Cell Structure Chapter 10
1/43
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
44 Terms
Cellular Respiration
Harvests energy remaining in pyruvate and NADH from glycolysis
Uses an external electron acceptor to oxidize substrates completely to CO2
In aerobic respiration the terminal electron acceptor is oxygen and the reduced form is water.
Where is Mitochondria found?
All aerobic cells of eukaryotes
Both chemotrophic and phototrophic cells
Where there is the greatest need for ATP
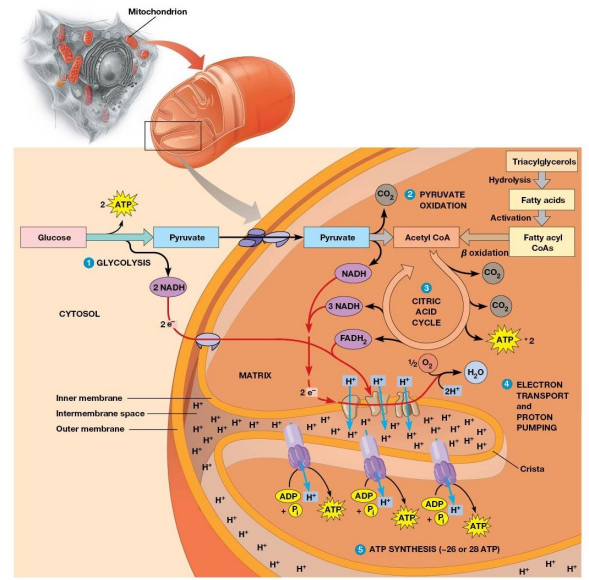
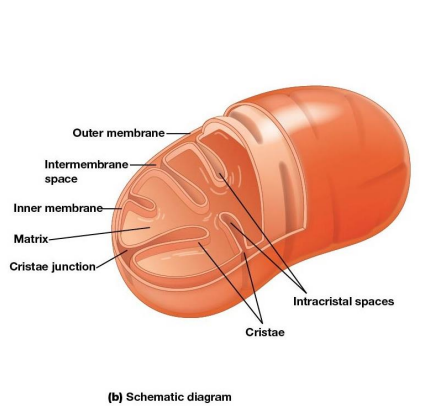
Mitochondrial Structure
Distinctive feature - both outer and inner membranes
Outer membrane contains porins
intermembrane space between inner and outer membrane
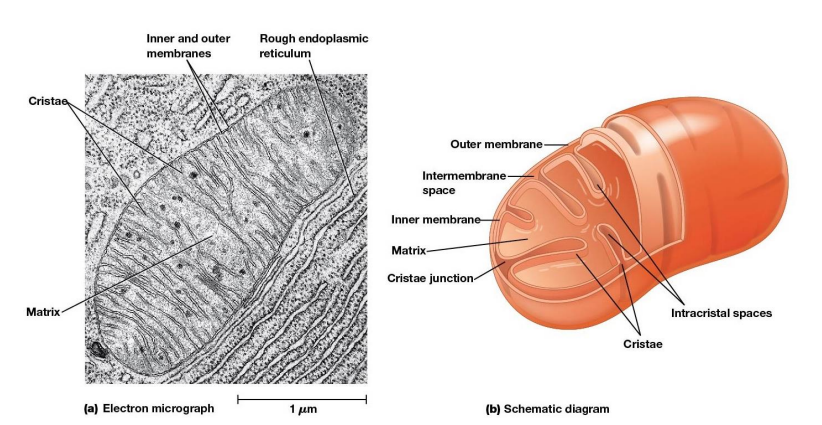
Inner Membrane
Impermeable to most solutes
2 Separate Compartments:
Intermembrane space
Mitochondrial matrix (interior)
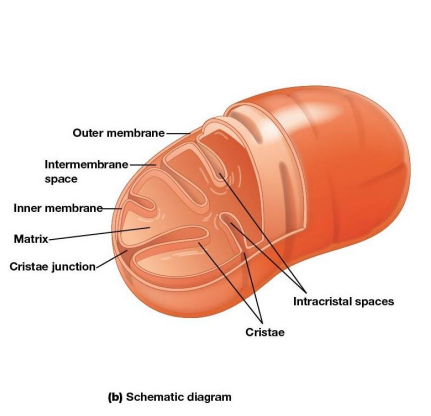
Cristae
Infoldings of the inner membrane meant to increase surface area and provide more space for electron transport to take place.
Note: Most proteins needed for respiration are imported into the mitochondria
Transit Sequences
Targeting signals located on the N terminal of a polypeptide
Transit peptidase
Enzymes that remove the transit sequence once the polypeptide has arrived
Transport Complexes
Proteins are unfolded for transport into the mitochondria
Ports
TOM (translocase of the outer membrane)
TIM (translocase of the inner membrane)
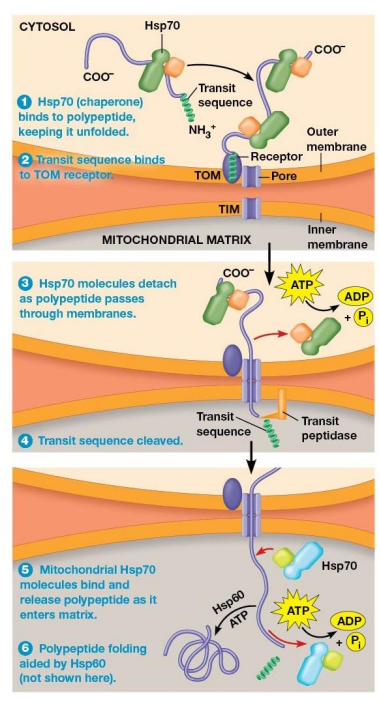
Transit Sequence Receptors
Component of transport complex that recognizes transit sequences
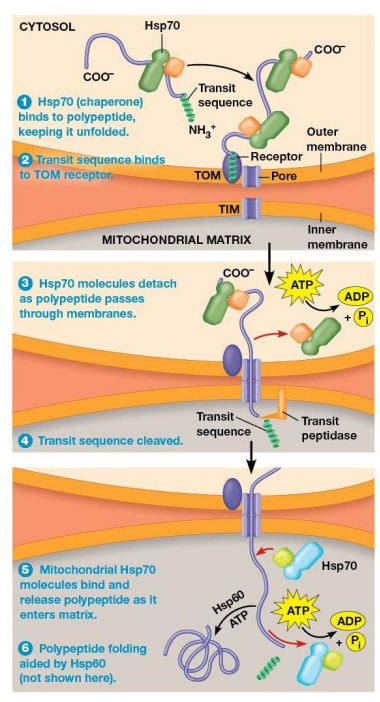
Chaperone Proteins
Bind polypeptides targeted to the mitochondria to help maintain the unfolded state.
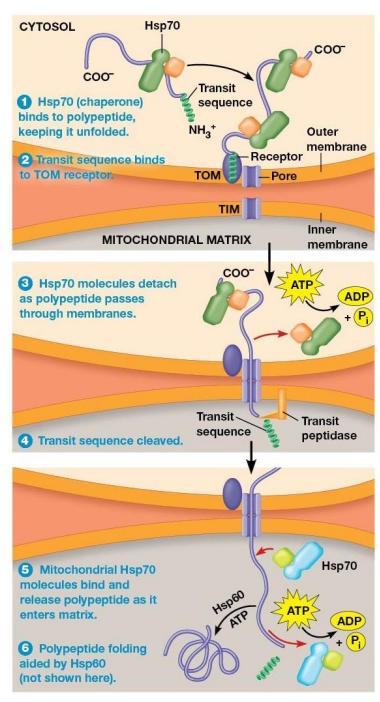
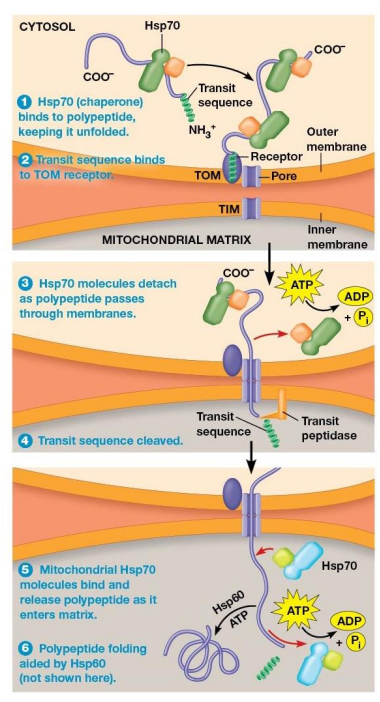
Import of Polypeptides into the Mitochondrial Matrix Mechanism
Hsp70 chaperone protein bind to polypeptide and help to unfold it
TOM transit sequence receptor binds the N-terminus of the polypeptide
Chaperone proteins released and ATP is hydrolyzed as polypeptide moves through the TOM and TIM pores
Transit Sequence removed by transit peptidase in the matrix as soon as the transit sequence enters matrix
Mitochondrial Hsp70 chaperone proteins bind polypeptide as it enters the matrix.
Often, mitochondrial Hsp60 proteins bind the polypeptide and assist in proper folding
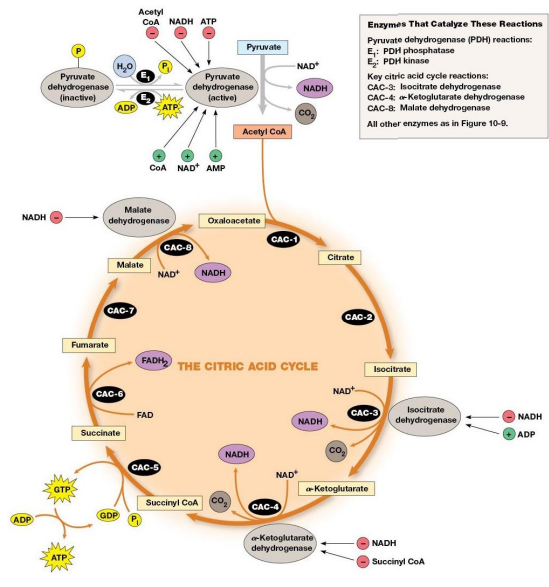
Citric Acid Cycle (TCA, Krebs Cycle)
Citrate is an important intermediate
Cleaving off carbons one at a time to release CO2 and making NADH
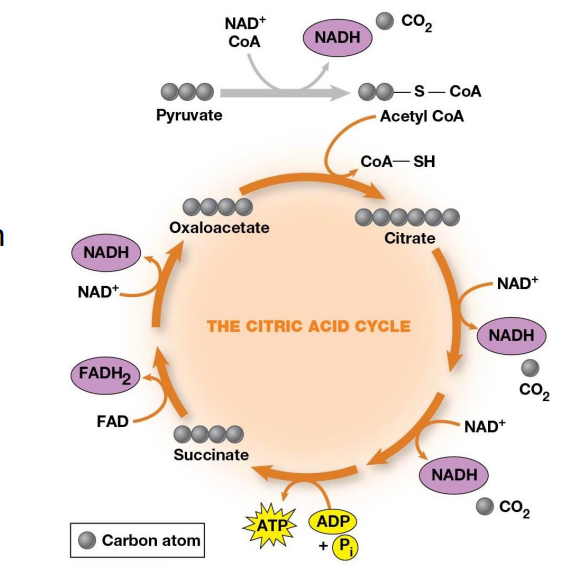
Overall Cycle
2 Carbons enter
Release of 2 CO2 and the regeneration of oxaloacetate
Electrons are accepted by coenzymes
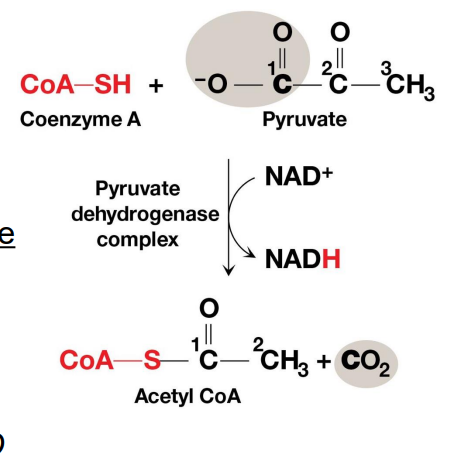
Bridging Reaction
At the inner mitochondrial membrane a specific symporter transports pyruvate into the matrix along with a proton.
Then, pyruvate is converted into acetyl-CoA (By PDH), releasing CO2 and generating NADH
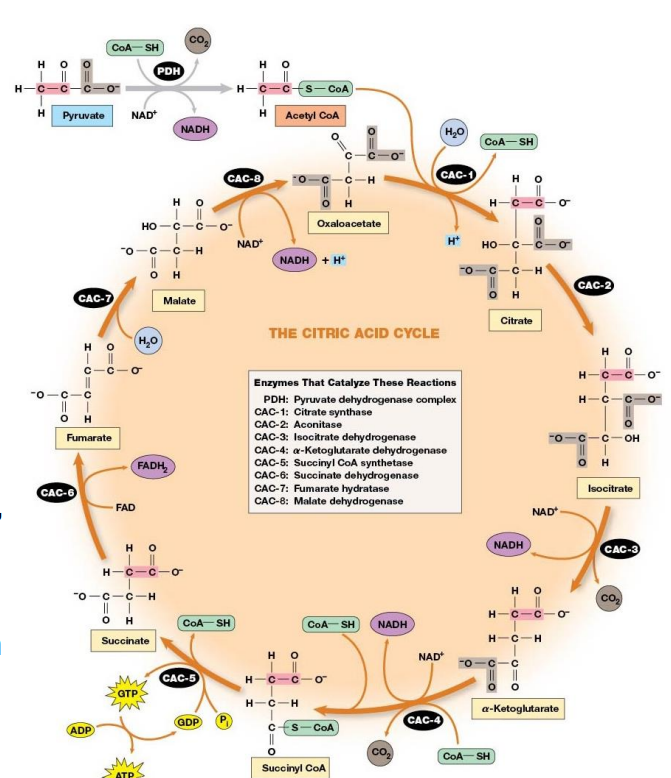
TCA Cycle
Start with 3 carbon compound.
Cleave off one CO2 in bridging reaction
Add other 2 to 4 C compound to make their cleavage easier
2 decarboxylations
3 NADH produced, 1 FADH2, and 1 GTP
CoA is coenzyme and co-substrate in bridging reaction and in cycle.
WATCH SOME VIDEOS TO EXPLAIN
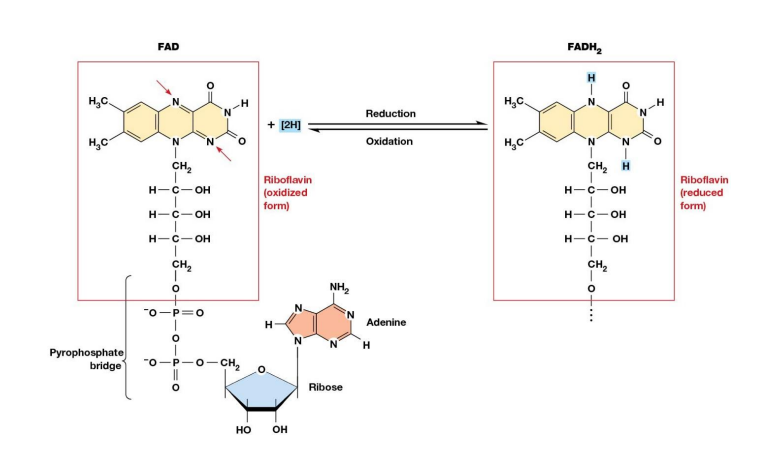
Structure of FAD and its Oxidation and Reduction
Citric Acid Cycle Summary
AcetylCOA + 3NAD+ + FAD + ADP + Pi → 2CO2 + 3NADH + FADH2 + COA - SH + ATP
Citric + Glycolysis + Pyruvate Decarboxylation
glucose + 10NAD+ + 2FAD + 4ADP + 4Pi → 6CO2 + 10NADH + 2FADH2 + 4 ATP
(Remainder of energy of the original glucose stored in NADH and FADH2)
Allosteric Regulation for Cycle
Most of the control of the cycle involves regulation of 4 key enzymes by specific effector molecules.
Effector molecules may be activators or inhibitors
Substrates and Products of TCA
Substrate: CoA, NAD+, FAD, ADP
Products: NADH, FADH2, CO2, ATP
Regulators and Activators
NADH, ATP, and acetyl CoA are allosteric inhibitors of enzymes in this cycle (2 products)
NAD+, ADP, and AMP each activate at least one regulator enzyme in this cycle (2 substrates)
Electron Transport
DEF: Transfer of electrons from reduced cofactors (NADH, FADH2 ) to oxygen
fundamentally linked to ATP generation
Electron Transport Chain
DEF: a multistep process involving an ordered series of reversibly oxidized electron carriers functioning together
Contains integral membrane proteins that are found in the inner mitochondrial membrane (or plasma membrane of bacteria)
Properties of the Respiratory Complexes
Complexes I, III, IV are found in the inner mitochondrial membrane
Complex II involved in succinate oxidation
For each pair of electrons transported through complexes Ⅰ, Ⅲ, and Ⅳ, 10 protons are pumped from the matrix into the intermembrane space.
Complex 1
Transfers electrons from NADH to CoQ and is called the NADH coenzyme Q oxidation complex (NADH dehydrogenase)
Receives electrons from NADH → bound FMN cofactor → Fe-S center → Mobile pool of CoQ
2 electrons transferred, 4 protons pumped
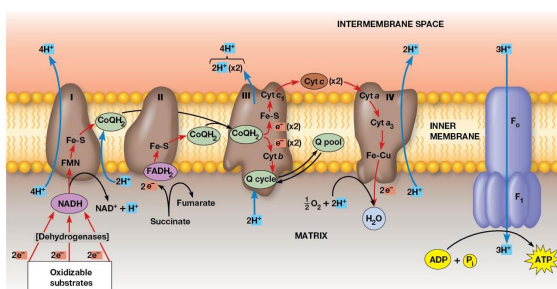
Complex 2
Transfers electron from Succinate → FAD. Electrons in FADH2 are transferred through Fe-S centers → CoQ
Complex called Succinate dehydrogenase
No protons pumped during this reaction
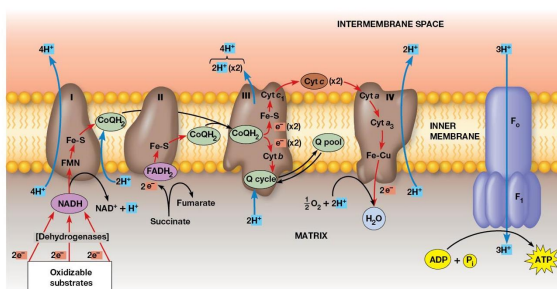
Complex 3
2 cytochromes are prominent components
Accepts electrons from CoQ and transfers them to cytochrome C
2 Electrons transferred, 4 protons are pumped across the membrane.
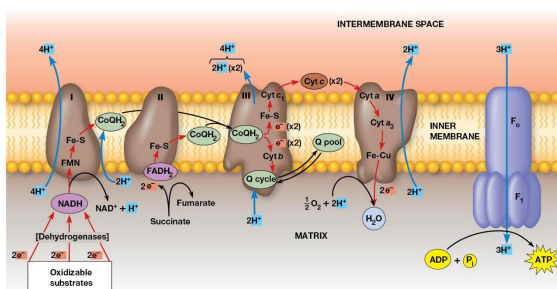
Complex 4
Electrons transfer from cytochrome c to an Fe atom in the home A cofactor of cytochrome a then to cytochrome a3. There are 2 copper atoms which each receive an electron.
4 electrons are needed to reduce O2 → H2O
2 Protons pumped for each electron pair
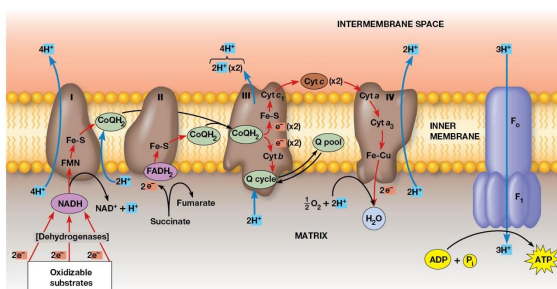
Cytochrome C oxidase
Terminal oxidase, transferring electrons directly to oxygen
Cyanide and Azide ions are poisons because they block electron transport
Genes encoded by Mitochondria DNA
Complex I, II, III, IV
tRNAs
Mitochondrial rRNA
Byproducts of Redox Reactions
Complex I and III can result in incomplete reduction of oxygen
Generates toxic superoxide anion O2- or hydrogen peroxide H2O2 which age cells.
Electrochemical Proton Gradient
Electron transport chain generates it
Drives ATP synthesis
Established by the directional pumping of protons across the membrane in which electron transport is occurring
ATP synthesis is coupled to electron transport
Chemiosmotic coupling model
Essential feature: link between electron transport and ATP formation is the electrochemical potential across a membrane
Created by pumping of protons as electrons are transferred through complexes
Transfer of 2 electrons from NADH is accompanied by the pumping of a total of 10 protons
Number of ATP Generated
2.5 - 3 ATP per NADH oxidation
1.5 - 2 ATP per FADH2 oxidation
F1Fo Complex
F1Fo ATPase generates ATP by coupling H+ transport with ATP synthesis
Both complexes attached and embedded in inner membrane
Fo acts as a proton translocator, the channel through which protons flow across the membrane
Fo provides channel for exergonic flow of protons
F1 carries out ATP synthesis powered by proton gradient
Together they form a complete ATP SYNTHASE
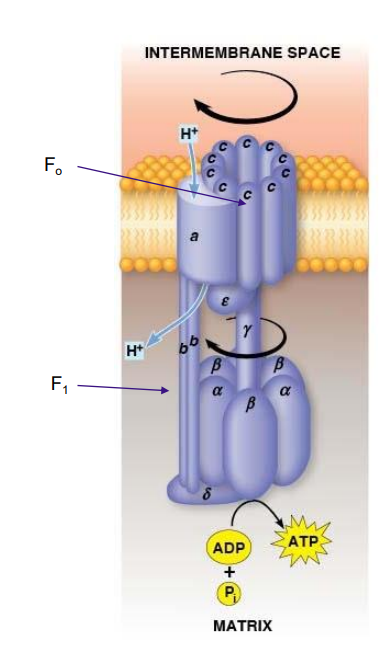
Fo Structrure
two b subunits and ten c subunits
a and b subunits are static
c subunits - form a ring that acts as a gear and can rotate
a subunit - proton channel
2 b subunits - form the stator stalk which connects the FoF1 complexes
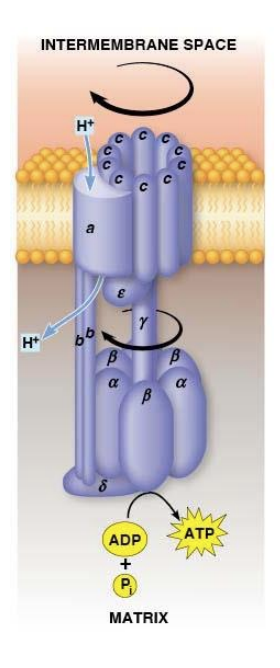
F1 Structure
three α and three β subunits, plus one δ, one γ, and one ε subunit
ATP is synthesized by a ring of three αβ complexes.
The δ subunit anchors the α3β3 catalytic ring to the b2 stator stalk of Fo
The mobile component of F1 is made up of the γ and ε subunits. They attach and move with the c subunits.
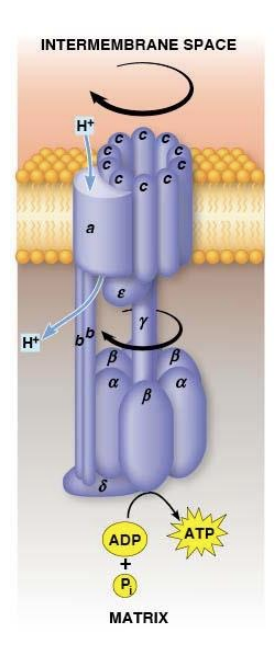
F1Fo Function
Protons move through Fo channel → c10 ring rotates spinning the γ subunit within the α3β3 catalytic ring → ATP synthesis by the catalytic ring
Binding Change Model
proposed that each of the three β subunits of the F1 complex progresses through three different conformations
Three Conformations
L (loose), binds ADP and Pi loosely
T (tight), binds ADP and Pi tightly and catalyzes the formation of ATP
O (open), little affinity for either substrates or product
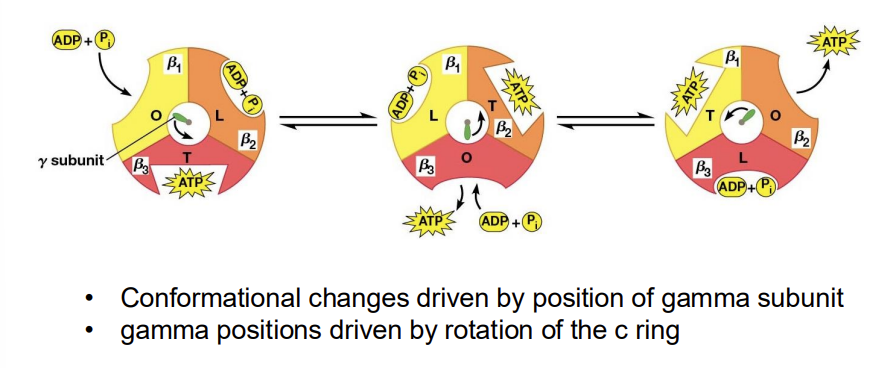
Binding Change Structures
Each β subunit passes through the O, L, and T conformations as the γ subunit rotates 360 degrees.
In Fo, the 10 c subunits each have an aspartate residue with an ionic bond to an arginine residue on the immobile a subunit.
Binding Change Mechanism
Proton taken in → neutralizes aspartate → disrupting ionic bond → rotating the C10 ring (and y subunit) one tenth turn
As the ring turns, the aspartate in the adjacent residue loses a proton and forms an ionic bond to arginine in the a subunit
As 10 protons pass through the membrane via the a subunit, the ring goes through one complete rotation
Maximum ATP per Glucose
Note: Typically less
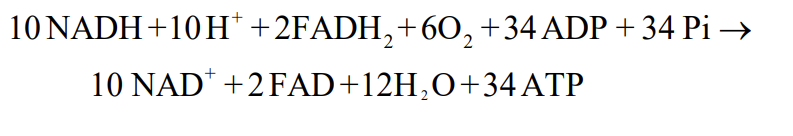
Why is it less?
Typically 30-32 ATP/glucose
Why?
H+ gradient used to exchange ADP and ATP into/out of the mitochondria
If NADH cannot enter matrix, an electron shuttle system will transport the electrons and H+ ions inward