Lecture 3 - DNA Replication
1/38
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
39 Terms
Stahl’s Experiment
e. coli is grown in a heavy nitrogen isotope so that its DNA becomes fully labelled with N15 then transferred to N14 (lighter isotope)
samples are collected after 1 and 2 generations of cell division
DNA was extracted and analyzed → found a shift in distribution of DNA weight to lighter side which means DNA is semi-conservative
Semi-Conservative
DNA replication is semi-conservative - newly replicated DNA has a new strand and an original strand
replication is bidirectional, both strands copied at the same time
Minimum Requirements for DNA Replication
original DNA strand - template
enzyme to polymerize the nucleotides into a strand of DNA - DNA polymerase
Problems with DNA Replication
DNA is a helix - solution is to unwind by helicase
template has antiparallel strands and polymerase can only catalyze in the 5’3 direction between OH and phosphate, respectively - solution is semidiscontinuous replication
DNA polymerases cannot initiate chain synthesis de novo - solution is to use a primer made by primase to initiate synthesis
Semidiscontinuous Replication
both strands are synthesized from 5’-3’, one continuously and the other in discontinuous fragments
3’-5’ leading strand is replicated continuously
5’-3’ lagging strand is replicated in short fragments called Okazaki fragments
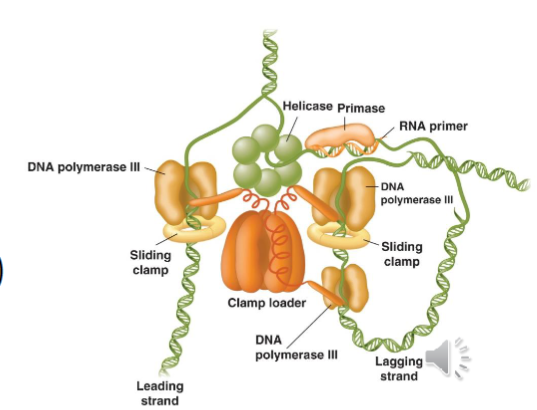
Replisome
multicomponent machine comprised of a helicase, primase, DNA polymerase, and 3’-5’ exonuclease
Intro to Bacterial DNA Replication
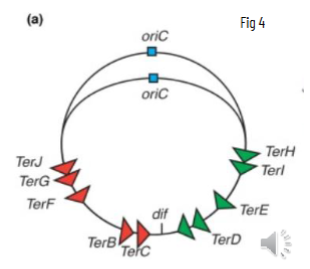
circular genome has a single origin of replication (OriC) and 10 termination sites
OriC is AT rich and termination site has 10 copies of 23 bp sequence specific replication arrest site, bound by Tus
bacteria have 5 DNA polymerases but not all are involved in replication
Bacterial Replication
DnaA (initiator) binds to OriC and melts the DNA
DnaB (helicase) encircles lagging strand and uses ATP hydrolysis to unwind the dsDNA
ssDNA is bound by SSBs to prevent breakdown by nucleases
primase, DnaG makes a 10-12 nucleotide RNA primer using DNA as a template
DnaG interacts with DnaB meaning primers can only be made at replication forks
sliding clamp sliding clamps beta are loaded onto the DNA by clamp loader. Clamp loader uses ATP
hydrolysis to load beta onto the DNADNA pol III are tethered to clamp loaded and also to DNA
2 of 3 pol III molecules are placed on lagging strand and take turns extending RNA primers into Okazaki fragments
replication fork advances and makes a DNA loop
Pol III is released after Okazaki fragment is made or by collision with previous fragment
new sliding clamp assembles on another RNA
primer, and DNA polymerases are associated
anew to make the next Okazaki fragmentsynthesis continues until polymerases meet each other at the Ter sites
after lagging strand is done pol I removes ribonucleotides of primer and replaces with complementary dNTPs
DNA ligase catalyzes formation of phosphodiester linkage between adjacent fragments
To Be Resolved After Bacterial Replication
removal of RNA primer
ligation of the DNA fragments
induction of supercoiling
DNA Polymerase I
made of 2 subunits:
subunit with 5’-3’ polymerase activity and 5’-3’ exonuclease activity called Klenow Fragment
subunit with 3’-5’ proofreading activity
Resolving Bacterial Supercoil
as replication fork moves forward, positive supercoiling happens ahead of the fork
freshly replicated strands are becoming negatively supercoiled
torsional strain is relieved by topoisomerase:
type I does not require ATP hydrolysis
type II requires ATP hydrolysis
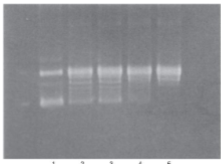
DNA relaxed by topoisomerase can be visualized on agarose gel → supercoiled DNA will move faster through the gel since it’s tightly packed
Type I Topoisomerase
works independent of ATP to unwind DNA
bacterial topoisomerase I can only relax negatively supercoiled DNA but eukaryotic can relax both negative and positive supercoils
in bacteria: binds negatively coiled circular DNA and unwinds DNA by making a transient single strand break in the phosphodiester bond but doesn’t make free ends
enzyme can then pass the unbroken ends between the cut strand and religates the DNA cut strand
Catenation
interlocking of circles of DNA like links in a chain
topoisomerase type II can unknot or decatenate entangled DNA molecules
Type II Topoisomerase
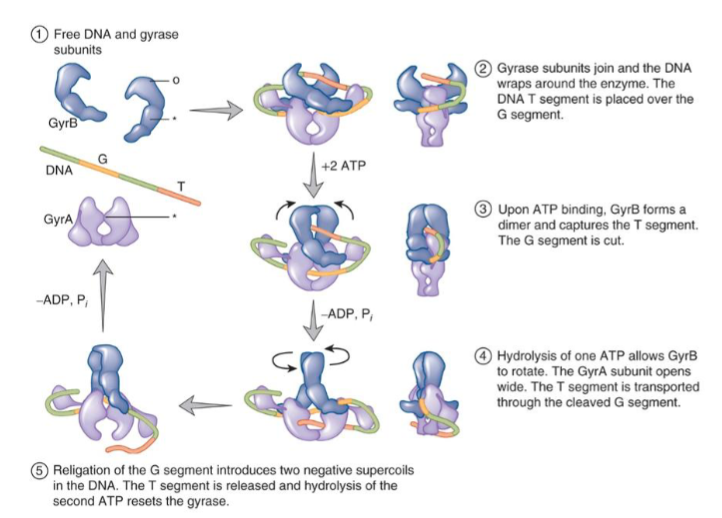
ATP dependent and can relax both positive and negatively supercoiled DNA; decatenate and unknot DNA
makes transient double strand breaks
can induce negative supercoils
makes a dimer when ATP binds, 2 ATP molecules are hydrolyzed to allow for rotation of the gyrase for cleavage and subsequent rotation for reset
Eukaryotic Replisome
linear genome with bidirectional replication means the ends are a problem
there are multiple origins of replication called origins (50000-80000 in humans)
yeast have autonomous replicating sequence but mammals don’t have a conserved consensus sequence
consensus sequence: ideal form of a DNA sequence found in slightly different forms in different organisms'
replication occurs in replication factories in the nucleus - more than 40 active forks clustered here in pulse chase experiments
The rate of genome replication in eukaryotes is affected by ________ and ________.
the number of origins used; the rate at which the origins are initiated
Active Replication Fork
synthesis is restricted to the S phase of the cell cycle
origin recognition is different from replication licensing: regulated formation and activation of prerreplication complexes by CDK to make sure DNA only replicates once per cycle
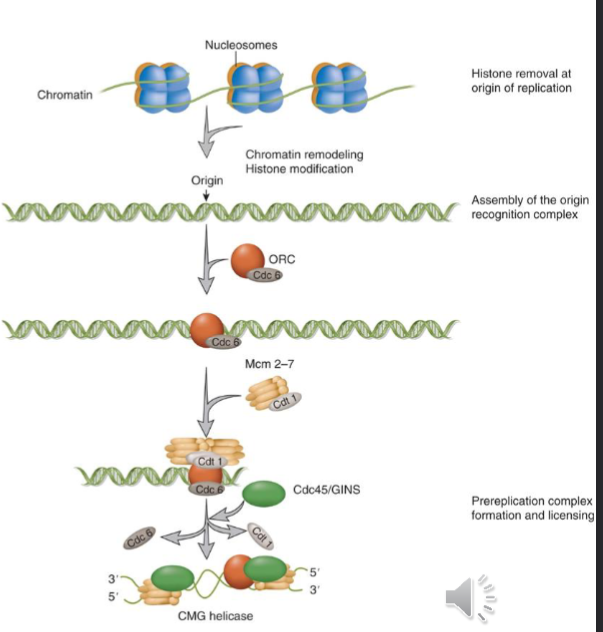
chromatin must be opened up and histones removed
origin recognition complex (ORC) and Cdc6 binds the origin and recruits a single Mcm2-7 helicase in a complex with Cdt1
other proteins like Cdc45 and GINS are loaded and completes pre-initiation complex
Cdc45, Mcm2-7, GINs are transformed into an active helicase called CMG that encircles the leading strands
Replication Licensing
Mcm2-7 acts as the licensing protein and a helicase in the CMG complex
ATP hydrolysis by ORC with the help of Cdc6 are needed to load mcm2-7 at the origin
mcm2-7 is only loaded on the origin when CDK activity is low
when CDK activity is high the licensed origins fire and CDK can phosphorylate Mcm2-7 and prevent it from being loaded at the origin
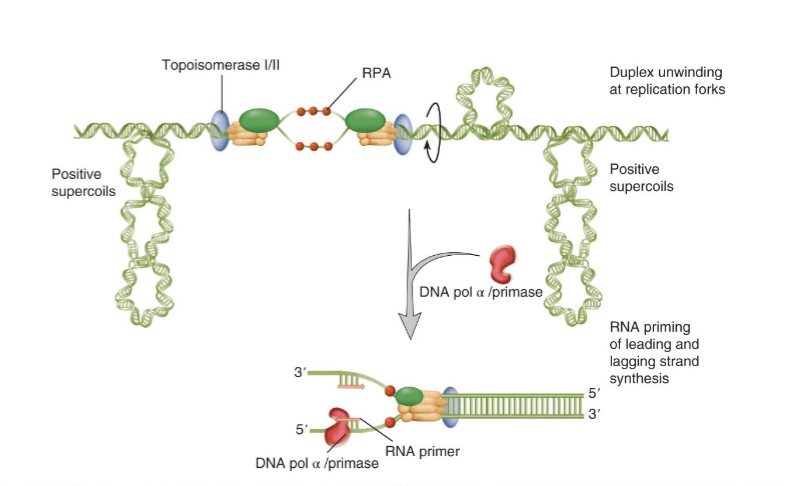
CMG helicase complex goes with replication fork and unwinds DNA ahead of the fork powered by ATP hydrolysis
ssDNA is bound by RPA as DNA is unwound which protects DNA from nucleases
positive supercoil ahead of fork and negative supercoil behind the fork need to be resolved by topoisomerase
Elongation in the Eukaryotic Replisome
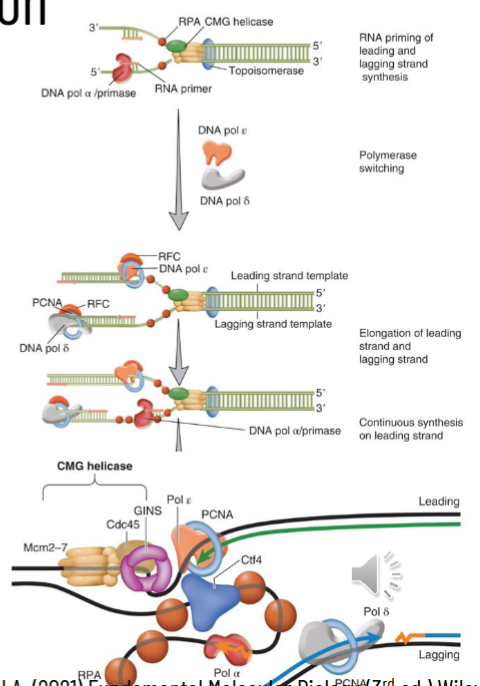
replication involves polymerase switching
once CMG helicase separated DNA, RNA primer has to be laid down
DNA pol-alpha/primase binds to initiation complex and makes a primer of 7-10 nucleotides of RNA and then 25-35 nucleotides of DNA → DNA part serves as initiator
DNA pol-alpha/primase switches to lagging strand for DNA pol-delta for replication; DNA pol-epsilon does replication on the leading strand (polymerase switching)
Elongation and Proofreading
PCNA: proliferating cell nuclear antigen is central to DNA pol switching
this is the sliding clamp for the eukaryotic replisome - gets loaded onto duplex DNA by clamp loader Replication Factor C (RPC) by ATP hydrolysis
PCNA works to prevent the DNA polymerase from losing its place at the replication fork and holds DNA complex together in front of the fork
Proofreading and Termination
DNA polymerases are high fidelity enzymes and proofreading 3’-5’ exonucleases in Pole (leading) and Pold (lagging)
abnormal geometry from mismatched base pair → steric hindrance that pauses pol and the exonuclease domain can excise the mismatched nucleotide before polymerase resuming elongation (slows down overall replication)
Termination and Maturation
replication terminates when the replisomes from neighbouring origins converge
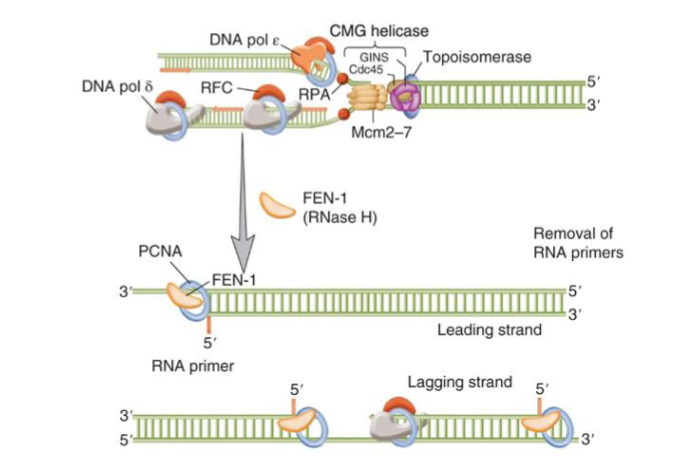
after termination, RNA primer must be removed, gaps filled in, Okazaki fragments joined
when DNA Pol-delta reaches the 5’ end of the preceding Okazaki fragment → strand displacement
flap endonuclease 1 removes the 5’ RNA flap
unreplicated DNA gaps are filled in by pol-delta on the lagging strand → double nick
ligase I joins the ends and distinguishes between RNA with substrates and dNTP is filled in based on the shape of the DNA
PCNA is needed for Okazaki fragment maturation and interacts with ligase I, FEN-1 and pol-delta
nucleosomes that were disassembled into H3-H4 tetramers and H2A-H2B dimers for replication reform behind the replication fork → histones are deposited as soon as there is enough DNA to reform nucleosomes
End Replication Problem
removing the final RNA primer from the lagging strand would leave an unreplicated portion of the DNA but there is no more DNA upstream for pol to fill the gap
left uncorrected, our DNA would get shorter after every round of division → chromosomal instability, loss of genetic information, cell death
Telomeres
telomeric DNA has arrays of TTAGGG repeats which makes a region of dsDNA ending in a single stranded G-overhang
telomeres and shelterin protein complex protect the ends of chromosomes preventing chromosomal end fusions and preventing the ends from being recognized as double strand breaks
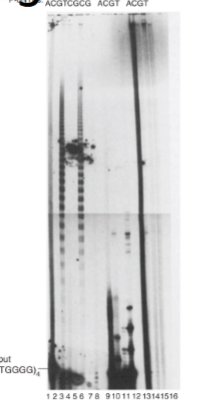
Identifying Telomerase
using cell-free extracts from tetrahymena thermophilia since they have a lot of telomeres
incubated with an oligonucleotide containing 4 repeats of TTGGGG (at end of chromosomes)
sequence was known but protein making telomeres was unknown
provided nucleotides (dNTPs radiolabelled or unlabelled)
after incubation, separated the products of the reaction by gel electrophoresis
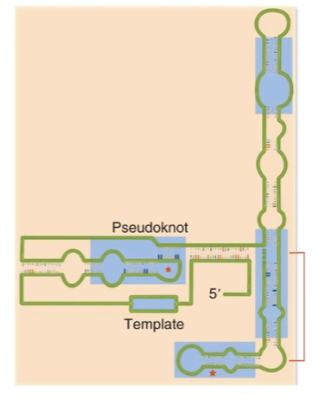
Telomerase
ribonucleoprotein complex has an RNA subunit and a protein subunit
protein component: reverse transcriptase called telomerase reverse transcriptase (TER)
reverse transcriptases make DNA from RNA
RNA subunit is a non-coding RNA called telomerase RNA component (TERC) or telomerase RNA (TER)
A critical feature of the RNA subunit of telomerase is a _____ structure.
pseudoknot
Cajal Bodies
non-membrane bound nuclear suborganelles made of proteins and RNA
plays a role in transcription and RNA processing
this is how telomerase exists when not copying telomeres
Telomerase Recruitment
250 copies of telomerase in each cell traverse the nucleus at 4 uM/s for telomeres
once a telomere is found, telomerase binds and is ready to go
Maintenance of Telomeres
telomerase RNA has a region of complementarity to the C strand so the RNA can then serve as a template for telomere repeat synthesis
telomerase extends the template for the lagging strand (adds nucleotides 5’-3’ on the 3’ overhanging strand)
Undergoes repeated translocation and elongation to create tandem arrays
results in the either the lagging strand
synthesis or t-loop formation
The C strand fill-in, in yeast and ciliate, is carried
out by the replication machinery including DNA pol
α/primase, DNA polymerase δ, RFC, and PCNA
C strand
shortened 5’ strand at the completion of the lagging strand
G strand
3’ 12-16 nucleotide long overhang on the leading strand
Regulation of Telomerase
activity must be regulated to prevent telomeres from getting too long or too short
shelterin: 6 proteins important for protecting chromosome ends and may play a role in preventing telomerase access to telomeres by making a folded structure
t-loop: invasion of double stranded telomeric DNA by the single stranded G strand
g-quadruplex: can also block telomerase activity
if there is an overly long telomere, limiting abundance of shelterin lets TZAP bind long telomeric repeats excise the T-loop
Telomere Maintenance in Drosophila
no telomerase activity → ends are maintained by addition of retrotransposons to ends
Alternative Lengthening of Telomeres (ALT)
another method of maintaining telomeres
observed in telomerase negative tumours where telomerase isn’t used
double stranded breaks at the telomeres triggers “break induced telomere synthesis”
Cells and tissues differ in the amount of _____ activity.
telomerase; specialized cells have less telomerase activity, highly repetitive cells have more telomerase activity
potential consequence in progressive shortening of telomeres by Hayflick Limit (limits # of doublings a somatic cell can do)
when cells reach their limit they enter an irreversible state of cellular aging called senescence and cells no longer divide
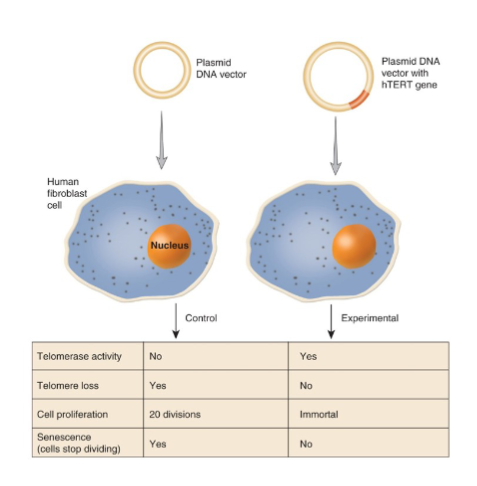
Telomerase in Aging and Cancer
length of telomeres can act as molecular clock where shortening can trigger aging
telomerase reactivation is considered a fountain of youth
most cancer cells have reactivated telomerase and others may use ALT pathway to maintain the telomere lengths
effect of experimental activation of telomerase on normal human somatic cells (telomerase reactivated
Rolling Circle DNA Replication
extrachromosomal DNA in bacteria has to be replicated (plasmids)
gene amplification events also use this
dsDNA is nicked on the positive strand by endonuclease
free 3’ end of nicked strand = primer for host polymerase to start replicating leading strand with negative strand as the template
5’ end of positive (outer) strand is displaced as negative strand gets copied
unit length of new positive strand is made and the displaced parental parent strand is cleaved off and ligated
replication continues and makes another positive strand with negative template
many ssDNA are made that can be a template for lagging strand synthesis and needs a primer
Rolling circle DNA replication is _____ and needs 2 separate steps for ______ and _____ strand synthesis.
asymmetric; leading; lagging