Human Bio Test 2
1/129
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
130 Terms
Protein Synthesis
Genome
total genetic info of organism
3 × 109 base pairs of DNA in diploid human cell
Gene
stretch of DNA that codes for a protein
resides on locus (specific location on chromosome)
~20,000 genes on human genome
genes are only small fraction of total genome
Genetic Code
DNA instructions that specifies for amino acid sequence of a protein (polypeptide)
Central Dogma
pathway of information from DNA → protein
Transcription (in nucleus): DNA → mRNA
Translation (in cytoplasm): mRNA → protein via amino acids
Types of RNA
mRNA: messenger RNA
coding RNA
tRNA: transfer RNA
non-coding RNA (20 tRNA for 20 amino acids)
carry amino acids to ribosomes to add to polypeptide chain during translation
rRNA: ribosomal RNA
forms small subunit of ribosomes
joins with proteins (large subunit) to make ribosome
Control of Gene expression
gene is expressed when protein is synthesized from it
all somatic cells contain the same genetic information → potential to express all genes
pattern of gene regulation distinguishes cell types
tightly regulated process
when DNA is more loosely bound with histones, the gene is more likely to be expressed
methylation adding methyl group to DNA, regulating how tightly bound DNA is to histones
more methyl groups binds the DNA tighter to histones, lowering the likelihood of gene expression
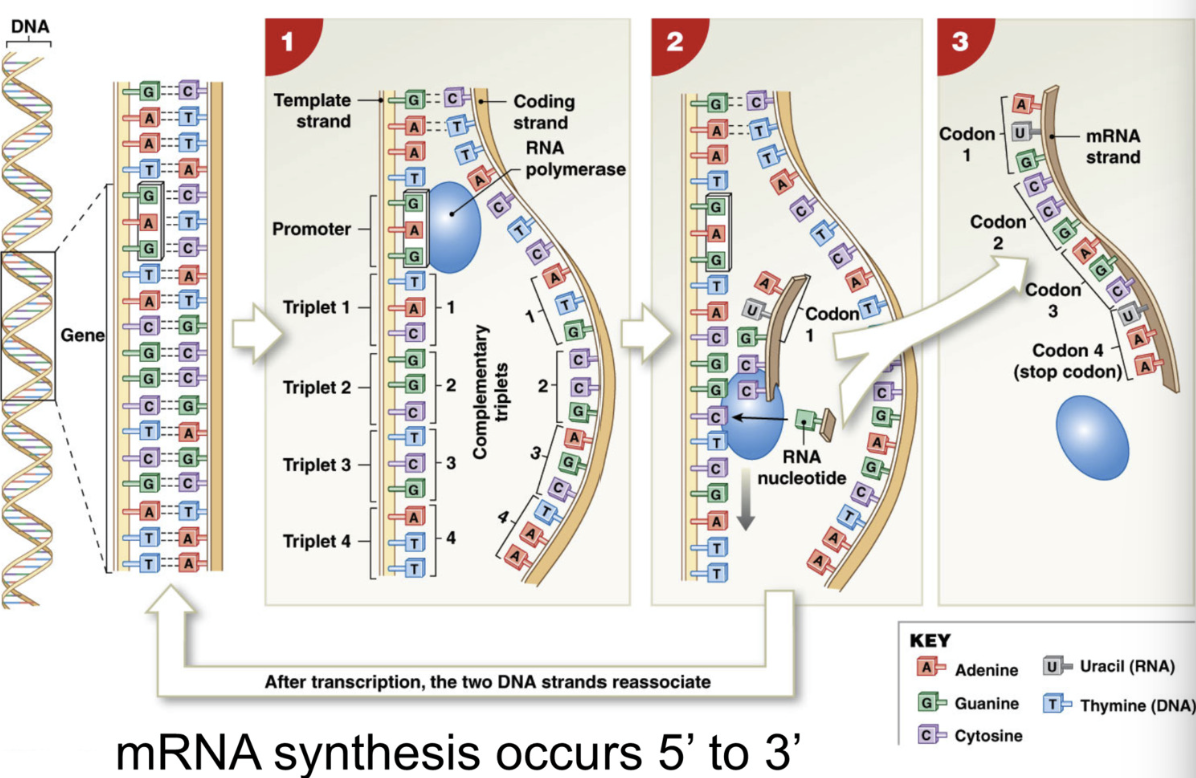
Transcription (in nucleus)
DNA → synthesis of mRNA
RNA polymerase = enzyme that synthesizes mRNA from DNA
attaches to promoter of DNA, reads template strand
assisted by transcription factors = protein that guides enzyme (RNA polymerase) to correct gene
Helicase = enzyme that unwinds DNA coding strand + template strand
pairs DNA with complimentary RNA base
DNA G → RNA C
DNA A → RNA U
DNA C → RNA G
DNA T → RNA A
NOTE: resulting mRNA should look similar to the CODING strand of DNA
Transcription stops when RNA polymerase reaches terminator sequence = sequence of nucleotides that end transcription
5’ → 3’
helicase unzips DNA → transcription factors guide RNA polymerase to promoter of template DNA strand → RNA polymerase pairs DNA w/ complimentary RNA base → transcription stops at terminator sequence
mRNA Processing
transcription produces pre-mRNA (immature)
pre-mRNA → mRNA
3 major edits happens
add 7-methylguanosine cap to 5’ end of mRNA
function: allows mRNA to pass through selective nuclear pores and be identified by ribosomes as mRNA in cytoplasm
add (lots of) poly-A tails to 3’ end of mRNA
buffer to prevent degradation of mRNA in cytoplasm by mRNA degradative enzymes
Processing of pre mRNA → mRNA (nucleus)
transcription produces pre-mRNA (immature)
3 major edits happens to mRNA before it leaves the nucleus
At 5’ end of mRNA: 7-methylguanosine cap added
function: allows mRNA to pass through nuclear pores and be identified by ribosomes as mRNA in cytoplasm
at 3’ end of mRNA: poly adenine (poly-A) tail added
function: acts as buffer to prevent degradation of mRNA in cytoplasm by RNA degrading enzymes
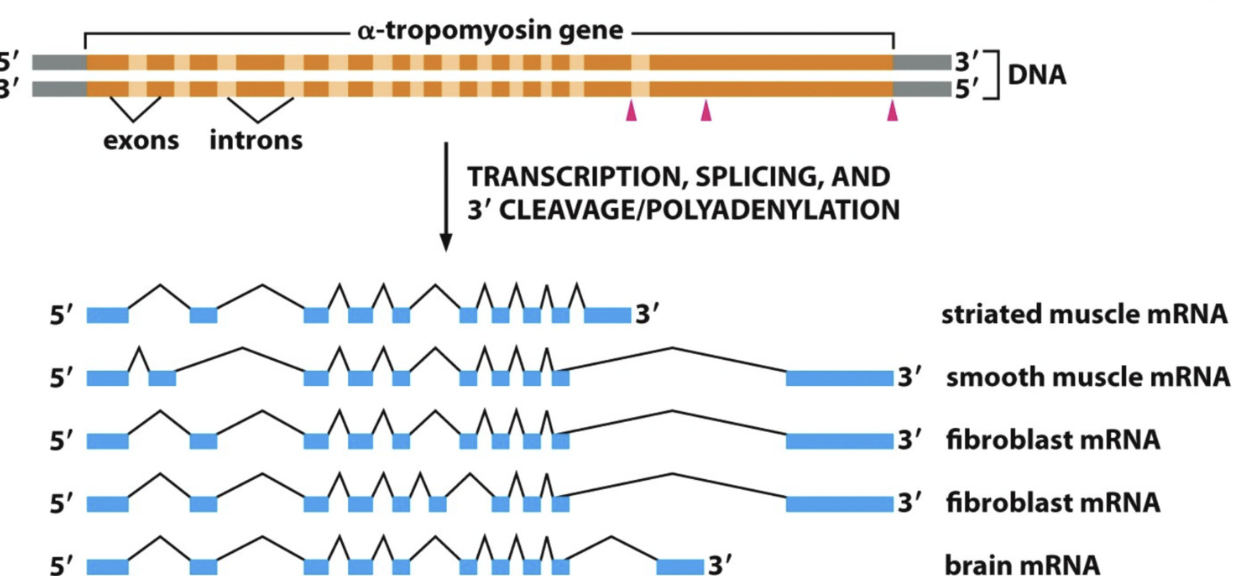
Splicing
removal of INTRONS (noncoding segment to be deleted) and join EXONS (coding segment for protein)
→ mature mRNA: 5’ cap, exons, poly-A tail
Alternative Splicing
Generating diverse protein products from single gene
removal of introns and rejoining of exons
different introns and exons spliced for different cells
“splice variants” expressed in different cells/tissues
response to different signals at different steps of development
(in nucleus)
Translation
Translation of mRNA sequence → polypeptide
Codon: triplet of nucleotides within mRNA which codes for an amino acid
translation of mRNA → polypeptide requires effort of mRNA, tRNA, and rRNA
AUG = methionine (START CODON)
almost every protein starts with methionine
ribosome recognizes start codon and begins to produce protein
don’t need to memorize codons
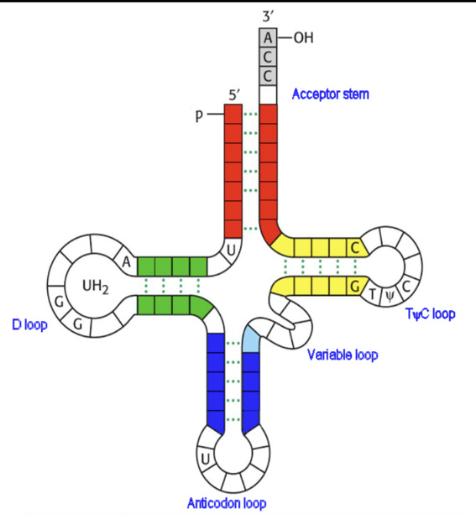
Transfer RNA (tRNA)
function: match amino acid to codon sequence of mRNA
clover leaf shaped non-coding RNA
Parts:
acceptor stem: end that binds to amino acid (contains 1 of 20 amino acids)
anticodon loop: has triplet nucleotide sequence that is complementary to codon of mRNA
tRNA has 20 amino acids and works with ribosomes
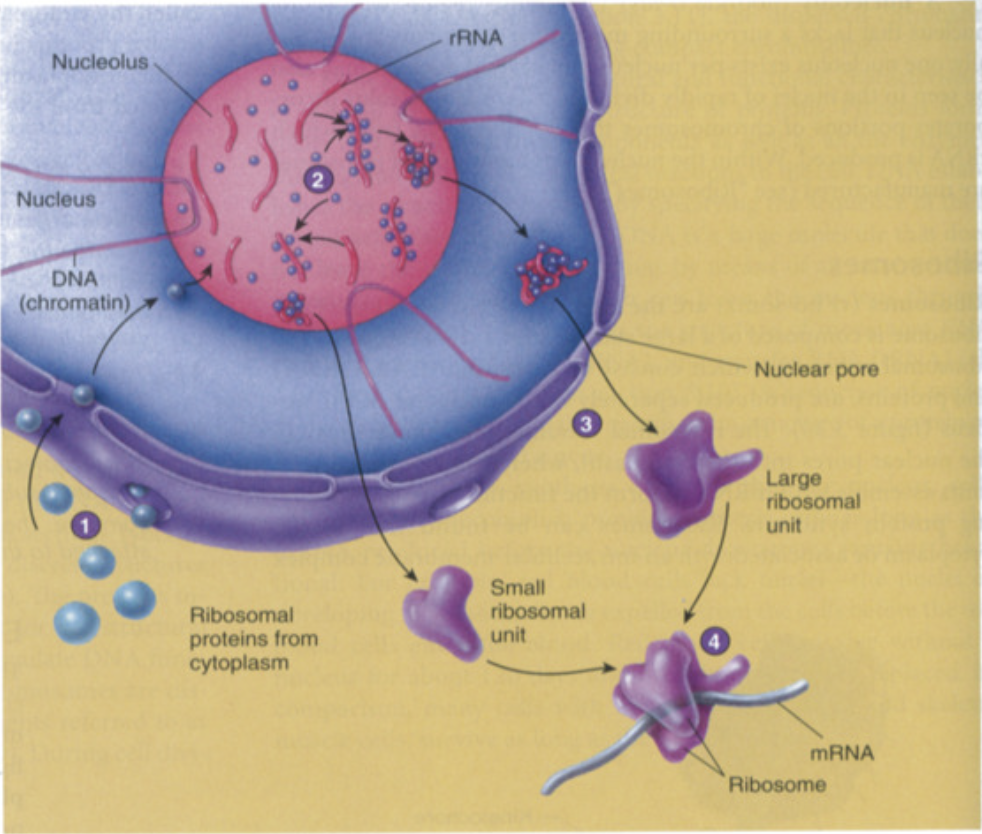
Ribosomal RNA (rRNA)
function: forms complex with ribosomal proteins to form ribosomal subunits
synthesis of ribosomal subunits
ribosomal proteins made in cytoplasm through translation and sent into nucleolus
rRNA made in nucleolus + ribosomal proteins (made in cytoplasm) = ribosomal subunits (made of both rRNA and proteins)
ribosomal subunits (large subunit + small subunit) assembled in nucleolus
small + large subunits leave through nuclear pores separately into cytoplasm and only come together during translation
in cytoplasm, during translation, small + large subunits combine to create ribosome
3 major binding sites of ribosome:
mRNA binding site
tRNA binding sites
P site: peptidyl tRNA → 1st tRNA binding site
A site: aminoacyl → site where tRNA molecule brings new AA
Ribosomal Anatomy
1 mRNA binding site, 2 tRNA binding sites
P site: peptidyl tRNA
first site where tRNA binds
A site: aminocyl tRNA
site where tRNA molecule brings in new amino acids
binding of tRNA to A site weakens bond between amino acid and tRNA in p-site
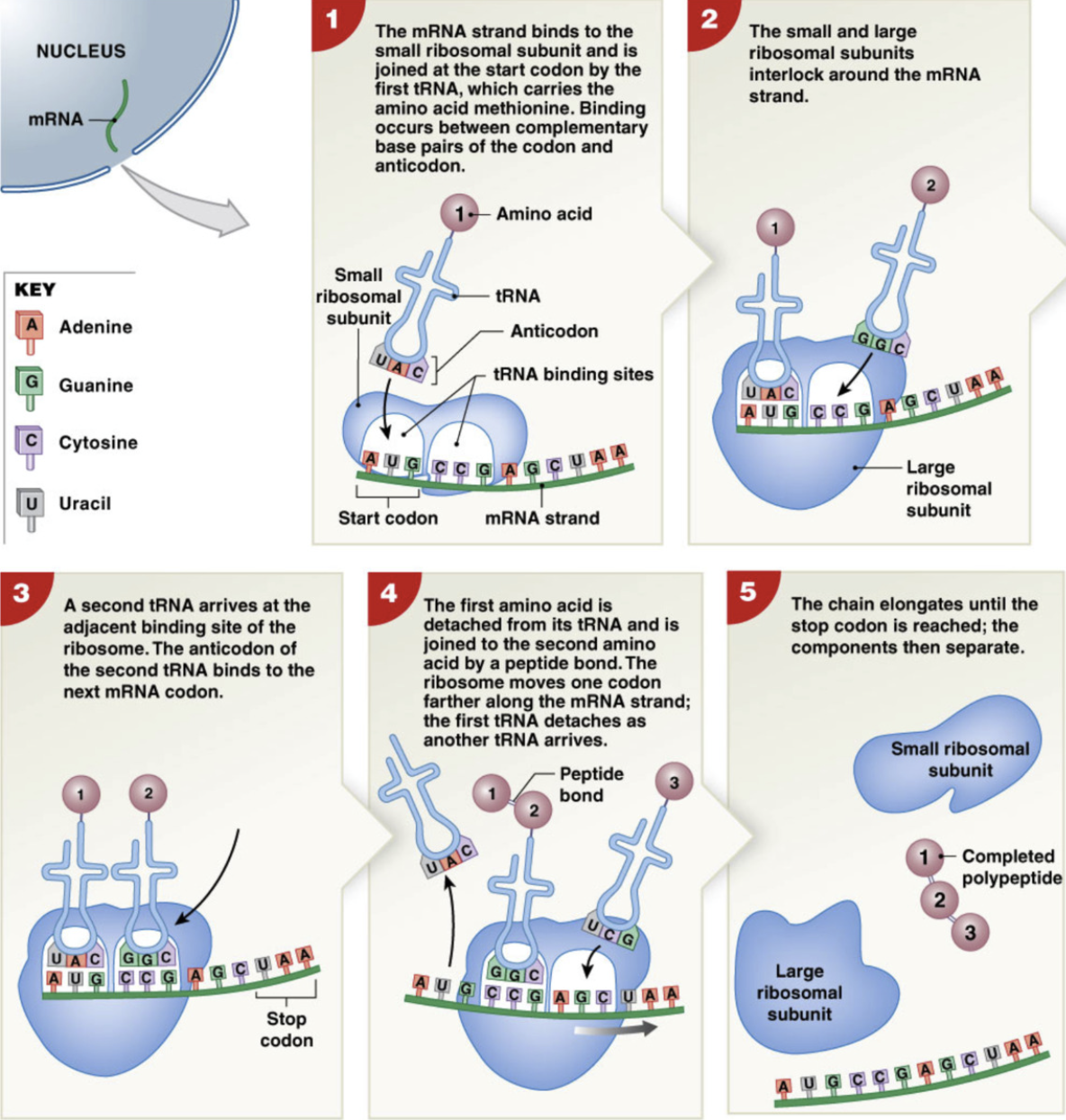
Steps of translation
the 5’ cap of mRNA is recognized by small ribosomal subunit
start codon AUG is aligned with the future P site location
tRNA binds anticodon to codon of mRNA and brings methionine amino acid via acceptor stem
Once anticodon of tRNA and codon of mRNA are bound together, large ribosomal subunit binds to small ribosomal subunit, completing ribosome → complete P and A site
new tRNA binds to the next codon in the A site
weakens bond between amino acid and tRNA in P site
the first amino acid is detached from tRNA and joined to second amino acid via peptide bond
the ribosome moves down a codon and the first tRNA is detached, moving the second tRNA into the P site, freeing the A site for another tRNA
process repeats, elongating chain until the stop codon is reached
polypeptide chain exits through E site (exit site) and ribosomal subunits separate
detached tRNA bind to new amino acids to be recycled.
Human Genetic Makeup
humans are 99.9% genetically identical to each other
~1 in every 300 base pairs is a variable region
singular nucleotide polymorphism (eg 25% have A at the site, 75% have a G)
substitution
deletion
insertion
this particular pattern at these variable sites makes us genetically unique
mutations = when variability interferes with resulting protein function
inherited mutation: passed down from parents
spontaneous mutations: errors in DNA replication
induced mutations: exposure to mutagen (eg UV light, radiation, tobacco)
Cellular Metabolism
all chemical reactions that occurs in an organism
usually energy
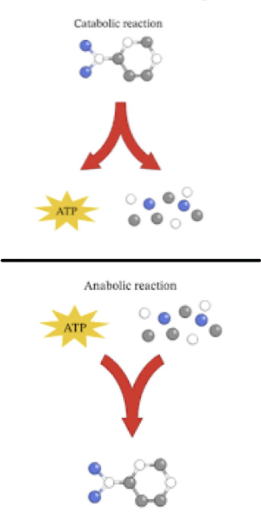
Metabolism
all chemical reactions that occur in organism
2 types:
Catabolism
breaking of molecules to release energy
ex. hydrolysis
Anabolism
building of organicc molecules by using energy
ex. dehydration synthesis
Coenzymes
organic type of cofactors
remove H+ ions from organic substances
binds to active sites of enzymes to catalyze reactions
NAD+ and FAD
energy carriers → transport 2e-
Act as intermediaries
accept e- from one molecule → transfer to another molecule
remove H+
oxidation reacton: reaction resulting in losing electron
reduction reaction: reaction resulting in gaining electron
OiL: Oxidation lose
RiG: Reduction gain
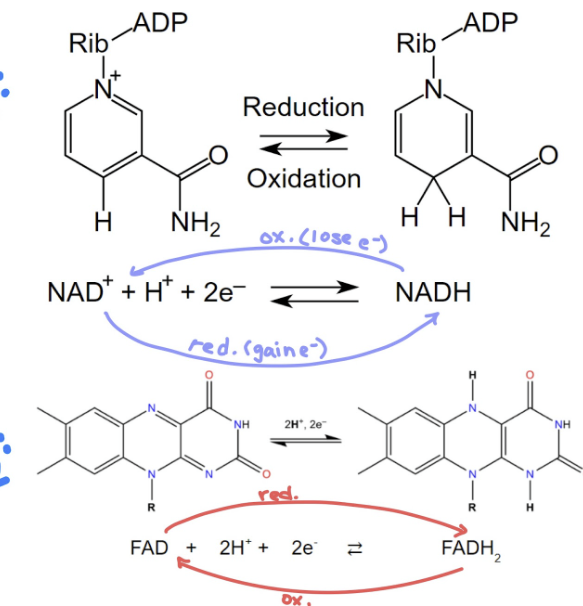
Coenzymes Redox Reactions (KNOW)
both NADH and FADH2 are 2 e- carriers
NADH:
NAD+ + H+ + 2e- < == > NADH
FADH2
FAD + 2H+ + 2e- < == > FADH
Carbohydrate Metabolism
Glucose catabolism (breakdown) → primary ATP production process
generates ATP and other high energy compounds
glucose + oxygen → carbon dioxide + water + ATP
anaerobic reaction: glycolysis
occurs in cytosol
no O2 required
small amounts of ATP produces
aerobic reaction: cellular respiration
occurs in mitochondria
uses O2
produces bulk ATP
Glycolysis
breakdown of 6 carbon glucose → two 3-carbon pyruvic acids
pyruvate = ionized pyruvic acid
occurs in cytoplasm
factors required:
glucose molecules
cytoplasmic enzymes
ATP and ADP
inorganic phosphates
NAD+ (coenzyme)
invest 2 ATP
produce 4 ATP (net gain 2 ATP)
cccccc
in cytoplasm, phosphate is added to glucose (taken from ATP→ADP) “glucose 6 phosphate”
pcccccc (-ATP)
2nd phosphate group is added to other end (ATP → ADP)
pccccccp (-ATP)
molecule is split into two 3-carbon molecules
pccc pccc
inorganic phosphate group from cytosol is added to other end of each molecule. 2NAD+ → 2NADH (NAD+ from mitochondria removes 2e- and H+ and is sent back to the mitochondria for Electron Transport Chain)
pcccp pcccp (+2NADH)
phosphate removed from each 3-carbons molecule producing 2 ATP (2ADP→2ATP)
pccc pccc (+2ATP)
both molecules’ phosphates are rearranged releasing 2 H2O molecules
p p (+2 H2O)
ccc ccc
last phosphates are removed from both molecules producing 2ATP (2ADP → 2ATP). 2 pyruvate remain (used in either citric acid cycle or fermentation)
ccc ccc (+2 ATP +2 pyruvate)
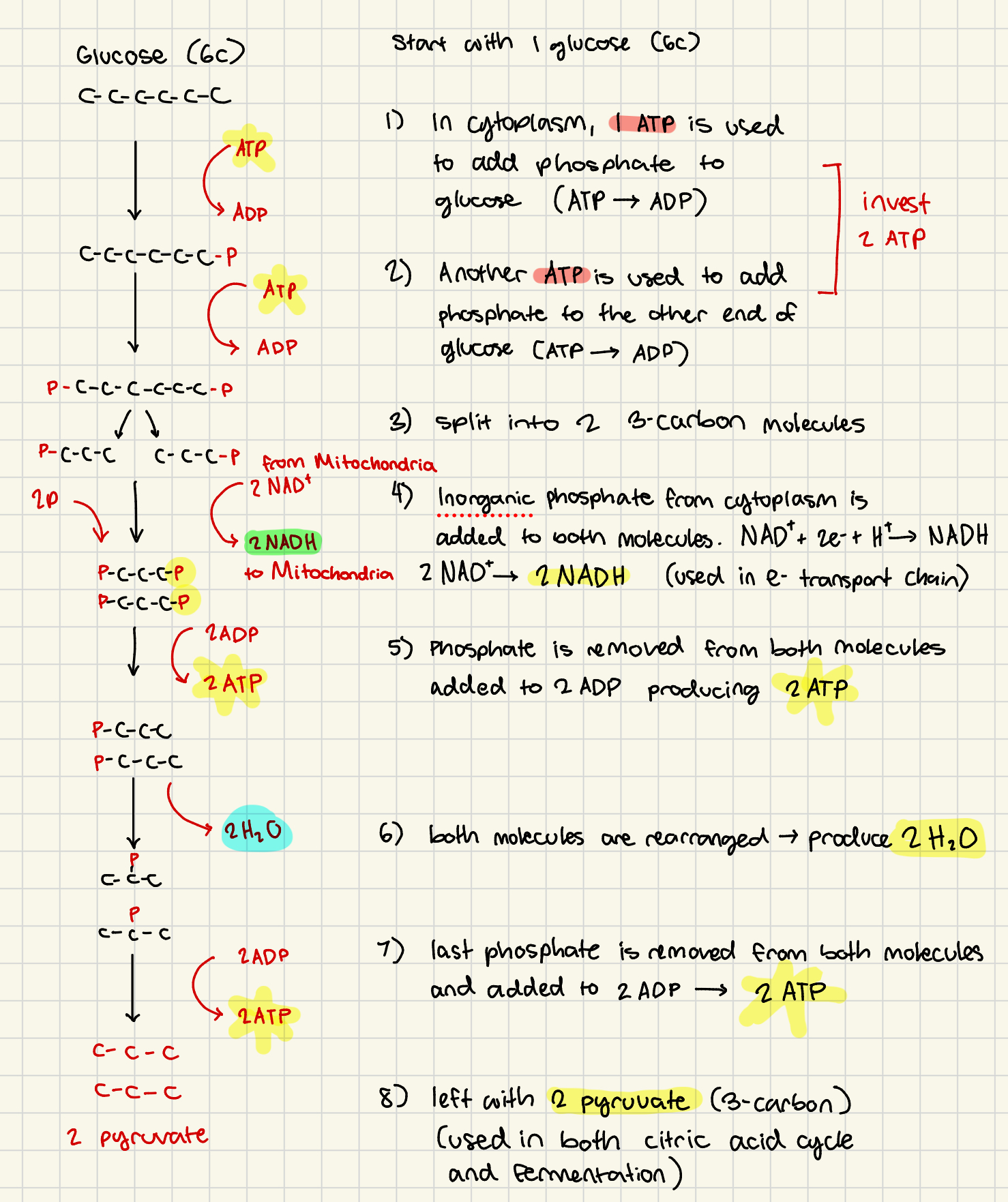
glycolysis products + location
location: cytoplasm
2 H2O
4 ATP (net 2 ATP)
2 NADH (used later in electron transport chain)
2 Pyruvate (used in either fermentation or citric acid cycle)
Aerobic Respiration
ATP production in mitochondria
mitochondrial absorb and break down pyruvate requiring oxygen
2 phases:
Citric Acid Cycle (in mitochondrial matrix)
coenzymes transfer e- → ETC
Electron Transport Chain (inner mitochondrial membrane)
e- passed down protein cascade producing H+ (proton) gradient (ADP→ ATP)
Mitochondria
Mitochondrial Membranes
Outer membrane
large diameter pores open to ions and small organic molecules (pyruvate can pass into inter membrane space )
Inner Membrane
contains carrier proteins for ETC
move pyruvate → mitochondrial matrix
Intermembrane Space
separate outer and inner membrane
Aerobic/Cellular Respiration (DRAW OUT!)
in presence of oxygen, mitochondrial absorbs and break down pyruvic acid molecules
Mitochondria
outer membrane: large diameter pores open to ions and small organic molecules
inner membrane: has carrier proteins, moves pyruvic acid into mitochondrial matrix
Part 1: Citric Acid (Krebs) Cycle
1 pyruvate (3 carbon) →1 carbon lost as CO2
Acetyl CoA (2C) → combines with oxaloocetic acid (4c)
ketoglutaric acid (5C) → 1 lost as
Succinctyl CoA (4C)
Oxaloacetic acid (4C)
Part 2: Electron Transport Chain
FADH2 and NADH deliver H+ and e- to enzymes in inner mitochondrial membrane
NADH donates e- to FMN (protein complex) then sent to coenzyme Q
FADH directly to CoQ
CoQ releases protons into intermembrane space, passes e- to series of cytochromes → protein surrounding pigment like copper or iron
e- passes through system losing energy
Oxygen accepts e- and combines with H+ to form H2O
electrons of ETC lose energy as they pass from coenzyme → cytochrome
Citric Acid (Krebs) Cycle (part 1 of cellular respiration)
for 1 pyruvate (glucose → 2 pyruvate → double products)
within mitochondrial matrix
Intermediate step: pyruvate oxidation
Pyruvate (3 C) is oxidized (NAD+ → NADH), loses 1 C as CO2
produces Acetyl CoA (2C)
Citric Acid Cycle
Acetyl CoA (2C)
→ combines with Oxaloacetic acid (4C) by coenzyme A
Citric Acid (6C)
→ loses 1 C as CO2
→ oxidized (NAD+ → NADH)
Ketoglutaric Acid (5C)
→ lose 1 C as CO2
→ oxidized (NAD+ → NADH)
Succinyl CoA (4C)
reconfigured many times into
→ 1 NADH
→ 1 FADH2
→ 1 GTP from GDP (donates P to ADP → ATP)
Products
3 CO2 (discarded into plasma as waste)
4 NADH (go to ETC)
1 FADH2 (go to ETC)
1 GTP (→ 1 ATP)
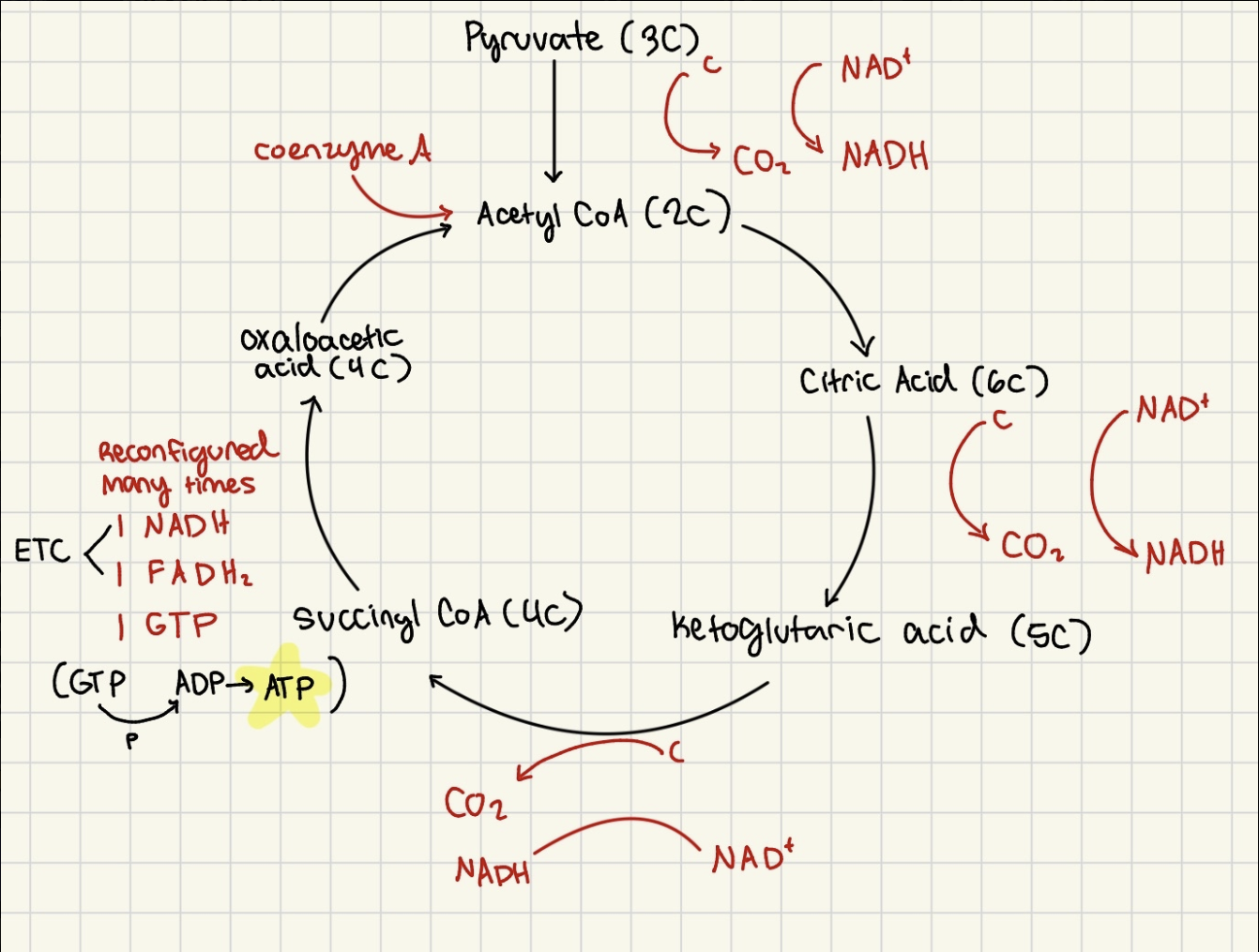
Citric Acid cycle Products + location (for 1 glucose/ 2pyruvate)
mitochondrial matrix
6 CO2 (including production of acetyl CoA)
H+ ions and e- transferred to 8 NADH and 2 FADH2 (go to Electron Transport Chain)
2GDP→2GTP (donates phosphate to 2ADP→ 2ATP)
Electron Transport Chain
NADH and FADH2 deliver H+ and e- to inner mitochondrial membrane
NADH transports H+ and e- → FMN → Coenzyme Q (CoQ)
FADH transports H+ and e- directly → CoQ
CoQ releases H+ into intermembrane space and pass e- to cytochromes (protein surrounding pigment which binds with e-)
cytochrome chain transports e- and uses e- to pump H+ into intermembrane space via H+ ion pumps (loses energy) → steep proton gradient in intermembrane space
Oxygen acts as last e- acceptor, combining with H+ to form H2O
chemiosmosis: H+ diffuses back into mitochondrial matrix via H+ Ion channel, and KE powers ATP synthase to make ADP→ 32ATP
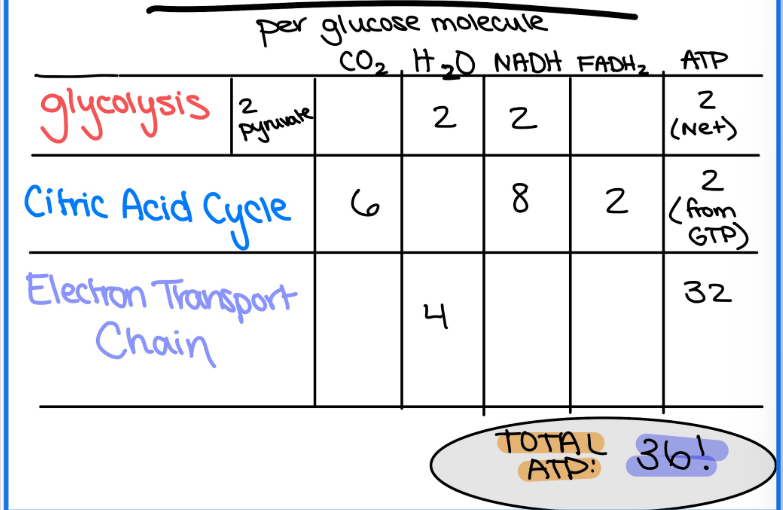
Summary of Products
Glycolysis (in cytoplasm):
2 NADH
2H2O
2ATP (net) 4 total
2 pyruvate
Citric Acid Cycle (in mitochondrial matrix) (for 1 glucose)
8 NADH
2 FADH2
6 CO2
2 ATP (from GTP)
Electron Transport Chain (in inner mitochondrial membrane) (for 1 glucose)
4 H2O
32 ATP
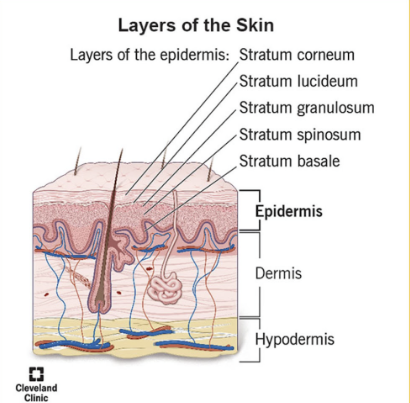
Integumentary System
Components
Most Superficial
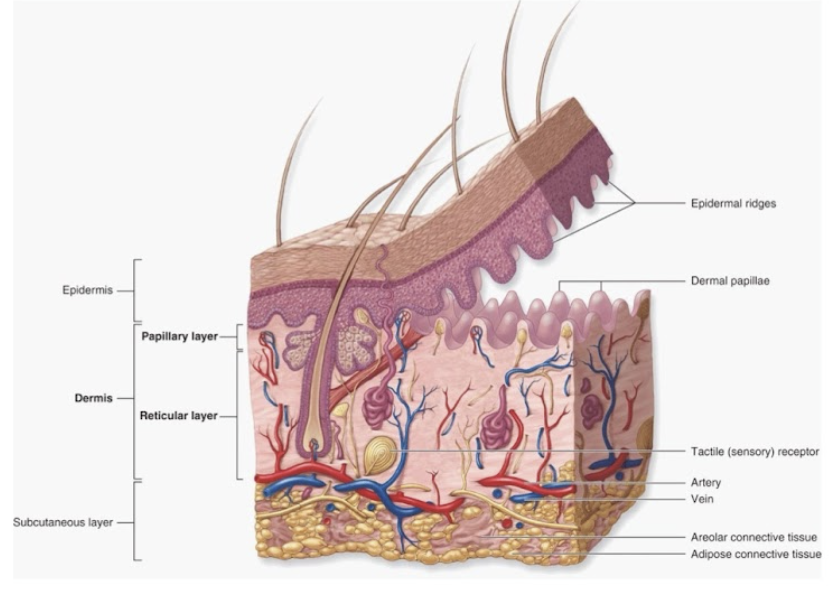
Epidermis:
4-5 layers of stratified squamous epithelium
Dermis: 2 layers
papillary layer → papillae, areolar tissue
reticular layer → dense connective tissue
Hypodermis (not apart of integumentary system)
underlying subcutaneous layer of adipose + areolar connective tissue
Integumentary system function
Protection
physical barrier
immune functions: keep pathogens out
protects from UV damage: melanocytes
Body Temperature Regulation
regulation of conductive/convective heat loss
conductive: transfer of heat from one solid to another
convective: transfer of heat from liquid to gas (air)
blood vessels to skin constrict/dilate to regulate blood flow to skin
skin blood flow regulated primarily by neural mechanisms
more blood to skin = more heat transfer
less blood to skin = heat conservation
regulation of evaporative heat loss
neural regulation of activity of sweat glands
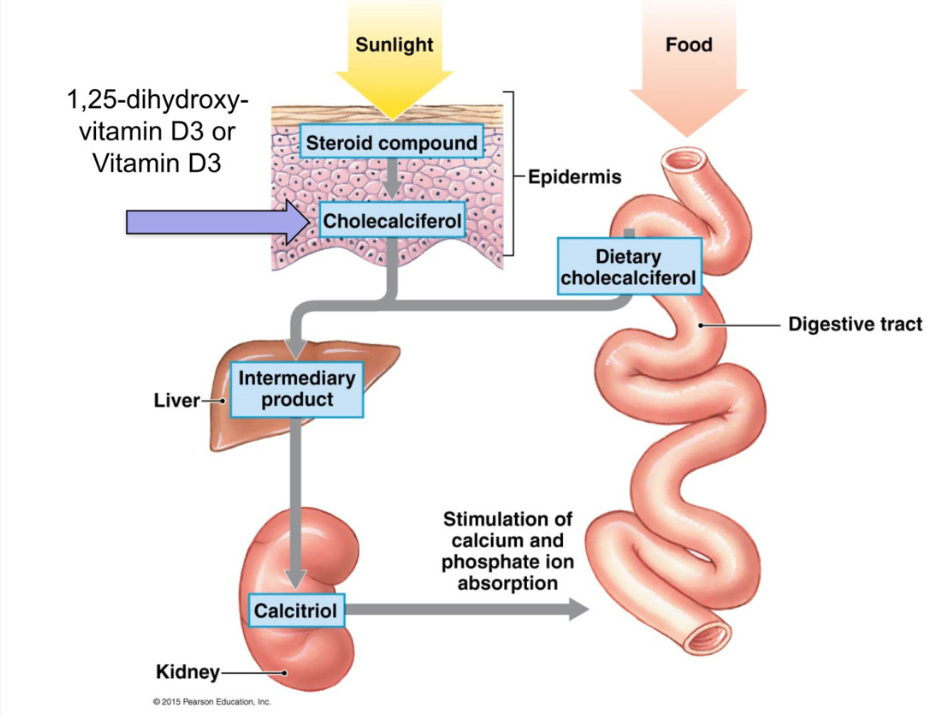
Vitamin D3 Synthesis
sunlight converts cholesterol in epidermis/diet → cholecalciferol (inactive D3)
sent to liver for modification
sent to kidney to form → calcitriol (active D3)
calcitriol stimulates Ca 2+ and PO43- absorption from digestive tract (increase # transport proteins)
Sensation
mechanoreceptors, thermoreceptors, nociceptors
excretion
glands
Neural regulation
negative feedback loops
hypothalamus = control center
Function of Intergumentary system Vitamin D-3 Synthesis
sunlight converts cholesterol in epidermis to cholecalciferol (inactive) (or absorbed from diet)
sent to liver for modification
sent to kidney to form calcitriol (active)
calcitriol stimulates Ca2+ and PO4³- absorption from digestive tract (increased # of transport proteins
Functions Sensation and excretion
sensation → mechanreceptors, thermo receptor
Epidermis: outermost layers
stratified squamous epithelium
ranges in thickness:
thin skin → 0.1-0.15 mm
thick skin (globular)→ 0.5-4.5 mm
eg soles of feet, palms of hands
separate from dermis by underlying basement membrane
avascular → no blood vessels = less metabolic demand
epidermal ridges project into dermis → increase surface area for attachment + fluid movement
Epidermis: Cell Types
Keratinocyte (Stratum Corneum)
most abundant cell type
synthesize and accumulate the protein keratin
produce lamellar granules ( lipid secretions) → waterproofing
put fat into skin to waterproof
Langerhans cells (dendritic cells) (stratum spinosum)
immune response against microbes and cancers via phagocytosis
found in stratum spinosum
Stem Cells (basal cells in stratum basale)
deepest layer of epidermis (stratum basale)
differentiate into keratinocytes
Melanocytes
produce melanin (pigment)
star shape wrap around keratinocytes
located in stratum basale
Merkel Cells (tactile) (stratum basale)
tactile
mechanoreceptors that make contact with sensory neurons to elicit sensation of light touch
tactile cell + nerve ending = merkel’s disc
found in stratum basale
Epidermis of skin has 4-5 layers ( DRAW OUT, know each cell and what layer its found in)
deep
Stratum Basale: single layer of cuboidal/ columnar cells
source of cell renewal (stem cells)
location of melanocytes and Merkel cells
further from stratum basale = less active because its farther from blood supply
Stratum spinosum: many layers (8-10) of keratinocytes. Cells flattening
Langerhans cells located here
Stratum granulosum: 3-5 cell layers of keratinocytes
transition between metabolically active cells and superficial dead keratinized cells
stratum lucidum: single layer of densely packed dead keratinocytes present in thick skin
layer of attachment
thick skin experiences more friction than thin skin, needing extra structural support from protein structures holding the skin in place
stratum corneum: 15-30 layers of fully keratinized dead keratinocytes (lacking nuclei)
Superficial
Layers of Epidermis
Superficial:
Stratum Corneum
15-30 layers of fully dead keratinized keratinocytes (lose nuclei
keratinocytes
Stratum Lucidum
single layer of densly packed keratinocytes for support (only in thick skin)
Stratum Granulosum
3-5 layers of keratinocytes transitioning from metabolically active → dead
flatten as cells move up
Stratum Spinosum
8-10 layers of keratinocytes, cells flatten
langerhans cells here
Stratum Basale
single layer of cuboidal/columnal cells
source of cell renewal (stem cells), melanocytes, merkel cells
most metabolically active = closer to basment membrane
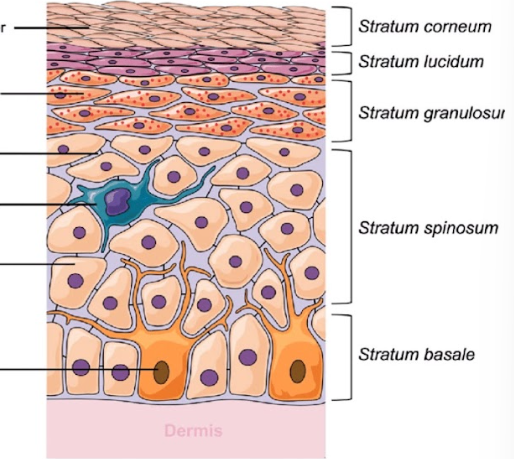
REMEMBER ORDER OF LAYERS
Come Let’s Get Sun Burned (superficial to deep)
Melanocytes
located in stratum basale
contain melanosomes
synthesis of the pigment melanin from amino acid tyrosine
delivered intact to neighboring keratinocytes
granules help to protect epidermal cells from DNA damage via UV light
eventually degraded by lysosomes in keratinocytes
exposure to UV light → production of melanin → tanning
Factors affecting skin color
epidermal pigmentation
melanin:
yellow-red = pheomelanin (pigment
brown-black pigment = eumelanin (pigment
differing rates/amounts/type/distribution of melanin = differences in skin color
Note: within individual, areas darker pigmentation due to differing densities of melanocytes
Blood Flow to skin: pigment in blood → hemoglobin
oxygenated = bright red
deoxygenated = dark red/purple
redness/flushing = increased blood glow to skin
Erythema: localized area of redness due to excess blood in dilated vessels
Cyanosis = bluish coloring
Dermis (2 layers)
between epidermis and hypodermis
Papillary layer
dermal papillae
vascular areolar tissue
capillary beds for blood e xchange
Reticular layer (NOT reticular tissue)
deeper layer of dense connective tissue with abundant collagen and elastin fibers
Location of accessory structures:
blood vessels
lymphatic vessels
nerves and sensory receptors
hair follicles
glands
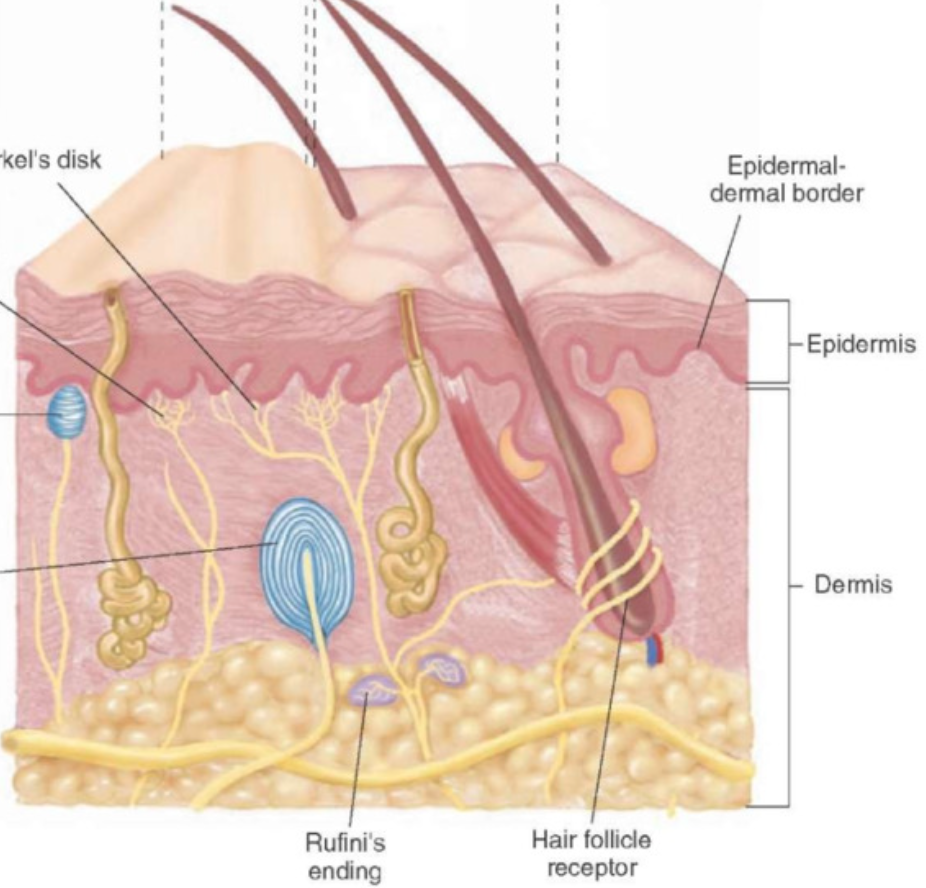
Accessory Structures: sensory receptors
Mechanoreceptors (5 types) :
Merkels discs: sensitive to fine tough and pressure, stratum basale
Meissner’s corpuscle: (concentrated in glabrous skin, nonhair skin) sensitive to fine touch and pressure; dermal papillae
Ruffini’s ending: sensitive to skin distortion and pressure; deep dermis
Pacinan (lamellated) corpuscle: sensitive to deep pressure and vibration; deep dermis/hypodermis
nerve endings surrounding hair root: sensitive to hair movement
all sensitive to touch but diff location
Nociceptors: sensitive to painful stimuli, free nerve endings
Thermoreceptors: sensitive to temperature, free nerve endings
glamorous skin (non hairy skin) = touch sensitive, palms, feet, lips, genitalia,
higher layer = lighter touch
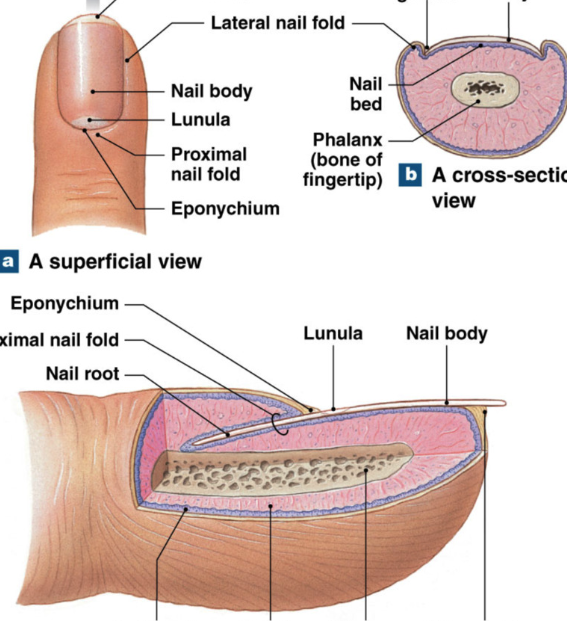
Accessory Structures: nails
nails =dense layers of dead, heavily keratinized, keratinocytes
function: protection, limit distortion, tools
Nail root: deep epithelial fold; contains stratum basale that gives rise to the nail
Nail body: visible portion of nail
Nail bed: epidermal layer just below body
Lunula: base of nail with no blood vessels (white)
Eponychium: cuticle, extension of stratum corneum from root over base of nail
Accessory Structures: hair
hair: dead keratinized cells, which project above surface of skin
produced in organs within the dermis called hair follicles
hairy skin has either:
vellus hair: small, short, delicate
terminal hair: large, course, and usually pigmented (distribution of melanin into hair as its produced)
follicle: specialized imagination of epidermis within surrounding connective tissue
base of follicle:
hair papilla: indentation in connective tissue (blood vessels) most active because they are closest to blood vessels
hair bulb: surrounds papilla and is site of production
hair root: anchors hair, extends from base
Follicle: specialized invagination of epidermis in surrounding connective tissue sheath
base of follicle
hair papilla: indentation in connective tissue (blood vessels)
hair bulb: surrounds papilla and is site of production
Hair root: anchors hair, extends from base to halfway to skin surface (hair cells active)
Hair shaft: extends from halfway point to surface (hair cells dead)
arrector pili → muscle that stands hair up, attached to sensory nerve endings.
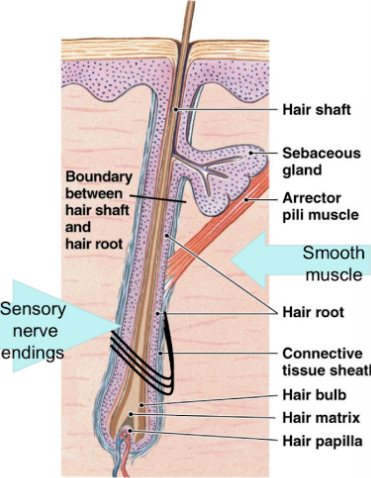
Hair growth
hair grows continually: 0.33 mm/day (scalp)
hair matrix
within hair bulb
site of epidermal stem cells (divide, pushed up, keratinized)
medulla (inner), cortex (middle), and cuticle (outer)
inner → outer layers = less → more keratin
keratinization complete at border of root and shaft
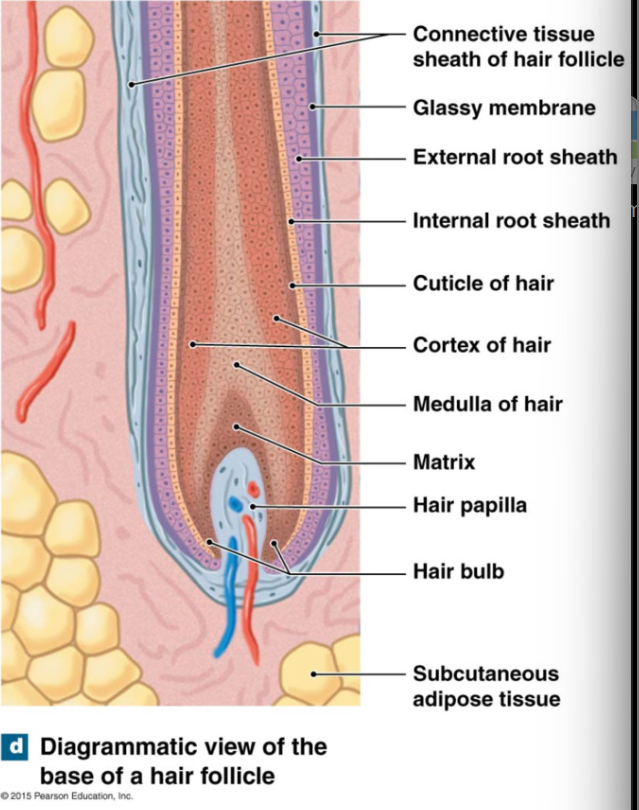
epithelial cells that line follicle are arranged in layers (not part of hair, surround and support hair)
inner to outer
internal root sheath (not part of hair, support hair in epithelial cells)
external root sheath
glassy membrane
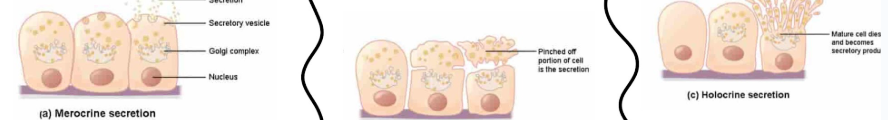
Accessory structures: secretion modes
merocrine secretion: uses exocytosis to discharge secretory vesicles at apical surface of gland cell
apocrine secretion:
apical (top) end breaks off, shedding cytoplasm and secretion
cell survives and regrow
holocrine secretion:
superficial gland cell bursts and leaks stuff
cell dies
replaced via stem cells
Accessory Structures: Glands
Sebaceous glands/follicles
holocrine secretion (cell explodes) called sebum: complex lipid mixture
function: waterproofing, moisturizing, antimicrobial action
Sweat glands (both MEROCRINE):
Apocrine Glands:
armpits, groin, nipples
MEROCRINE SECRETION
secrete onto hair follicles
scent
emotional + sexual stimuli
bromhidrosis: stinky BO → bacteria break down secretions from apocrine glands
Eccrine Glands (found throughout body)
merocrine
watery secretion: small amount of electrolytes and waste products
regulated by nervous system (thermoregulation, excretion)
Function of Bone
Support, movement, and protection
basic body shape
supports body weight
protects vital organs (eg lungs, heart)
movement (skeletal muscle pull bone → movement)
Metabolic function
Hematopoiesis: forms blood cells in red bone marrow
Storage of minerals and lipids
calcium salts, phosphates
lipids in yellow bone marrow
vitamin D3 production → increase uptake of phosphates and calcium ions to bones
Structure of Bone (98% extracellular matrix connective tissue)
Compact vs. Spongy
Compact: outside, dense
Spongy: inside, “cobweb”
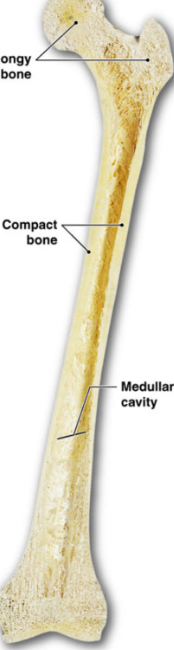
Long vs. Flat
Long: all limbs, hallowed chamber, no spongy
Epiphysis
ends of bone
spongy bone
Diaphysis
long shaft
no spongy bone
medullary cavity: hollow insides
Flat: skull and ribs
interior is spongy bone
Bone (Osseous Tissue)
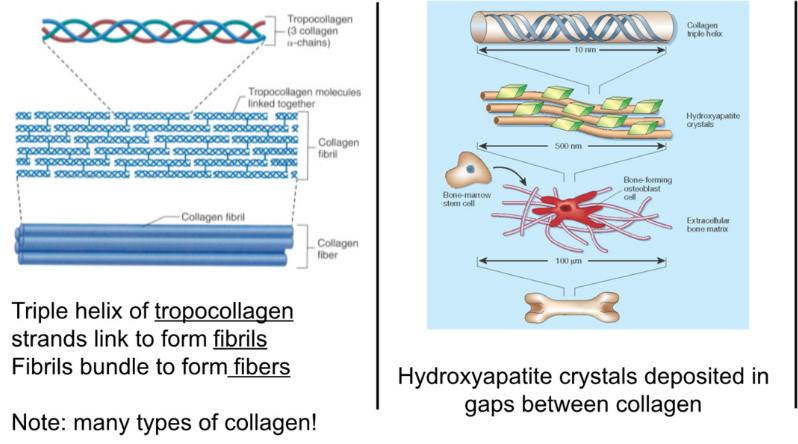
Specialized cells dispersed in hardened extra cellular matrix
ECM:
1/3 collagen → strength and flexibility
triple helix of tropocollagen link → fibrils → fibers, form framework
2/3 calcium → hardness, brittle, resist compression, interact to form hydroxyapatite crystals (calcium and phosphorus)
crystals deposit between gaps in collagen
ground substance (interstitial fluid) → proteoglycans (type of glycoprotein—sugary proteins) and glycoproteins
sugar groups are polar→ hydrophilic → keep water in place
Types of bone cells
bone cells make up <2% of bone
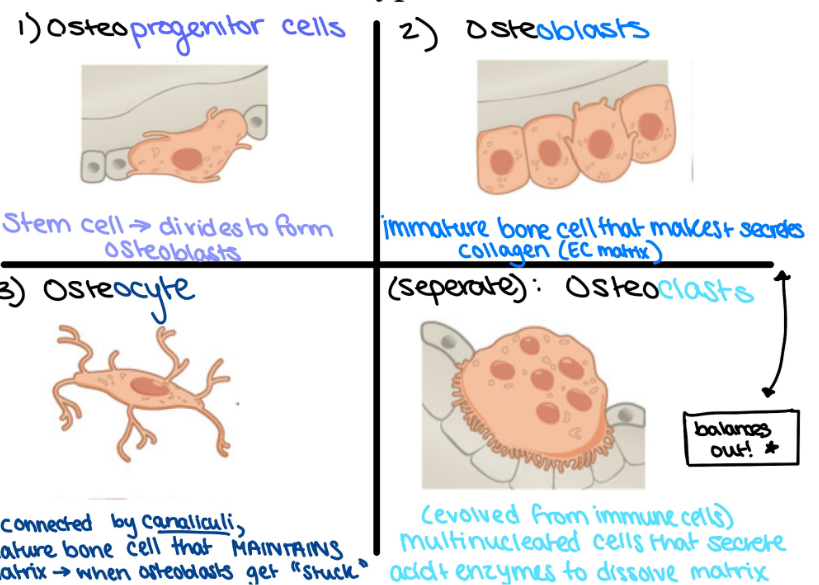
Osteoprogenitors
stem cells → divide to form osteoblasts
Osteoblasts
immature bone cell that produces collagen (ECM)
Osteocytes
mature bone cells, regulate matrix → release chemicals
connected by canaliculi → channels allowing for exchange of nutrients, waste, and oxygen
Osteoclast (from immune cells, DIFFERENT LINEAGE)
multinucleated
breaks down and recycle bone matrix via acid and enzymes
balance osteoblasts
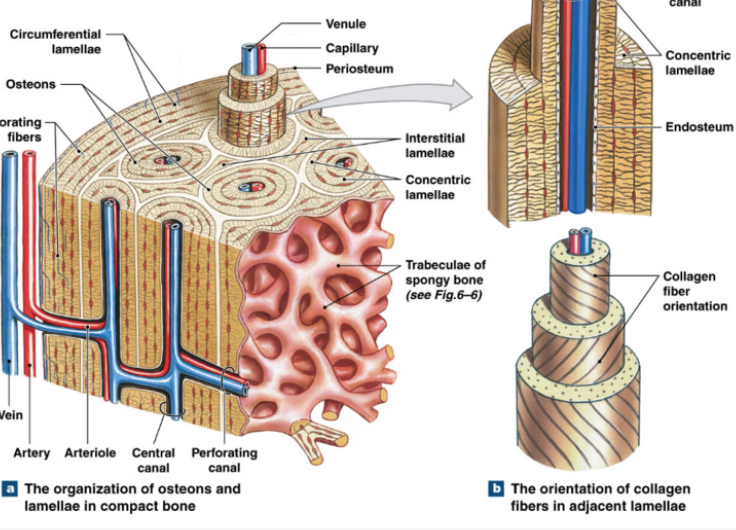
Compact (Dense) Bone
Function: protection, support, resist stress
dense matrix
arranged around blood vessels
Osteon (haversian system): cylindrical unit of circular layers of lamellae + osteocytes trapped between layers and in lacunae
canaliculi connect haversian canal and lacunae for nutrient and waste exchange
Haversian Canal: center of osteon, canal for blood vessels
Perforating Canal: transverse blood vessel canals connecting haversian canals
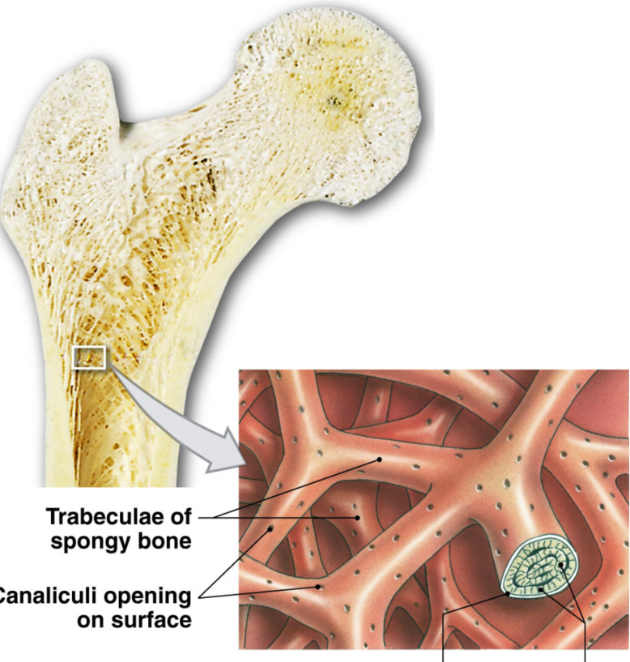
Spongy (Cancellous) Bone
Function: support, store bone marrow
less dense matrix
open network
trabeculae → interconnecting bundles of spongey bone
thin with lamellae and osteocytes between layers (no central blood vessel)
canaliculi → pores for osteocyte nutrient exchange
endosteum interior layer covering bone
contains bone marrow in gaps of trabeculae → hematopoiesis
Bone Marrow
Surround trabeculae of spongy bone
Red Bone Marrow
site of hematopoiesis → produces blood cell
found in interior of flat bones, epiphysis of long bones (and in medullary cavity in children)
Yellow bone marrow
site of storage of adipocytes
found in medullary cavity
red bone marrow as child → yellow bone marrow as adult
Covering of Bone
Periosteum
membrane that covers outer layer of bone except articular surfaces
attachment points (tendons, ligaments)
2 layers
inner cellular layer of osteoblasts and osteoprogenitor cells (for bone replenishment)
outer fibrous membrane (fibroblasts in fibrous matrix)
isolates and protects bone
Endosteum
incomplete cellular covering on interior surface of bones
covers trabeculae and lines Haversian canals
endothelial (epithelial) cells mixed with osteoprogenitor cells and largest concentration of osteoclasts
important for growth and remodeling
Bone Development (Ossification)
2 types of bone formation
Intramembranous ossification
Endochondral ossification
both types
bone matrix initially laid as osteoid (organic proteins + collagen) by osteoblasts → subsequently mineralized
bone produced first in disorganized fashion → woven bone
woven bone is remodeled around blood vessels to form organized bone
Intramembranous Ossification
bones forming from membrane
intramembranous bones:
bones of skull and clavicle
formed directly from mesenchyme: sheet like embryonic tissue giving rise connective tissue
begins 8 weeks gestation, complete after 2 years (for brain growth)
fontanels = soft spots between bones of skull (fibrous membrane connecting cranial bones)
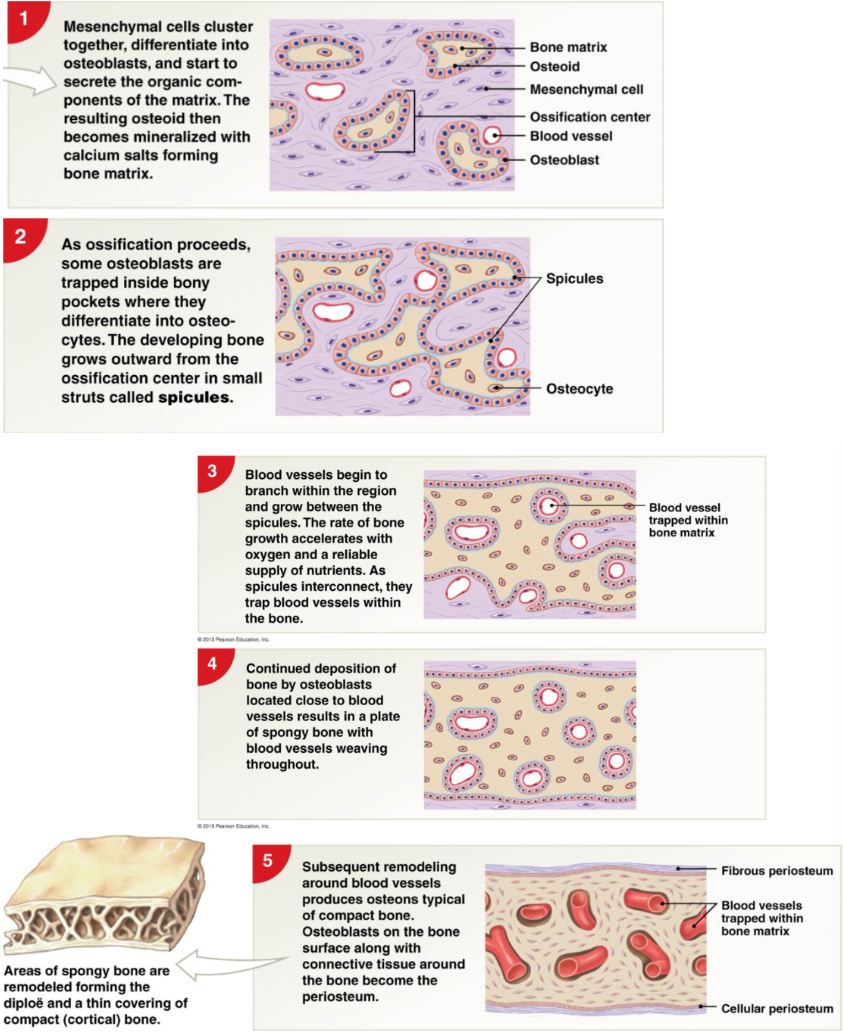
Steps of intramembranous ossification
starts 8th weeks of embryonic development
mesenchymal cells cluster, differentiate osteoblasts → osteoblasts produce collagen to form osteoid (collagen + organic proteins) → mineralized with calcium salts forming bone matrix
as ossification proceeds some osteoblasts are trapped in boney pockets where they differentiate into osteocytes. developing bone grow outwards from ossification center in struts called spicules
Blood vessels begin to branch within region and grow between spicules → spicules connect and trap blood vessels in bone
deposition of bone by osteoblasts near blood vessels → plate of spongy bone and woven blood vessels WOVEN BONE
remodeling of blood vessels → osteons, osteoblasts on bone surface + connective tissue around bone → periosteum
Endochondral Ossification (cartilage)
bones formed from cartilage
begins 8-12 weeks into gestation → early adulthood
every bone other than intramembranous bone
hyaline cartilage model → convert into bone
formation of bone INDIRECTLY FROM MESENCHYME
mesenchymal cells deposit cartilage to model growing bone → replaced with both tissue
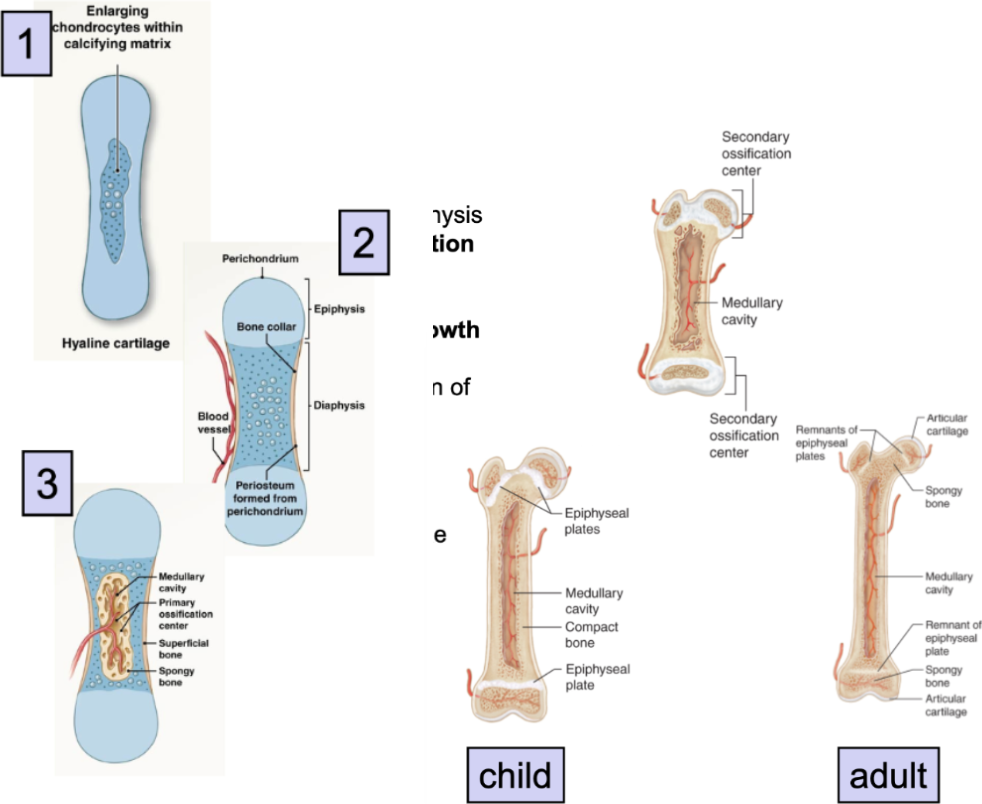
Endochondral Ossification steps
formation of hyaline cartilage models
mesenchymal cells cluster → develop into chondroblasts
chondroblasts (immature cartilage cells) produce hyaline cartilage matrix
chondrocytes enlarge and stimulate formation of calcified cartilage
formation of bony collar (solid bone)
blood vessels grow around cartilage model
outer ring of cells differentiate into → osteoblasts →
osteoblasts produce bone collar
Vascular Invasion (hollow out bone for medullary cavity)
blood vessels, osteoblasts, osteoclasts → invade center
primary ossification center forms
osteoblasts replace calcified cartilage → bone (solid bone)
osteoclasts eat center of bone → medullary cavity
Elongation (overlap with step 3)
blood vessels invade epiphysis → form secondary ossification centers
hyaline cartilage plate at metaphysis (growth/epiphyseal plates) persists after birth
allow for further bone elongation after birth
Epiphyseal Plate ossification
post puberty, epiphyseal plate ossifies and lengthening stops → becomes epiphyseal line
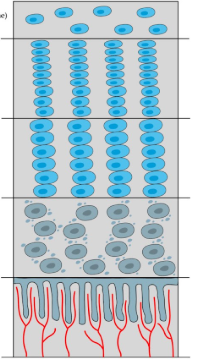
Bone Growth at Growth plate
begins at epiphysis
first layer: reserve zone
non-proliferative (non-dividing) chondroblasts (operate as stem cells)
second later: zone of proliferation
growth factors stimulate rapid proliferation/ mitosis of chondroblasts
cartilage matrix begins forming
when chondroblasts are completely surrounded by cartilage matrix in lacunae → diff into chondrocytes
third layer: zone of hypertrophy
chondrocytes mature and enlarge
matrix expands
fourth layer: Zone of Calcification
chondrocyte death → emptying of lacunae
calcification of matrix
osteoblasts invade and deposit bone matrix on calcified cartilage: convert cartilage → bone
end near diaphysis
cartilage growth rate = osteoblast bone production rate → at puberty, osteoblast conversion rate overtakes cartilage growth rate, and replaces all cartilage→ growth stops
Bone Remodeling
continual process of bone matrix turnover throughout life
partially replaces matrix, but leaves whole bone intact
regeneration of damaged bone → adaptive response
metabolic function: calcium + phosphate regulation, releases old minerals and deposits new minerals
site specific: areas with great friction eg femur head
osteoblasts: form new matrix proteins (balance osteoclasts)
osteoclasts: destroy old matrix (balance osteoblasts)
osteocytes: stimulate breakdown of old calcium crystals + formation of new ones
Factors Affecting Bone Growth
Vitamins
Vitamin C → required for normal collagen synthesis in osteoblasts
Vitamin D → promotes calcium + phosphate absorption from diet
deficiencies → fragile bones
Hormones
calcium regulating hormones
Parathyroid hormone: stimulates osteoclasts when not enough Ca in blood → break down bone for Ca
Calcitonin: inhibits osteoclasts when there is too much Ca2+ in blood
Growth hormone: stimulates cell growth and division
Thyroid hormone: stimulates cell metabolism and osteoblast activity
sex hormones
Androgens and estrogens at puberty → increase osteoblast activity → increase bone formation over rate of epiphyseal plate expansion (estrogen is faster) → stop growing faster
lack of estrogen at menopause → accelerates loss of bone
Fractures
Any crack or
Division of Nervous System
Central Nervous System (CNS): brain and spinal cord
Peripheral Nervous System (PNS): Neural tissue outside of CNS
nerves: carry information between CNS and body, connective tissue + axons
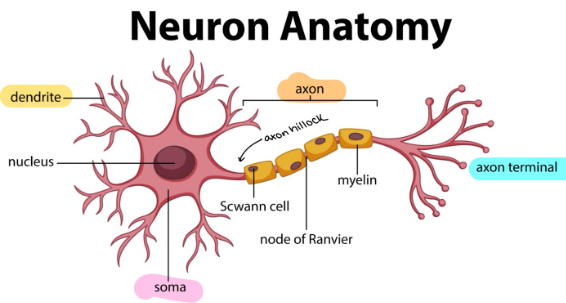
Prototypical Neuron Structure
Cells specialized for intercellular communication
dendrite: receives information
soma/cell body: site of nucleus, organelles, protein production, etc
axon hillock: generates action potential
axon: long, myelinated section in which action potential passes down → sends signals
Axon terminals/synapse: site of communication with next neuron
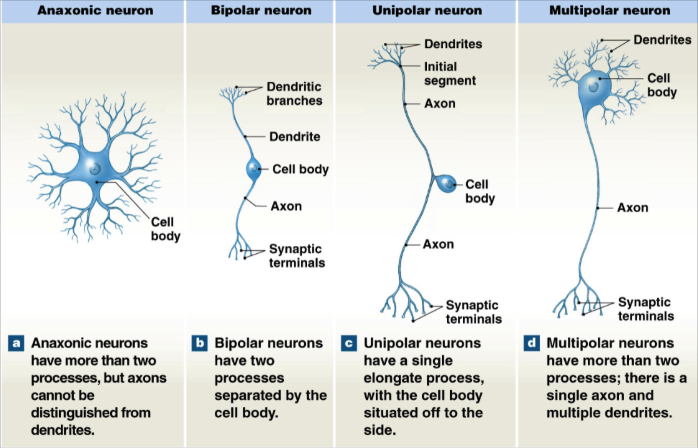
Anatomical Neuron Classification
Anaxonic neuron
more than 2 processes (axons and dendrites)
axons indistinguishable from dendrites
Bipolar neurons
2 processes separated by cell body
1 long dendrite and 1 long axon
Unipolar Neuron
single long axon process
cell body to the side
Multipolar Neuron
more than 2 processes
single axon, multiple dendrites
Functional Neuron Classification
Afferent neurons (sensory) accepting signals
carry information TOWARD spinal cord or brain
somatic: information about external world and body position
visceral: information about internal systems
Efferent neurons (motor) effect signals
carry information AWAY from spinal cord or brain to PNS
somatic: innervate skeletal muscles (voluntary)
visceral: innervate smooth muscle, cardiac muscle, glands (involuntary)
Interneurons
Communicate between neurons
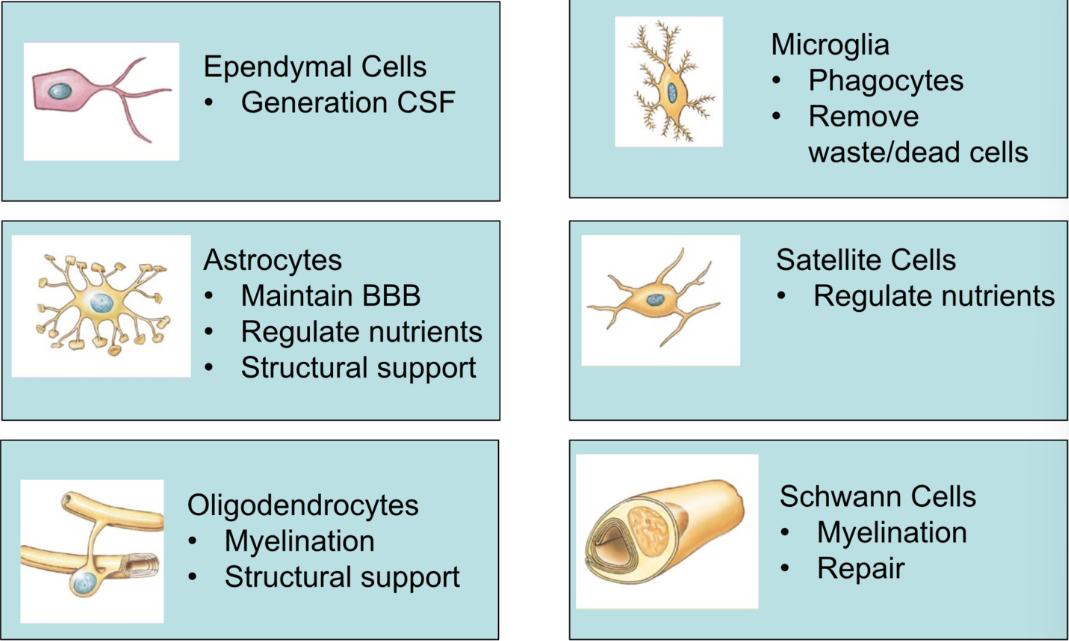
Neuroglia
Supporting Cells, ½ of neural tissue
Ependymal (CNS)
make and secret CSF (cerebral spinal fluid)
Microglia (CNS)
phagocytes
remove waste + dead cells
Astrocytes (CNS)
maintain BBB (blood brain barrier)
monitor what passes through
grab onto neurons and capillary beds of brain
structural support
regulate nutrients
Satellite Cells (PNS)
regulate nutrients in peripheral nervous system
monitor and support
Oligodendrocytes (CNS)
myelination
structural support
Schwann (PNS)
myelination + repair
Supporting Structures
cerebral spinal fluid CFS (from ependymal cells)
clear fluid in brain and spinal cord
function: protection + support, nutrients delivery for brain, remove waste, immune protection
constant circulation (drains into veins)
Blood Brain Barrier
highly selective permeable membrane
specialized capillaries and neuroglia that tightly regulate what moves into CSF from plasma
astrocytes bind neurons to capillary beds of brain → selective food exchange between blood and brain
Myelin (oligodendrites (CNS) and schwann (PNS))
insulating membranes surrounding SOME axons (white matter)
from specialized neuroglia: oligodendrites in CNS, schwann cells in PNS
increase in speed of action potential down axon with some gaps
white matter = myelinated axons
gray matter = unmyelinated axons + cell bodies
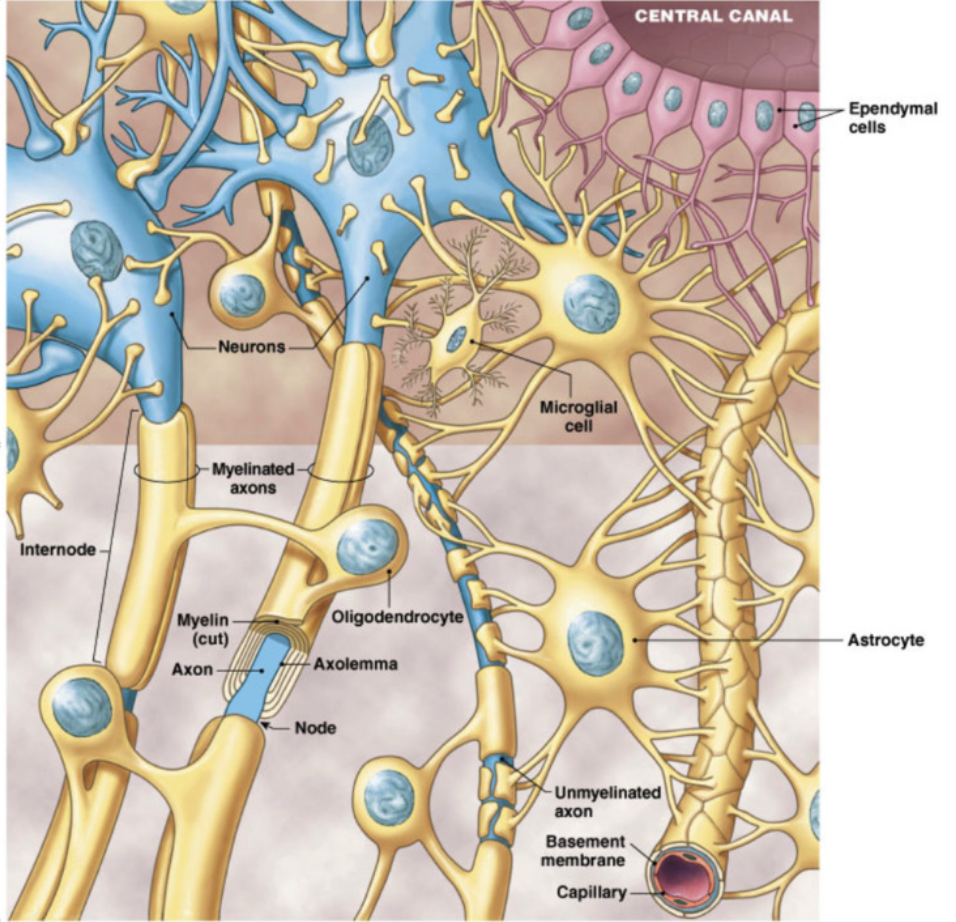
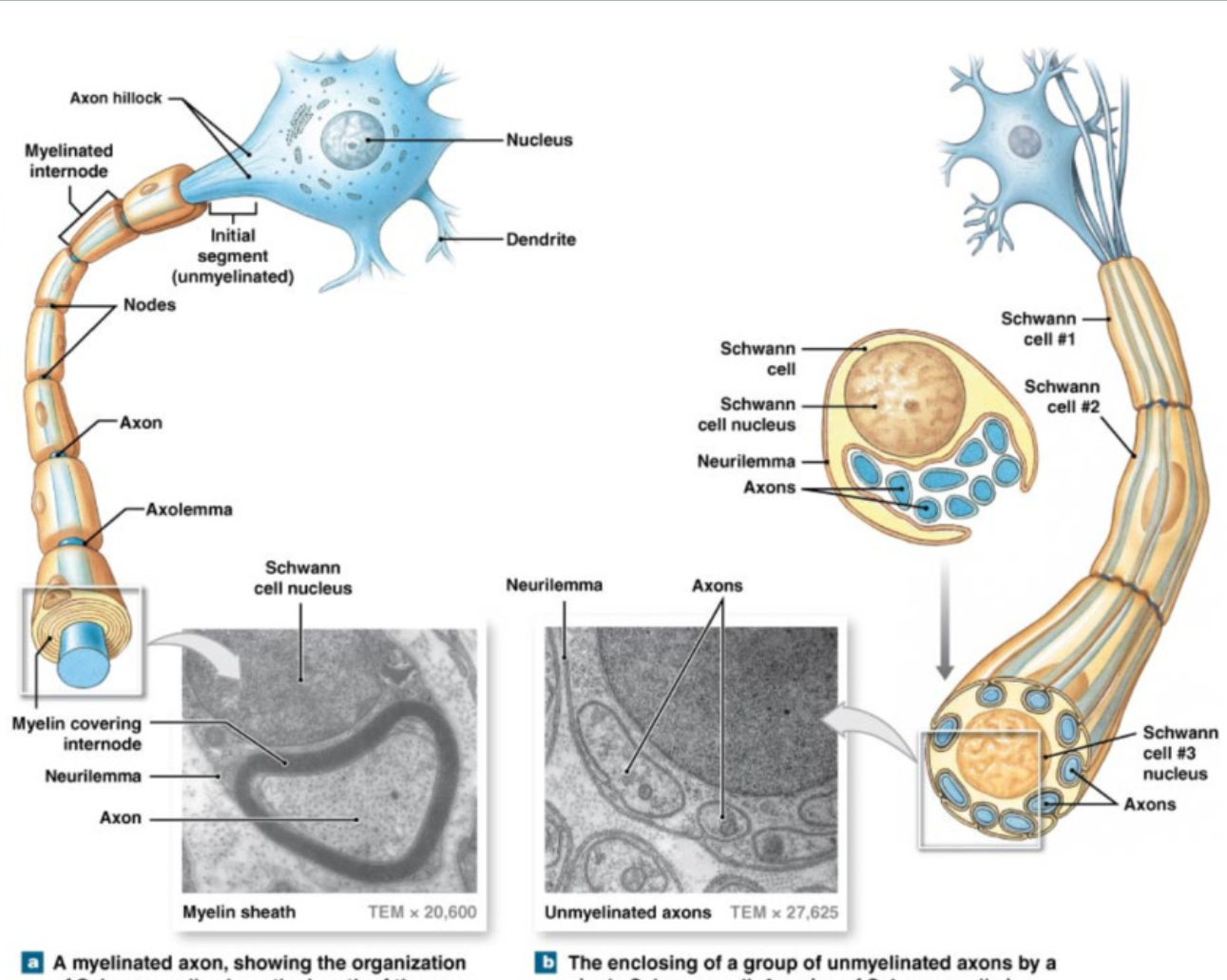
Schwann Cells and Axons (PNS)
White Matter
axons surrounded by myelin → prevent mixture with outside fluid
multiple Schwann cells
gaps = nodes of Ranviet
allow for exchange only at these points
Grey Matter
unmyelinated axons + cell bodies
covered by only 1 Schwann cell → can hold multiple axons
little covered, top of axons still exposed
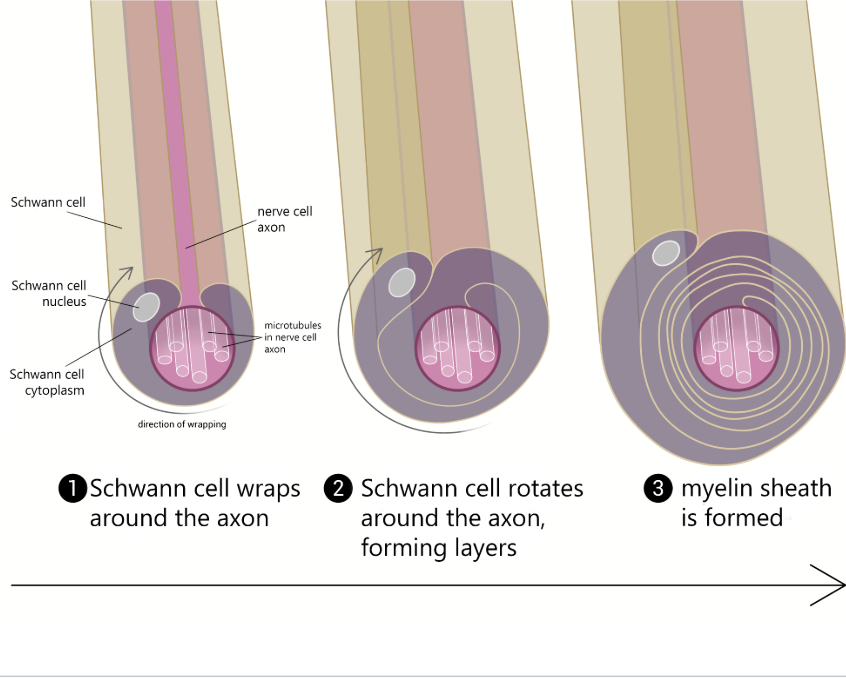
Myelination Process
Schwann cell wraps around axon, forms layers → myelin sheath
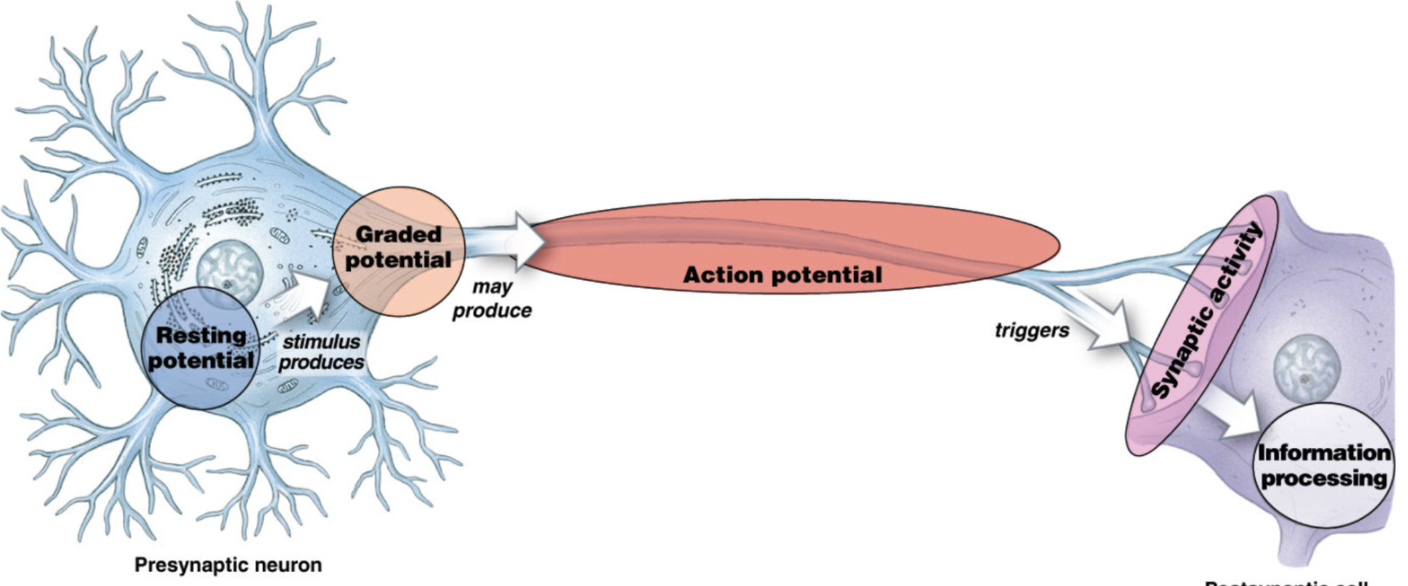
Neural Physiology
resting potential in cell body
graded potential: small localized change in potential, along dendrites → soma
axon hillock: enacts action potential at high enough graded potential
triggers synaptic activity (communication to target cell)
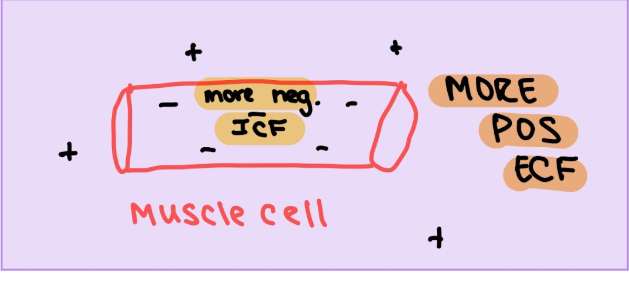
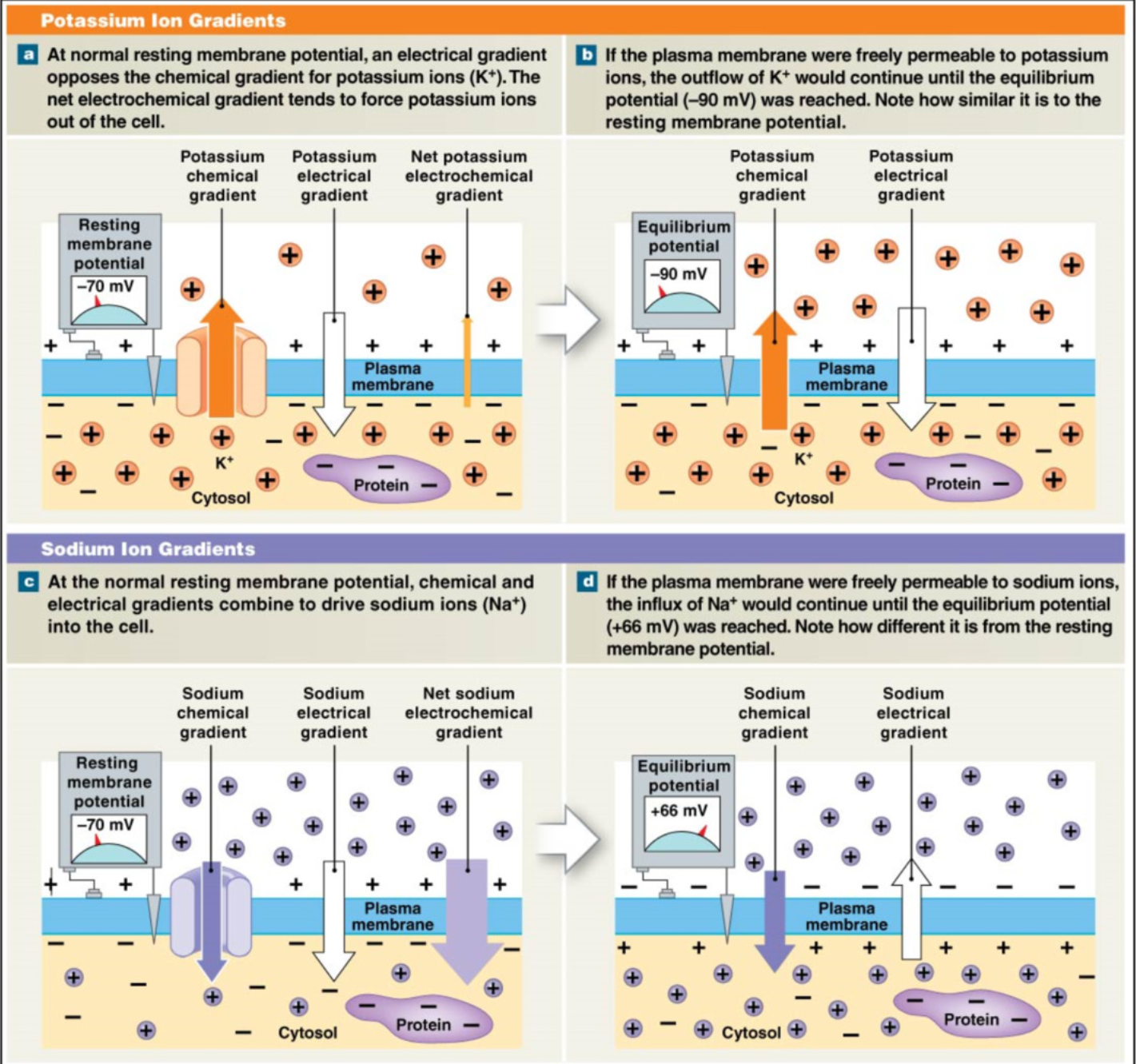
Membrane Potential
Voltage (V) = difference between electrical potential between 2 points (comparing inside to outside of cells, eg -50 mV is more negative on inside)
Current (I) = movement of charge too eliminate voltage
stronger with higher voltage
Resistance (R) = anything impeding movement of charge
I = V/R
Resting Membrane Potential (RMP)
-70 mV for typical neuron (cell is more negative because of proteins)
electrical potential diff across cell membrane during resting condition
3 contributing factors
passive chemical gradients
passive electrical gradients
active transport (K+/Na+ pump)
MOST IMPORTANT IONS in membrane potential =
K+ and Na+
K+ is much more permeable than Na+
Chemical and Electrical Gradients (Passive Transport)
K+
Salty bowl of special K → high K+ in cell →
Chemical Gradients: K+ wants to leave cell
Electrical Gradients: cell is more negative → K+ wants to enter negative cell
opposing gradients → net movement out of cell
K+ leak out cell through leak channels
free K+ movement → equilibrium = -90mV → K+ is much more permeable than Na+
Na+
chemical gradient: Na+ wants to enter cell (more concentration outside)
electrical gradient: Na+ is positive → wants to enter into negative cell
chemical + electrical gradient same direction → Na+ really wants to enter cell
if Na+ allowed to pass freely → equilibrium = +66 mV, → high Na+ resistance → only a little Na+ allowed to pass during RMP
at RMP → movement of ions → leak channels
K+ >>>> Na+
Active Transport: Na+/K+ Pump in RMP
primary active transport: move ions against passive gradient
transport 3 Na+ out, 2K+ in
restore resting membrane potential → remove Na+ leaking in and retrieve K+ leaked out
Changes in RMP
occur due to activation of
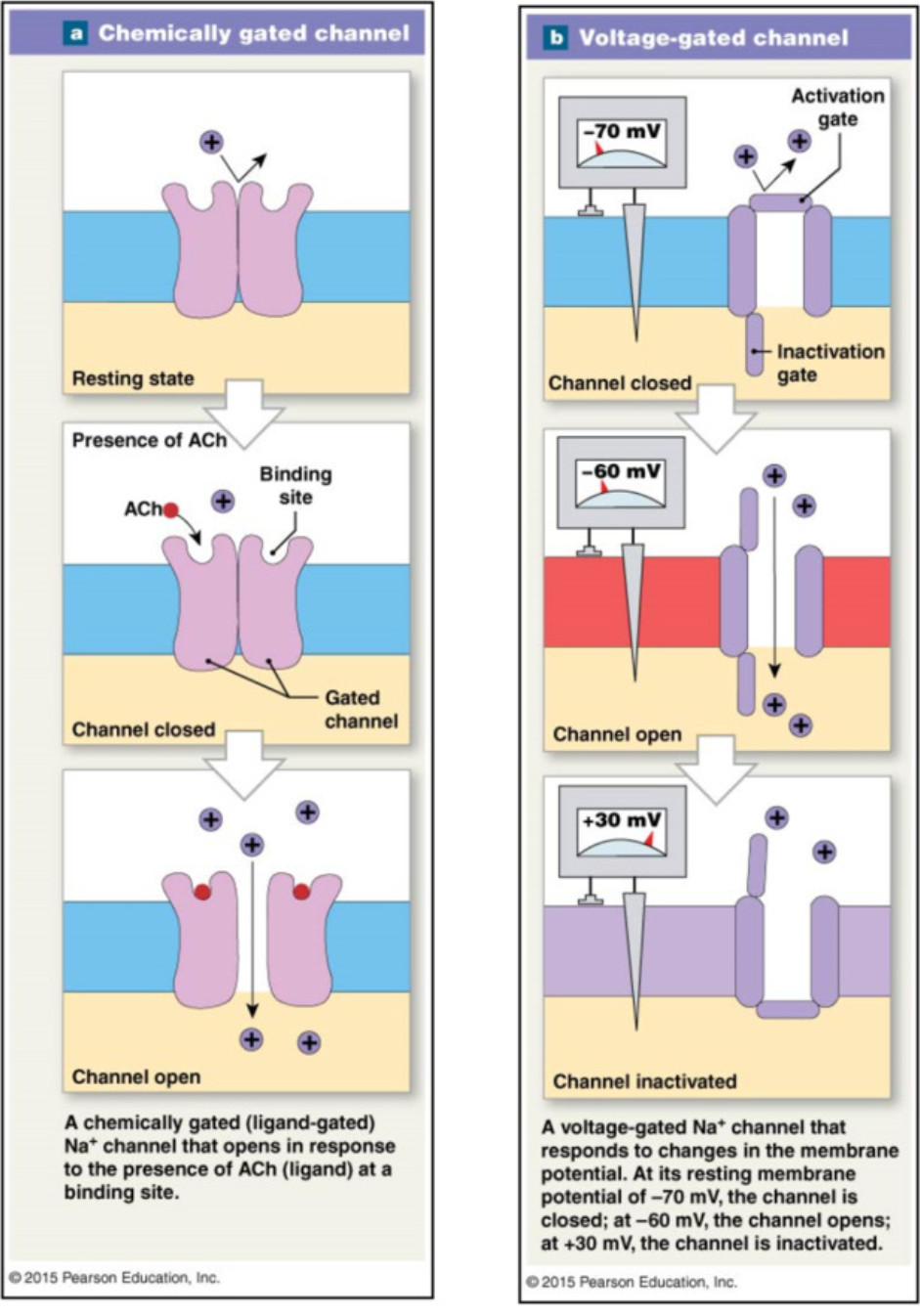
Chemically gated channels → graded potential
binding of chemical → open
Voltage gated channels → action potential
change in voltage → open
Changing Membrane Potential Terms
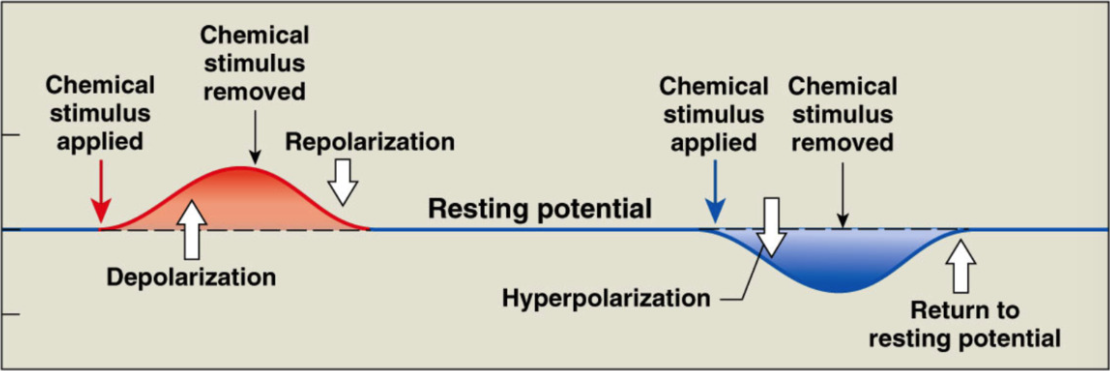
Polarization
inside of cell is negative to outside (RMP = -70mV)
Depolarization
process of making inside of cell less negative -70 → 0
Repolarization
process of returning membrane potential back to RMP + → -70mV
Hyperpolarization
process of making inside of cell MORE negative than RMP (< -70mV)
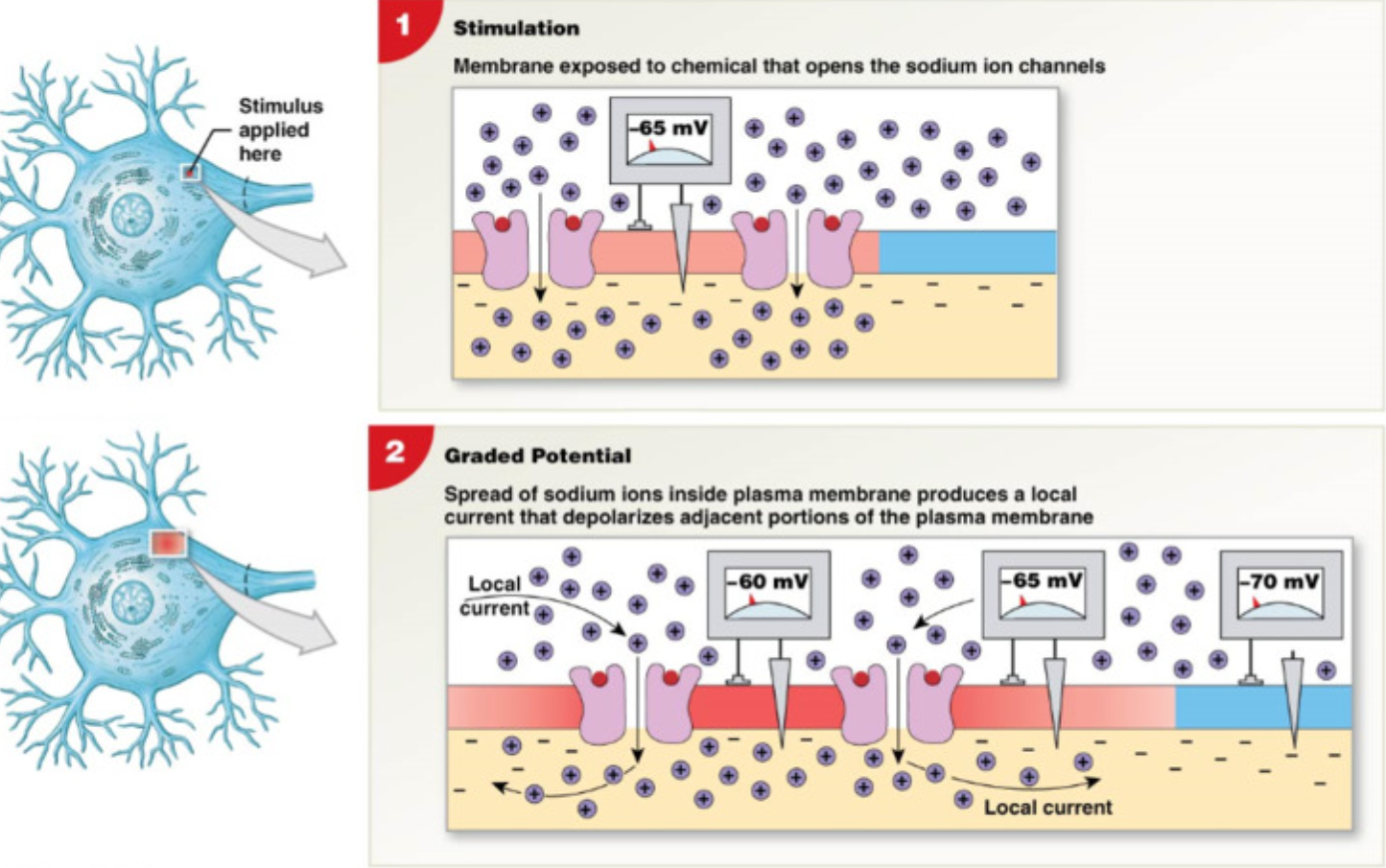
Graded Potential
Localized change in membrane potential (dendrites and soma)
Excitatory post synaptic potentials (EPSPs)
depolarizes (increase voltage >RMP)
open chemically gated Na+ channels → Na+ leak in → cell more +
increases likelihood of action potential
Inhibitory post-synaptic potentials (IPSPs)
hyperpolarizes cell (decrease membrane potential < -70mV)
opens chemically gated K+ channels → K+ leave cell → lose + charge → cell more negative
decrease likelihood of action potential
EPSPs and IPSPs balance → can cancel
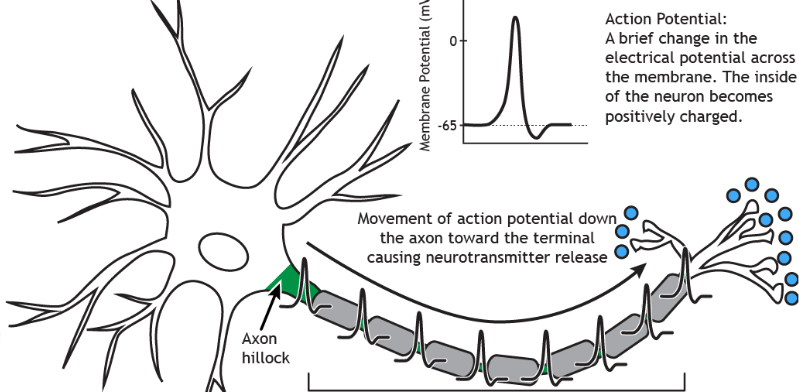
Action Potentials
Change in membrane potential down axon
graded potential from dendrites and soma
localized ion currents → travel down interior of soma to axon hillock (trigger zone)
summates all electrical signals
axon hillock reaches threshold (-60mV) → action potential is triggered
action potential → voltage gated channels (Na+ and K+) on axon
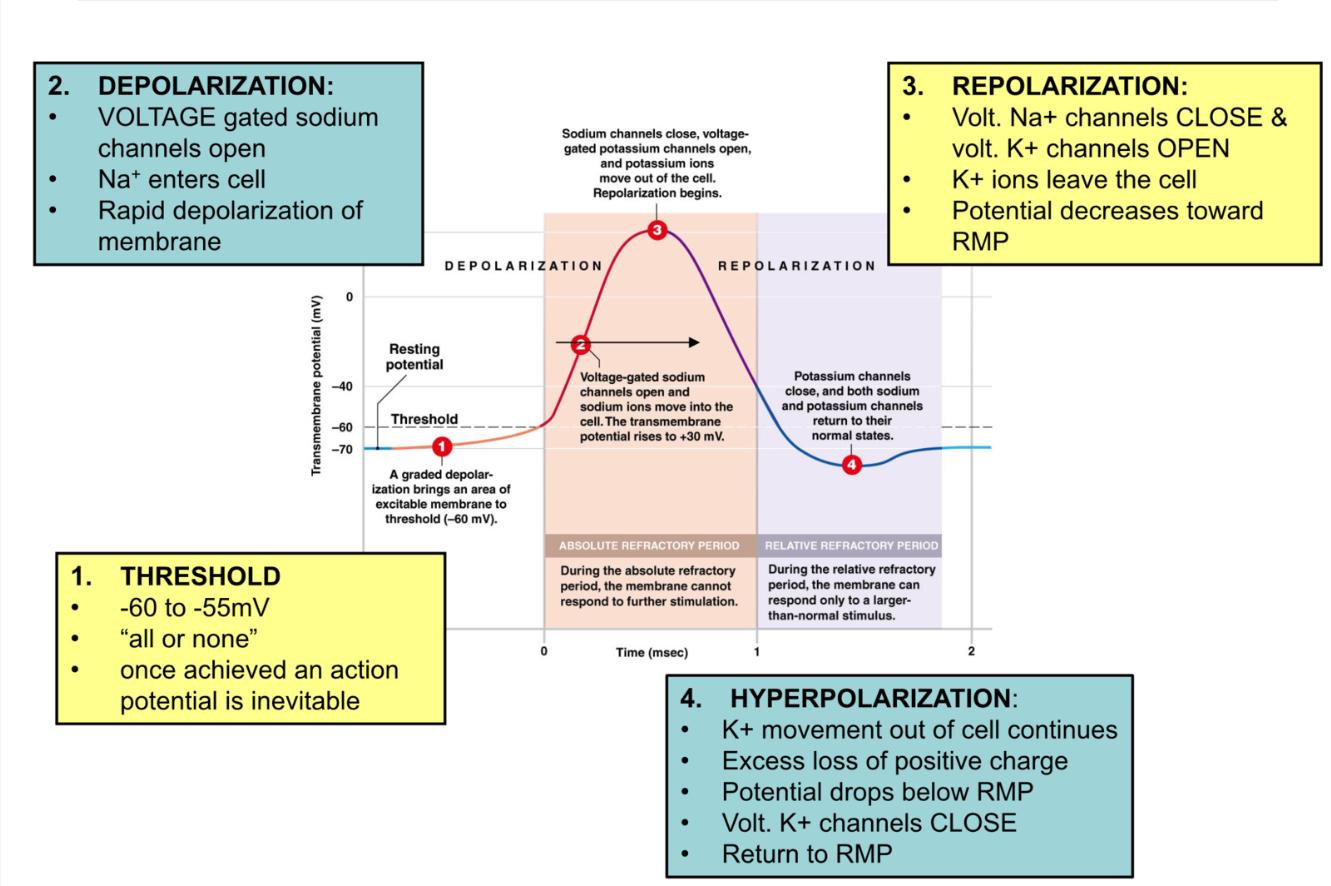
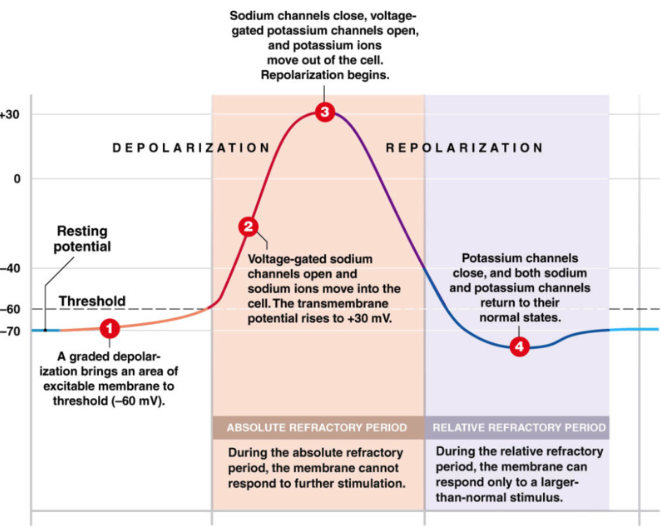
Anatomy of Action Potential
RMP (-70mV) → graded potential →axon hillock threshold (-60mV) → action potential activated
(Na+ active gate = closed, inactive gate = open, inactivates any stimulus response when closed)
Threshold
threshold = -60mV → inevitable action potential
Depolarization (more positive)
open voltage gated Na+ channels → Na+ enter → cell becomes more positive
rapid depolarization
active gate opens, inactive gate open, → Na+ gate open
Repolarization (more negative)
at +30mV voltage gated Na+ channels close (active gate open, inactive gate closed) → stop Na+ flow
voltage gated K+ channels OPEN → K+ leaves cell → cell becomes more negative towards RMP
Hyperpolarization (more negative than RMP)
excess loss of positive charge
around RMP voltage gated K+ channels begin to close but don’t fully close until -90mV
return to RMP
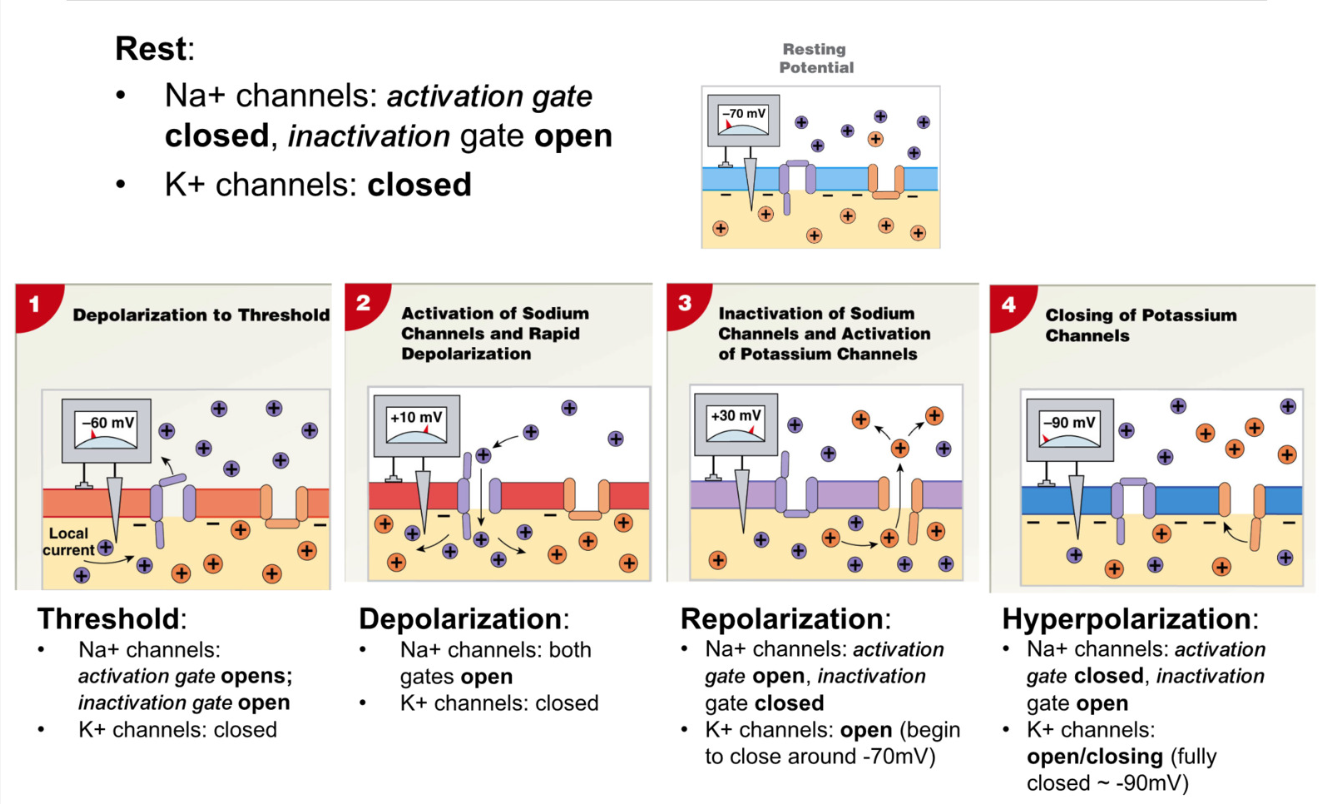
Voltage Gated Ion Channels
RMP
Na+ channels: activation gate closed, inactivation gate open
K+ channels: closed
Depolarization
Na+ channels: activation gate opens, inactivation gate closes
K+ channels: closed
Repolarization
Na+ channels: activation gate closes, inactivation gate closed
K+ channels: open
Hyperpolarization
Na+ channels: activation gate closed, inactivation gate opens (around -40mV)
K+ channels begin closing -70mV, fully closed at -90mV
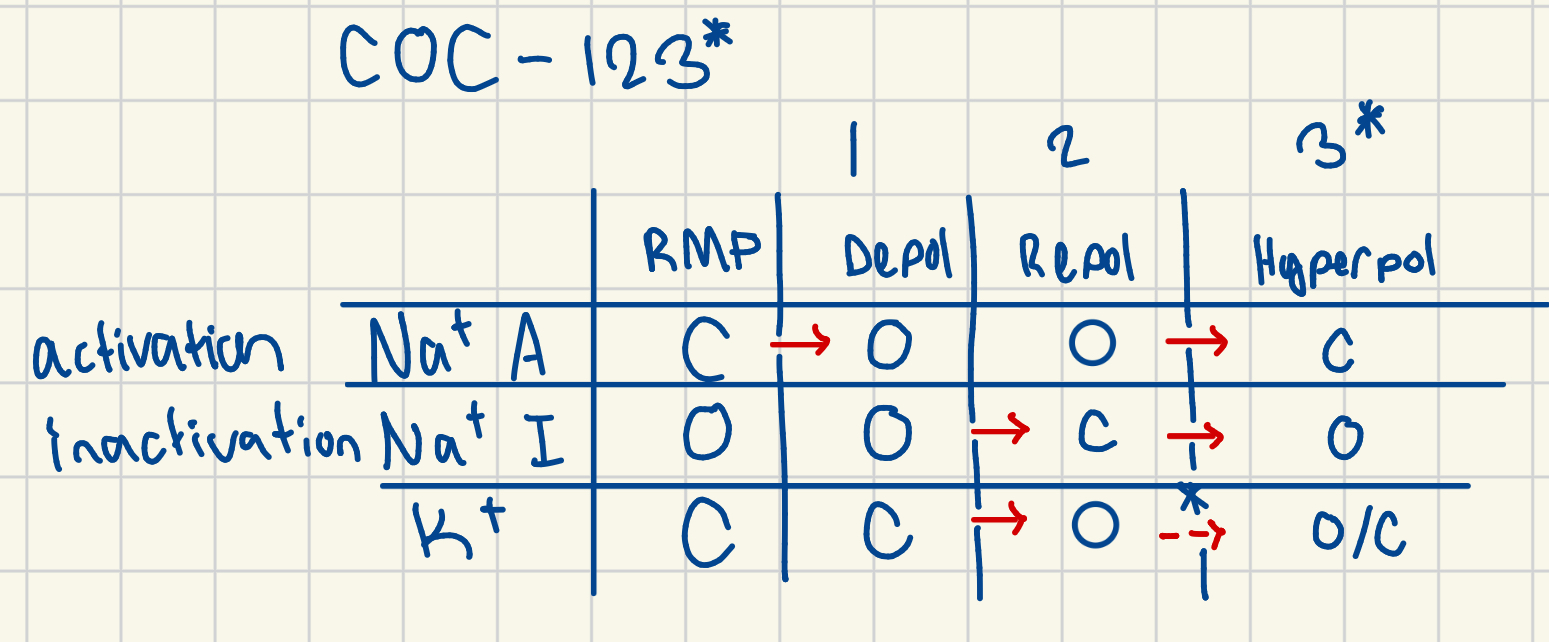
COC-123*
Refractory Period
Absolute Refractory Period
cell cannot fire another action potential
during:
Depolarization: voltage gated Na+ are already open (can’t open more)
Repolarization: voltage gated Na+ channels are already closed, inactivation gate closed → channel is inactivated and can’t open to stimuli
Relative Refractory Period:
cell can only be stimulated to fire action potential if depolarization event → GREATER than usual
during:
Hyperpolarization: more negative than RMP → need more EPSP to reach threshold
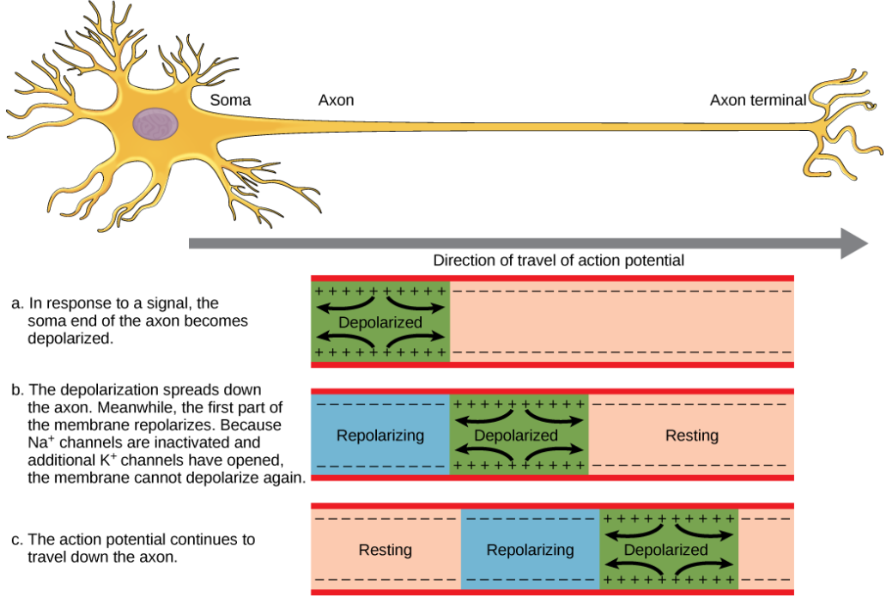
Continuous Propagation (movement of action potential)
unmyelinated axons (slow action potential)
membrane is depolarized in one region
passive Na+ current diffuses locally outside and increases adjacent region’s potential to threshold
adjacent region’s voltage gated channels open → action potential
depolarizes section by section down axon
unidirectional propagation of action potential: regions behind action potential are in refractory period and can’t be reactivated
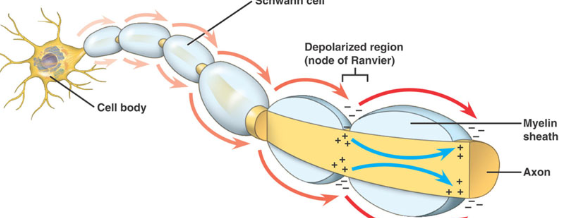
Saltatory Propagation (jumping)
myelinated axons (very fast action potential)
Na+ diffuses, but is contained by myelin, insulator prevents electric potential from being lost
high density of voltage gated channels at Nodes of Ranvier
Na+ current diffuses down interior axon until node of Ranvier → open gated ion channels → action potential simulated → boosts signal → jump from node to node
why is this faster?
Passive Na+ gradient currents are stronger + travel further
less time required to open voltage Na+ channels because potential is mostly conserved
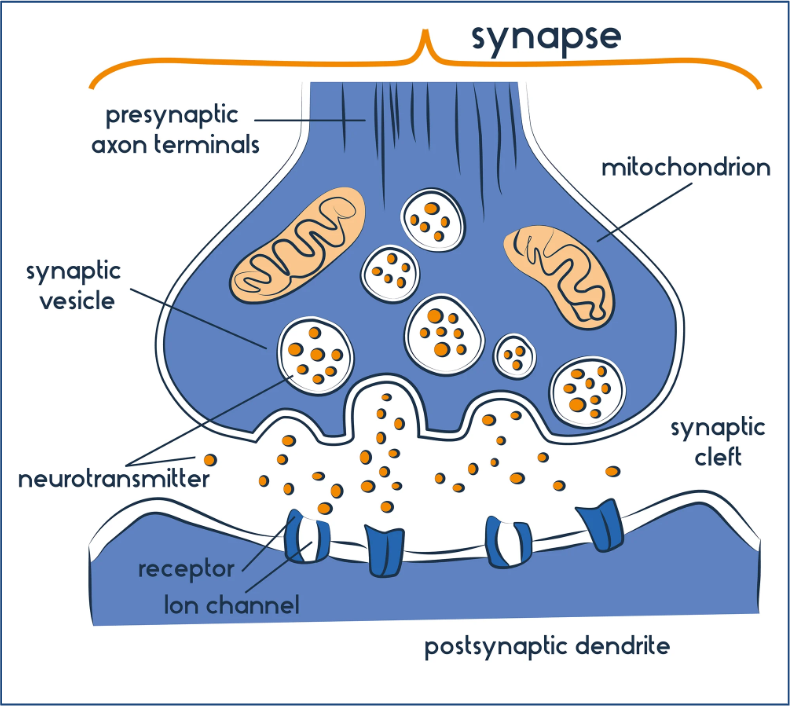
The Synapse
Site of neural communication (action pot. → neurotransmitter)
each synapse contains
Presynaptic cell: neuron
synaptic cleft:
Postsynaptic cell: target neuron or another type of cell
Action potential reaches synaptic terminal:
vesicles attach to presynaptic membrane → release neurotransmitter (nt) into synaptic cleft
neurotransmitter binds to receptors on postsynaptic membrane
may inhibit or excite postsynaptic cell
synaptic terminal can reabsorb and reuse neurotransmitters from cleft
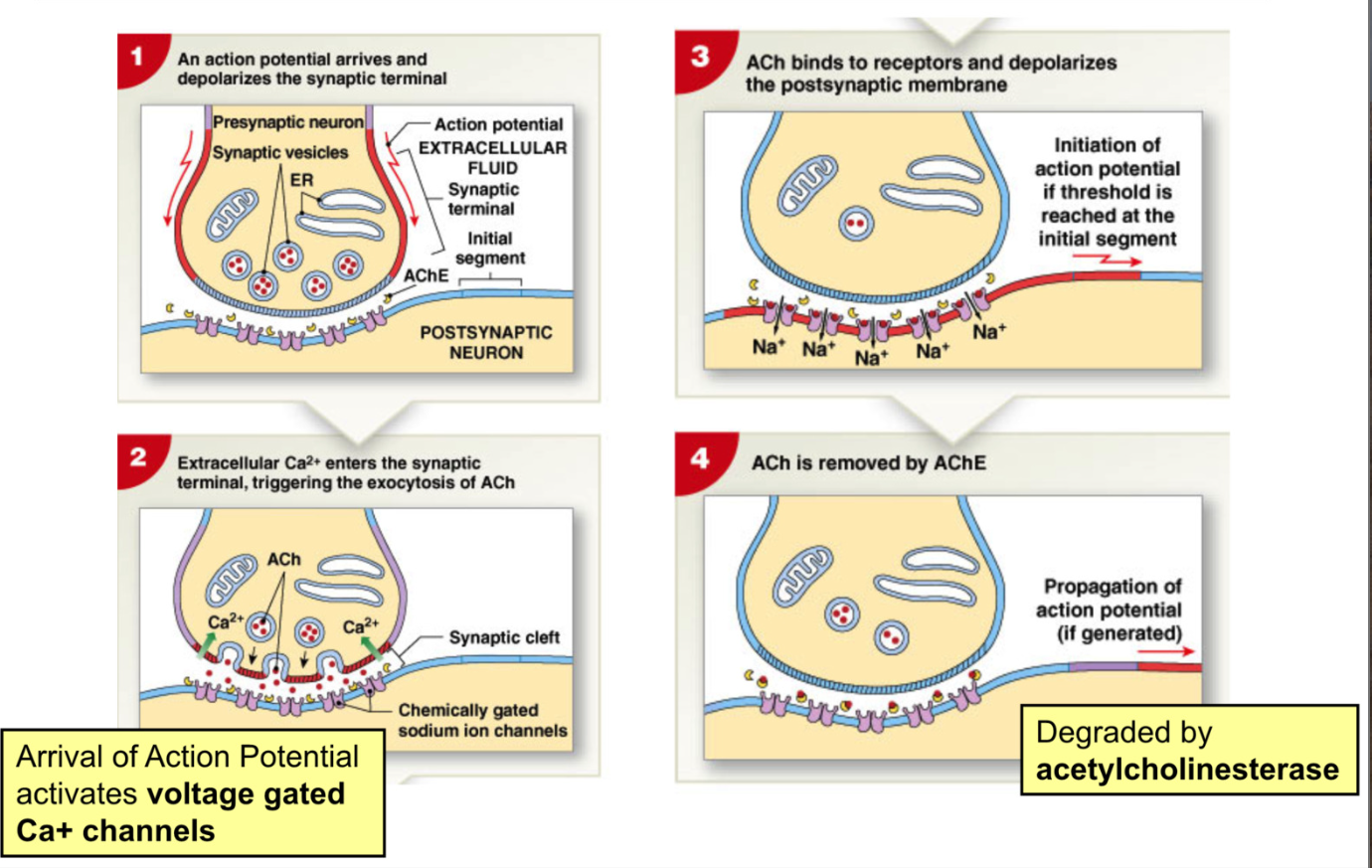
Synaptic Transmission process
Cholinergic Synapse: uses Acetylcholine (Ach) as active transmitter
Action Pot. arrives and depolarizes synaptic terminal
action pot activates voltage gated Ca2+ channels → triggers exocytosis of Ach
Ach binds to receptors and depolarizes postsynaptic membrane
Ach degraded by acetylcholinesterase
Common Neurotransmitters (KNOW)
Glutamate (glutamatergic synapse): Excitatory neurotransmitter → depolarizes post synaptic cell
Gamma-aminobutyric acid (GABA) (gabanergic synapse: inhibitory effect → hyperpolarization
Norepinephrine (NE) (adrenergic synapse): excitatory effect
Dopamine (dopaminergic synapse): excitatory or inhibitory effect depending on location
Serotonin (serotonergic synapse): excitatory effect → involved in attention and emotion
Nitric Oxide (NO) → vasodilation, released by neurons innervating smooth muscle associated with blood vessels
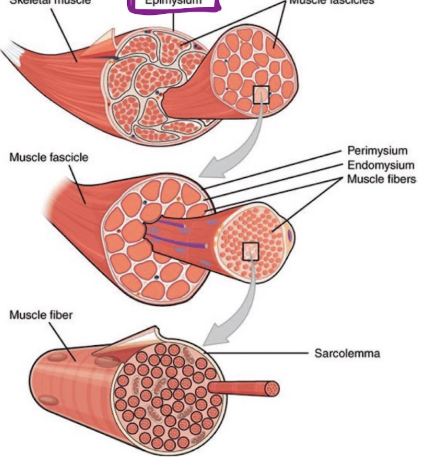
Connective Tissue
Muscle has 3 layers of connective tissue
Epimysium
dense connective tissue (collagen fibers) surrounding and separating the entire muscle
Perimysium
divides muscle into fascicles
collagen, elastin, site of blood vessels
Endomysium
within fascicle, surrounds individual muscle cells, called fibers
elastic tissue, contains nerve fibers and capillary beds
site of myosatellite cells (stem cells)
Development of Myocytes (muscle cells)
large multinucleated cells formed by fusion of many myoblasts
myosatellite stem cells → myoblasts
multiple myoblasts fuse to form myocyte (muscle fiber cell)
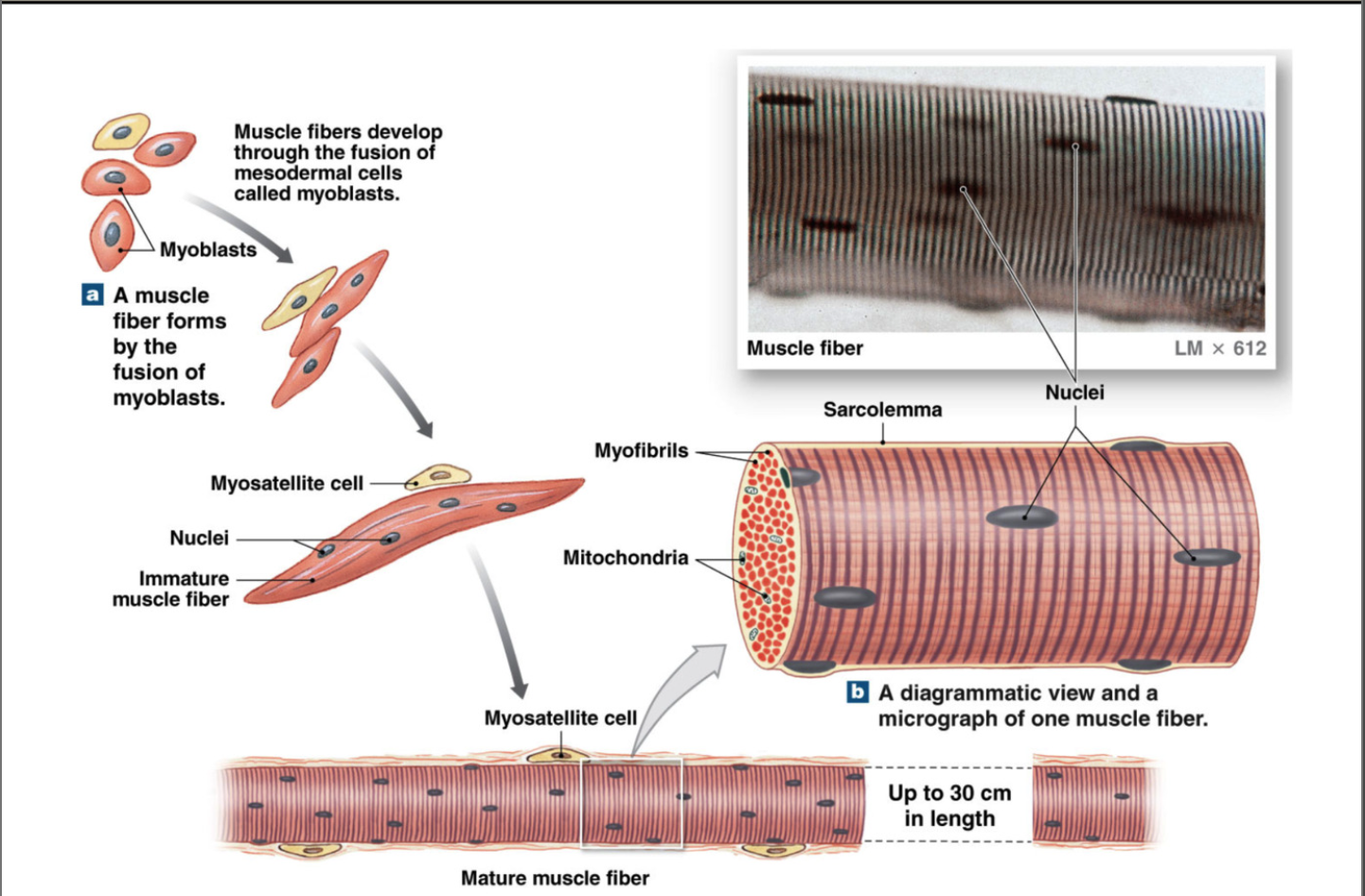
Structure of Muscle Fiber
muscle fibers made of many myofibrils (made of many repeating sarcomeres)cc
myofibrils made of many myofilaments: thin (actin) and thick (myosin) filaments
sarcoplasmic reticulum stores calcium
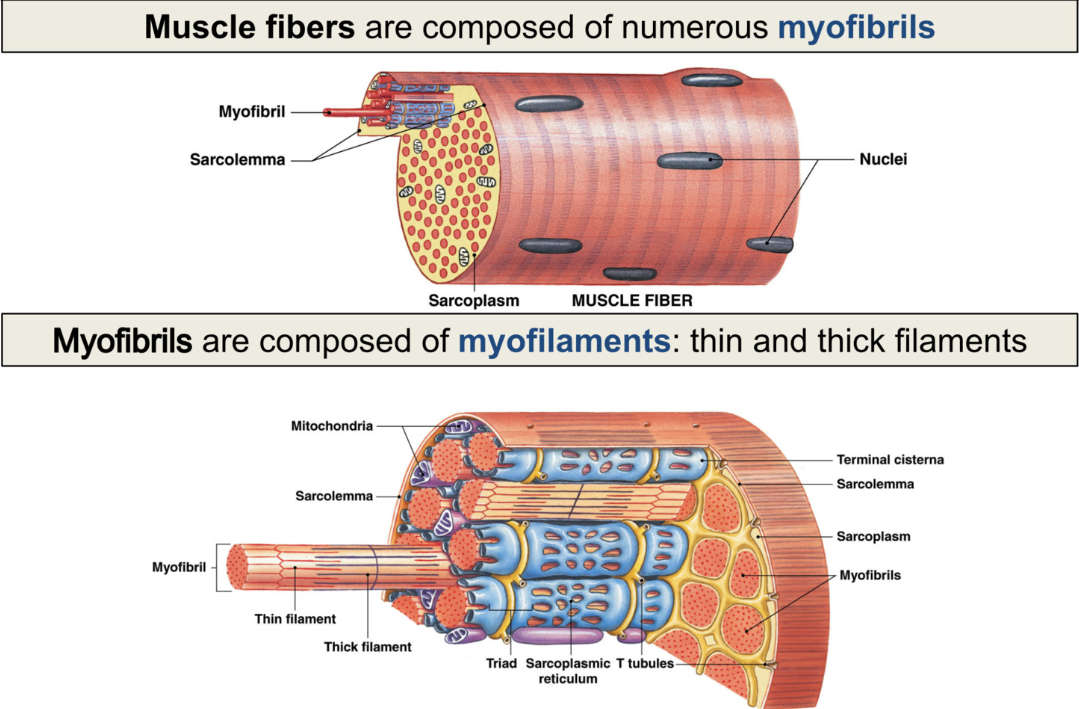
Sarcolemma
plasma membrane of muscle cell
surrounds sarcoplasm (cytoplasm of muscle cells)
polarized like neurons: have membrane potential
more negative inside cell
sudden change in membrane potential → contraction
ion movement across membrane (electrical impulse) → action potential
all parts of muscle cell must contract at same time