherited from the other parent
possesses 2 homologous pairs of each chromosome, one inherited from each parent
somatic cell
all the cells in a multicellular organism except for gametes (sperm and egg cells) and the cells that produce them
essentially, any body cell that is not a germ cell
make up the body tissues and organs
diploid
typically contain two copies of each chromosome, one inherited from each parent
mitosis
divide through mitosis, a process that produces two identical daughter cells
not reproductive
not involved in the production of new offspring
differentiated
are specialized and can perform different functions within the body
sex cell
a reproductive cell (sperm or egg) that contains half the normal number of chromosomes (haploid)
aka gametes - specialized cells involved in sexual reproduction
combine during fertilization to form a zygote with the full diploid number of chromosomes
haploid
have a single set of chromosomes, while other cells in the body have two sets
fertilization'
when a sperm and egg cell fuse, their haploid nuclei combine to form a diploid zygote
meiosis
are produced through meiosis, which reduces the chromosome number by half
gonad
a reproductive organ that produces gametes (sex cells) and hormones
females: ovaries, produce eggs and female hormones
males: testes - produce sperm and male hormones
testes
the paired male reproductive glands responsible for producing sperm and testosterone
ovary
the female reproductive organ responsible for producing eggs and estrogen & progesterone
synapsis
the pairing of homologous chromosomes during early meiosis
ensures that each resulting cell has the correct number of chromosomes after cell division
allows for the exchange of genetic material between homologous chromosomes, leading to the formation of tetrads, which then undergo crossing over
occurs in Prophase 1
tetrad
the structures formed by the paired homologous chromosomes after synapsis
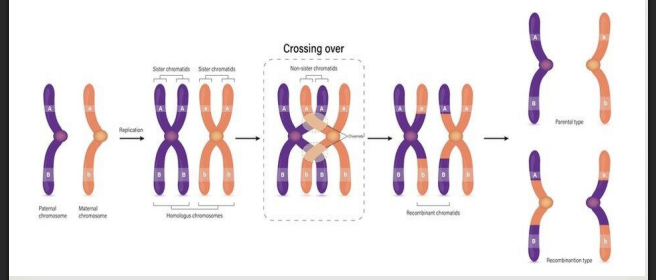
crossing over (genetic recombination)
the exchange of genetic material between homologous chromosomes that happens during/after synapsis which leads to increased genetic diversity in offspring
during Prophase 1 crossing over, a tetrad forms and sections of non-sister chromatids break off, switch places, and reattach
during Meiosis 1 Prophase 1 the homologous chromosomes line up closely in a process called synapsis
the 2 aligned chromosomes are called tetrads (four chromatids)
during Prophase 1, an enzyme called Recombinase acts as scissors and cuts sections from one chromosome and pastes it on another - the results are recombinant chromosomes (cut, switched, recombined)
* chiasma is the site of the crossing over (appears as an X)
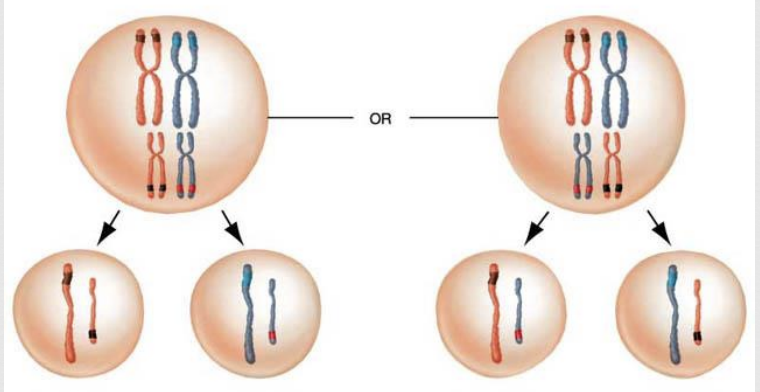
random alignment
the second mixing of genes that happens during Metaphase 1
during Metaphase 1 paternal/maternal pairs randomly position themselves along the midline of the cell
spermatogenesis
sperm producing process
in males, the spermatagonium are arrested before they begin Meiosis and will not start Meiosis until puberty
takes about 100 days from start to finish
millions of spermatagonium cells in the testes are in various stages of Meiosis on any given day
oogenesis
ova (egg) producing process
primary oocytes (immature eggs) in women are produced in the fetus but are halted at Meiosis 1 Prophase 1 until puberty
this is why women are said to have a limited number of eggs determined at birth
of the millions of primary oocytes present in a woman’s ovaries, only about 400 will mature during a woman’s lifetime
only 1 egg is needed so 3 additional egg cells (polar bodies) that are produced will eventually decay
polar bodies are smaller
all of the “good stuff” - cytoplasm, organelles, etc. - are found in the designated egg
the egg is produced in day 14-18 of oogenesis
roughly 12 days after ovulation, if the egg hasn’t been fertilized, the unfertilized egg and uterine lining in released in the menstrual cycle (monthly)
at menopause, hormones close down the process of oogenesis (Meiosis)
gamete
haploid reproductive cells produced as a result of meiosis
two types of animal:
sperm
ovum
zygote
the first cell in a new organism when a sperm and ovum (egg) cell fuse
sperm
haploid male sex cell
ovum (egg)
haploid female sex cell
polar bodies
the cells produced in oogenesis that are targeted to decay and not become eggs
mitosis
purpose:
organism growth
cell size limitation (SA:V ratio)
replace dead cells
repair damaged/injured tissue
done by: somatic (body) cells of multicellular eukaryotes
#divisions: 1
location: occurs actively in all cells of the body (except nerve cells and skeletal muscle cells)
sexual or asexual: asexual
#genetically identical cells produced: 2
Diploid cells are created; produces cells with full complement of chromosomes as parent cell
meiosis
purpose:
done by: sex cells (gametes)
#divisions: 2
location: occurs only in the reproductive organs (testes and ovaries)
sexual or asexual: sexual
#genetically identical cells produced: 4
Haploid cells are created; produces cells with half the chromosomes as the parent cell
* cells do not re-enter interphase between Meiosis 1 and Meiosis 2
* crossing over and random alignment cause genetic variation
* all genetic variability occurs in Meiosis 1
At the end of meiosis I, are the daughter cells 2n or n? How many chromatids does each chromosome have at that point?
at the end of meiosis 1. the daughter cells are haploid (n) and each chromosome still consists of two sister chromatids
At the end of meiosis II, are the daughter cells 2n or n? How many chromatids does each chromosome have at that point?
The daughter cells at the end of meiosis II are haploid (n) and each chromosome has one chromatid
how many homologous pairs are there in the human body?
23; the last determining biological gender
cytokinesis plant cell versus cytokinesis plant cell
plant cell
plant cells divide using cell plate formation = vesicles packed with cell wall building materials (from the Golgi bodies) form along the midline and fuse with one another forming the cell plate
animal cell
animal cells divide by partitioning their cytoplasm using a contractile ring mechanism = a thin band of contractile filaments just below the plasma membrane contracts forming a cleavage furrow, and then pinches the cytoplasm in two
Mitosis plant cell versus Mitosis animal cell
plant cell
animal cell
What does “reduction” mean in Meiosis?
In Meiosis, "reduction" refers to the reduction of the chromosome number from diploid (2n) to haploid (n). This is achieved through the two stages of Meiosis I and Meiosis II, where homologous chromosomes separate in the first division, and sister chromatids separate in the second division, resulting in four haploid daughter cells.
meiosis 1 (reduction division)
During Meiosis I, homologous chromosomes separate, resulting in two daughter cells each with half the number of chromosomes as the original cell.
meiosis 2 (equational division)
Meiosis II separates sister chromatids, similar to mitosis. While it does not further reduce the chromosome number, it does result in four daughter cells.
purpose
This reduction in chromosome number is crucial for sexual reproduction. When a sperm (haploid) and egg (haploid) fuse, they combine their genetic material to form a diploid zygote, ensuring the correct chromosome number for the next generation.
What role do spindle fibers play in Mitosis/Meiosis?
Spindle fibers, made of microtubules, are crucial for separating chromosomes during both mitosis and meiosis. They attach to chromosomes at the centromeres, ensuring each new daughter cell receives an accurate number of chromosomes.
mitosis: pull sister chromatids apart to opposite poles
meiosis: separate homologous chromosomes in meiosis I and sister chromatids in meiosis II
Human Genome Project (2003)
developed the complete genetic map (mapped the entire human genome)
found that not just a single gene was causing a disease but the WHOLE THING
helped determine what each of the chromosomes do
there are over 20,000 genes in the human genome that are responsible for traits or disorders
3 billion base pairs
nucleotide
monomer of nucleic acids (DNA) that contains:
a deoxyribose
a phosphate group
a nitrogen-containing base (Adenine, Guanine, Thymine, Cytosine)
Purine
the bigger, double carbon ring nitrogenous bases: Adenine & Guanine
Pyrimidine
the smaller, single carbon ring nitrogenous bases: Thymine & Cytosine
Base pairing rules
A pairs with T (2 hydrogen bonds)
G pairs with C (3 hydrogen bonds)
semi-conservative replication
half of original DNA is conserved in the new DNA molecule
DNA is 2 nucleotide strands held together by hydrogen bonds
hydrogen bonds between 2 strands are weak and easily broken
each single old strand then serves as a template for new complimentary strand
every cell has a big pool of free nucleotides available to join the original strand “template”
main enzyme actors in DNA replication
topoisomerase - untwists the double helix
DNA helicase - breaks hydrogen bonds between DNA strands
DNA primase - catalyzes the synthesis of a short chain of RNA because DNA can not initiate the synthesis of itself (this starter is called a “primer”)
DNA polymerase - joins free nucleotides into a new strand of DNA
DNA ligase - joins DNA fragments on the discontinuous strand
eukaryotic DNA replication steps
initiator proteins attach to DNA at certain sites on the chromosome (replication can typically start at multiple points for eukaryotes)
these proteins attract 2 key enzymes to the chromosome:
a) topoisomerase - unwinds the DNA
b) DNA helicase - breaks the hydrogen bonds joining the base pairs (this area where the strands are split is called the “replication fork”)
another enzyme, DNA primase, makes short RNA primers that attach to the DNA where new nucleotides will be added
DNA polymerase attaches to these primers and begins to assemble the complementary strand in “its” 5-3 direction
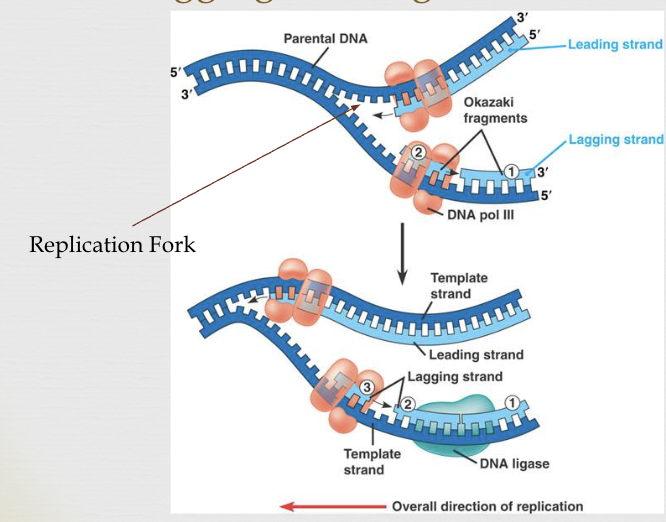
there is a continuous (leading strand) and a discontinuous strand (lagging strand)
the segments of the lagging strand (called “Okazaki fragments”) are joined by a third enzyme, DNA ligase
* helps maintain chromosome number
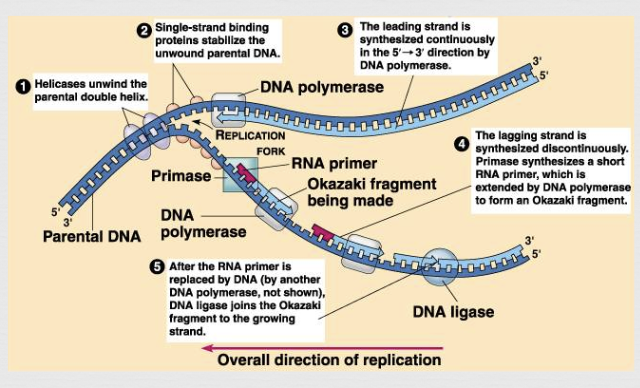
replication fork
area where continual separation of DNA strands occurs
leading (continuous) strand goes towards
lagging (discontinuous) strand goes away
mutation
mistake that occurs during DNA replication
those acquired during life can harm the organism
ie. skin cancer
inherited are already in cells, so gamete can pass them to offspring
ie. Huntington’s disease, sickle cell anemia, hemophilia
genes
heritable information in chromosomal DNA
Cell Cycle
a series of events from one cell division to the next
Interphase (95% time - 18-24 hours)
G1 (10-12 hours)
S (4-6 hours)
G2 (4-6 hours)
Mitosis (5% time - 0.5-1 hour)
Prophase
Metaphase
Anaphase
Telophase
Cytokinesis
* Eukaryotic cells spend 95% of their time in interphase so only a small portion of time in actual mitosis
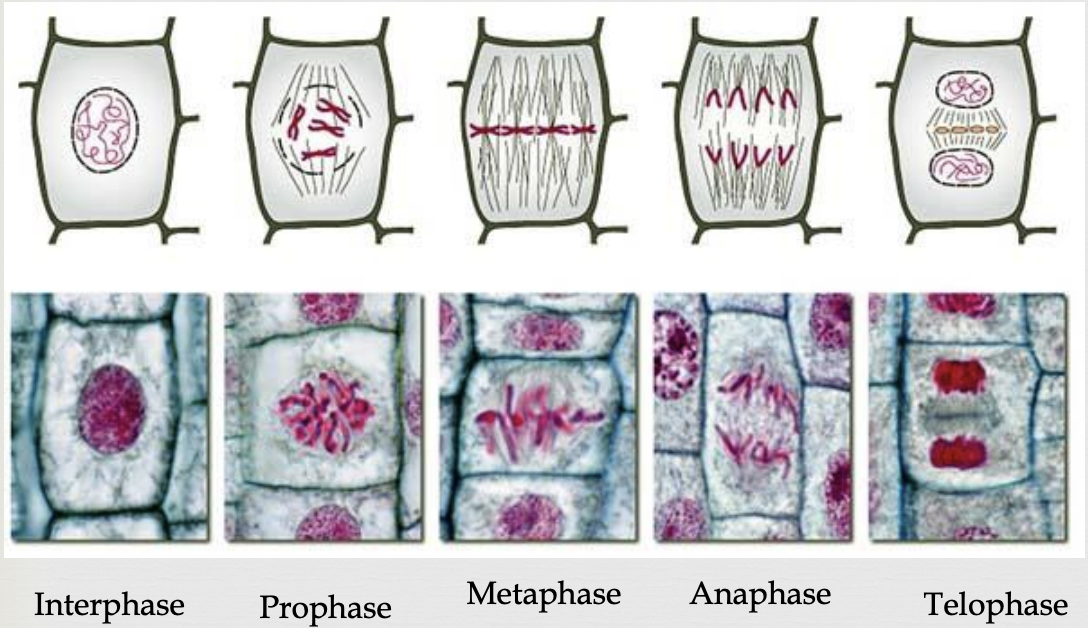
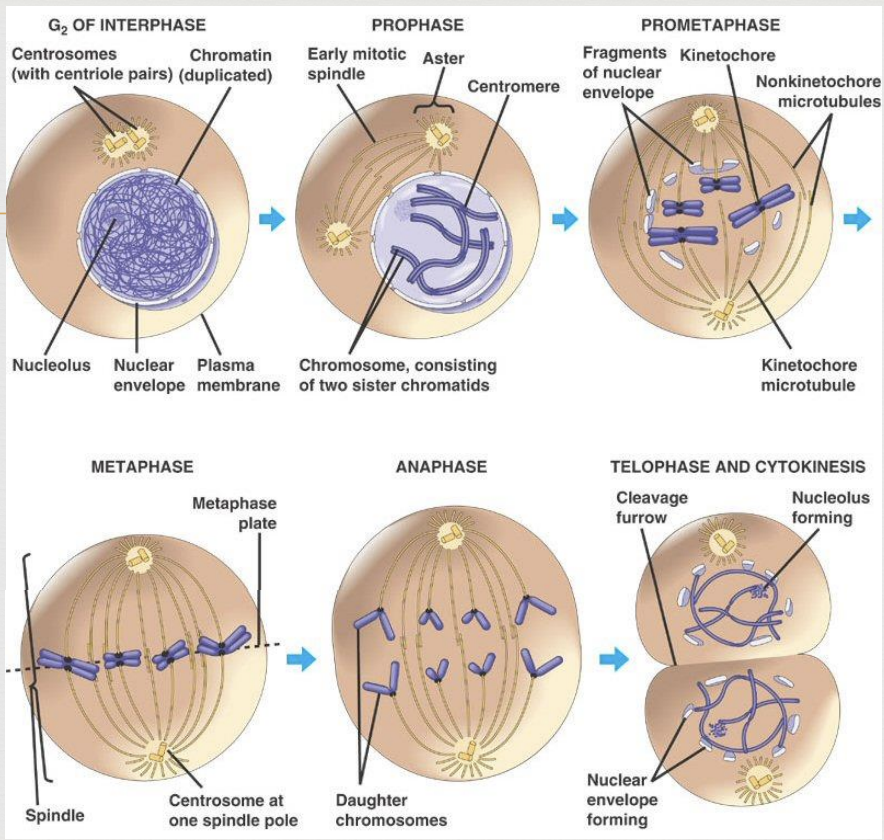
Mitosis Prophase
duplicated chromosomes begin to condense and become more visible
microtubules (spindle fibers) create a bipolar mitotic spindle that reaches from one pole to the other
nuclear envelope breaks up
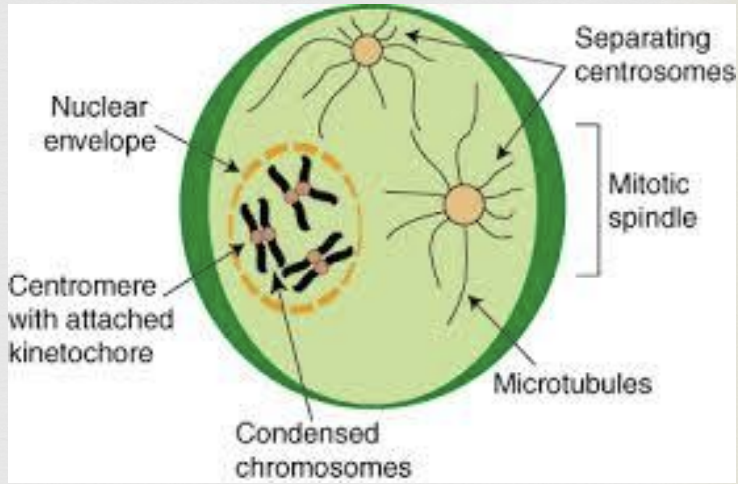
Mitosis Metaphase
microtubules (spindle fibers) move chromosomes to the center (metaphase plate) of the cell
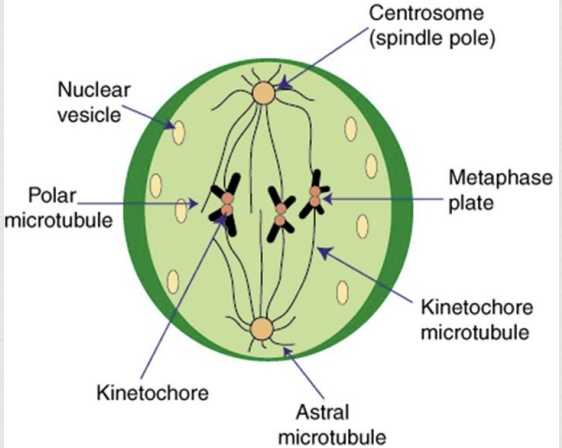
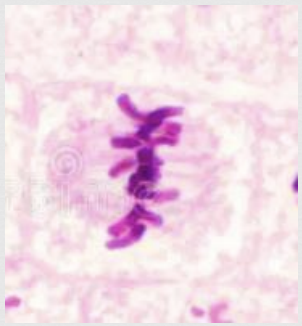
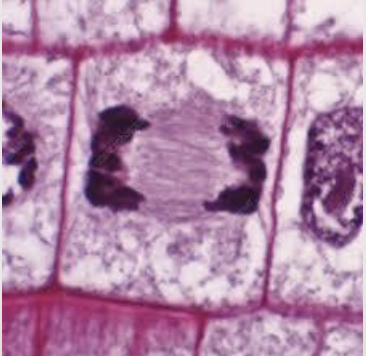
Mitosis Anaphase
sister chromatids split and become independent chromosomes
microtubules (spindle fibers) contract and drag sister chromosomes to opposite poles
once chromatids separate they are considered two separate chromosomes
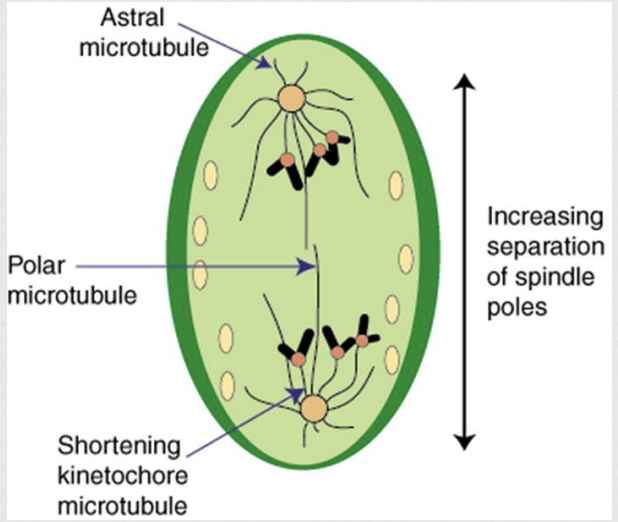
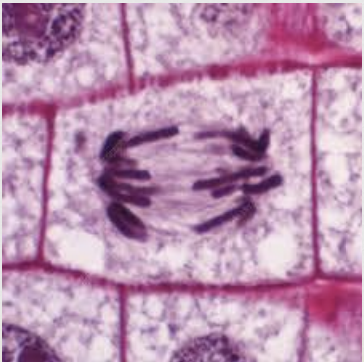
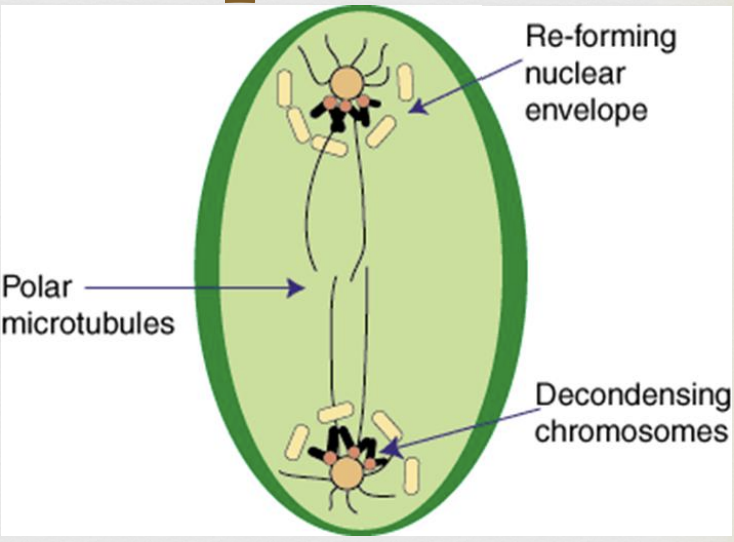
Mitosis Telophase
chromosomes begin to decondense
nuclear membranes begin to reappear
2 daughter cells form, each with the same chromosome number as the original parent cell
cytokinesis occurs (division of the cytoplasm)
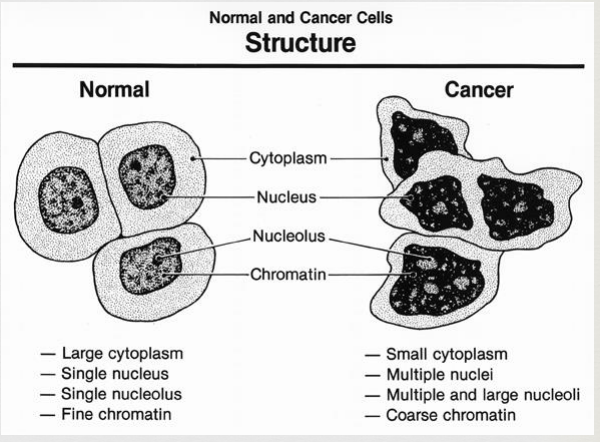
Cancer characteristics
abnormal growth and division
cytoplasm and plasma membrane become grossly altered
cytoskeleton becomes disorganized
membrane becomes leaky/missing proteins
cancer cells have a weakened capacity for adhesion
don’t stay anchored to the proper tissues (metastasis)
cancer cells can crowd out normal cells and also “infect” them, turning them into cancer cells
RNA primer
a short sequence of RNA that initiates DNA synthesis during DNA replication
serves as a starting point for DNA polymerase to add nucleotides to the template strand
are synthesized by an enzyme called primase
function
are essential for DNA replication because DNA polymerase, the enzyme responsible for synthesizing new DNA strands, cannot start adding nucleotides without pre-existing 3'-OH (hydroxyl) group to attach to the 3'-OH group at the end of the RNA primer provides this necessary starting point
complementarity
must be complementary to the DNA strand it will prime, meaning that its base sequence must match the base sequence of the DNA strand, according to the base-pairing rules (A-T) (G-C)
removal and replacement
once DNA synthesis is complete, the RNA primers are removed and replaced with DNA by specialized enzymes, ensuring a continuous DNA strand
importance
crucial for both leading and lagging strand DNA replication, especially on the lagging strand where multiple primers are needed to synthesize short Okazaki fragments
Topoisomerases
enzymes that play a crucial role in managing the topology of DNA, essentially untangling and relaxing DNA strands during processes (replication, transcription, and cell division) by temporarily cutting and resealing DNA strands
prevents DNA tangles and supercoils, which can hinder essential biological processes
DNA topology
are essential for maintaining the proper structure of DNA
replication & transcription
during replication and transcription, the DNA helix needs to be unwound and accessed, which can lead to tension and tangles → topoisomerases help to relieve this tension by introducing and resealing DNA breaks
cell division
during cell division, chromosomes need to be separated → topoisomerases play a role in detangling and separating replicated sister chromatids
Helicases
enzymes that unwind double-stranded nucleic acids, like DNA and RNA, by breaking the hydrogen bonds between base pairs
crucial for various cellular functions (DNA replication, repair, transcription)
unwinding DNA
act as molecular motos, using energy from ATP hydrolysis to “unzip” the DNA double helix
this unwinding creates exposed single strands of DNA, which are then used as templates for DNA replication and transcription
DNA replication
during DNA replication, helicases are essential for separating the two strands of the double helix, allowing DNA polymerase to synthesize new complementary strands
the process ensures that each daughter cell receives a complete copy of the genetic information
other cellular processes
involved in DNA repair, where they help to identify and remove damaged DNA
play a role in transcription, where they unwind DNA to allow RNA polymerase to access the DNA and synthesize RNA
DNA Polymerase
a vital enzyme that creates DNA molecules by assembling nucleotides, ensuring the accurate replication and maintenance of DNA
essential to DNA replication
usually work in pairs to create two identical DNA strands form one original DNA molecule
synthesizes new DNA strands by adding complementary nucleotides to a template strand, ensuring the accurate duplication of genetic information
DNA replication
builds new DNA strands by adding nucleotides, ensuring accurate replication of the genetic code
proofreading & repair
has proofreading capabilities, allowing it to detect and correct errors during replication, maintaining the integrity of the genome
Ligase
an enzyme that joins two DNA fragments together
required for the repair, replication, and recombination of DNA
DNA replication
crucial for joining together Okazaki fragments synthesized on the lagging strand
DNA repair
crucial for repairing breaks in the DNA backbone caused by damage or other processes
Semi-conservative replication
a DNA replication process where each new DNA molecule consists of one original strands and one newly synthesized strands
each daughter DNA helix contains half the original DNA, which is essential for maintaining genetic continuity
process
during replication, the double helix of DNA unwinds, and the two strands separate
each original strand then serves as a template for the synthesis of a new complementary strand
result
the end result is two new DNA molecules, each with one original strand and one newly synthesized strand
importance
ensures that genetic information is passed on accurately to daughter cells during cell division
Leading strand
in DNA replication, the DNA strand that is synthesized continuously in the 5-3 direction towards the replication fork
made in the same direction as the replication fork is moving
continuous synthesis - made without breaks or fragments
requires only 1 primer - a single RNA primer is needed to initiate synthesis
does not require ligase - since its synthesized continuously, it doesn’t require ligase to join fragments
efficient replication - the continuous nature of leading strand synthesis makes for faster and more efficient replication compared to the lagging strand
Lagging strand
in DNA replication, the DNA strand that is synthesized discontinuously, in short Okazaki fragments in the 5-3 direction away from the replication fork
made in the opposite direction as the replication fork is moving
discontinuous synthesis - must be made in short fragments
requires multiple primers - fragments are initiated by RNA primers and then extended by DNA polymerase leaving RNA primers that are later on removed and replaced with DNA by other enzymes
requires ligase - ligase join together gaps between fragments
inefficient replication - the discontinuous nature of lagging strand synthesis makes for slower and less efficient replication compared to the leading strand