BIO3710 Chapter 3 Bacteria Cell Structure and Cell Wall
1/98
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
99 Terms
most common cell shapes
cocci and rods, though less common are possible
determination of cellular arrangement
plane of division
separation after division
characteristics of cocci bacteria
sphere
diplococci (pairs)
streptococci (chains)
staphylococci (clusters)
tetrads (4 cocci in a square)
sarcinae (8 cocci in a cube)
Rods (Bacilli) characteristics
singe rod
coccobacilli (very short rods)
vibrios (comma-shaped rods)
Bacterial shape: Spirillum
rigid helix
Bacterial shape: Spirochete
flexible helix
Bacterial shape: Mycelium
filamentous, multinucleate
bacterial shape: pleomorphic
variable shape
smallest size bacteria
mycoplasma 0.3 um
average size rod
1.1 - 1.5 × 2-6 um (E. coli)
very large size bacteria
600 × 80 um (Epulopiscium fishelsoni)
organization of bacterial cell
cell envelope (3 layers)
cytoplasm
external structures
plasma membrane
selectively permeable barrier, mechanical boundary of cell, nutrient and waste support, location of many metabolic processes ( respiration, photosynthesis) detection of environmental cues for chemotaxis
gas vacuole
an inclusion that provides buoyancy for floating in aquatic environment
ribosomes
protein synthesis
inclusions
storage of carbon, phosphate, and other substances; site of chemical reaction (microcompartments); movement
nucleoid
localization of genetic material (DNA)
periplasmic space
in typical Gram-negative bacteria, contains hydrolytic enzymes and binding proteins for nutrient processing and uptake; in typical Gram-positive bacteria, may be smaller or absent
cell wall
protection from osmotic stress, helps maintain cell shape
capsules and slime layers
resistance to phagocytic adherence to surfaces
fimbriae and pili
attachment to surfaces, bacterial conjugation and transformation, twitching
flagella
swimming and swarming motility
endospore
survival under harsh environmental conditions
bacterial cell envelope
plasma membrane, cell wall, and surrounding layers
Fluid Mosaic Model of Plasma membrane
peripheral protein in plasma membrane
loosely connected proteins
integral protein of plasma membrane
embedded in membrane
hopanoid in plasma membrane
bacterial version of cholesterol
bacterial cell wall
rigid structure just outside plasma membrane
made of peptidoglycan
2 types (G+ and G-)
Functions of Cell Wall
maintain shape of bacteria
protect cell from osmotic lysis and toxic materials
can contribute to pathogenicity
peptidoglycan
meshlike polymer of identical subunits forming long strands
two alternating sugars in peptidoglycan
N-acetylglucosamine (NAG)
N-acetylmuramic (NAM)
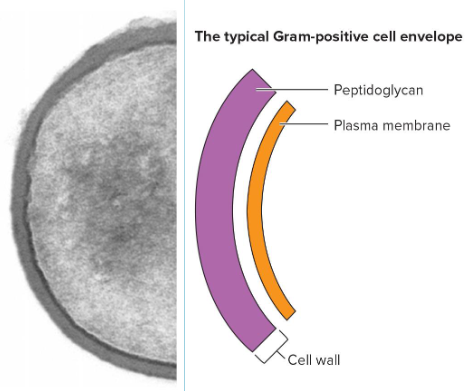
Gram-Positive (G+)
thick peptidoglycan
small or no periplasm
many contain teichoic acids
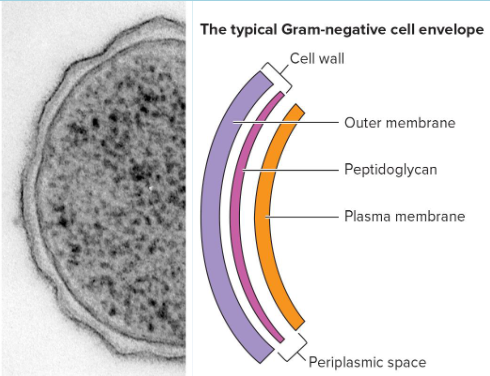
Gram-Negative (G-)
thin peptidoglycan
large periplasm
outer membrane of lipids, lipoproteins, and lipopolysaccharides (LPS)
cytoskeleton
homologues of 3 eukaryotic cytoskeletal elements in bacteria
cytoskeleton in eukaryotes
microfilaments (actin)
microtubules (tubulin)
intermediate filaments (lamin and keratin)
cytoskeleton in bacteria
Mreb and Mbl- maintain cell shape in rods
FtsZ- forms ring at center of dividing cell
CreS- forms curve shape of Caulobacter crescentus
Plasma membrane infoldings in Intracytoplasmic membranes
photosynthetic bacteria
highly respiratory bacteria
Anammoxosome in Planctomycetes
organelle serving as site of ammonia oxidation
bacterial cell inclusions
aggregation of organic or inorganic substances
granules, crystals, or globules
free in cytoplasm or enclosed in shell
storage or reduces osmotic pressure
quantity varies with nutritional status of cell
storage inclusion
store nutrient or metabolic end product
glycogen
long branched chain of glucose units
Poly-beta-hydroxybutyrate
carbon storage
form distinct bodies able to be viewed with light microscope
industrial use - biodegradable plastics
polyphosphate granules
phosphate storage
metachromatic granules- appear red or blue when stained with blue dyes
sulfur globules
storage reservoir for sulfur
gas vacuoles
provide buoyancy to aquatic organisms
regulate buoyancy to float at proper depth
Aggregates of gas vesicles
single protein subunits assembled into cylinder
impermeable to water
permeable to gases
Magnetosomes
found in aquatic bacteria
intracellular chains of magnetite (Fe3O4) particles enclosed in plasma membrane
tiny magnets used to orient to earth’s magnetic field
MamK cytoskeletal protein
helps form magnetosome chain
Ribosomes
found throughout cytoplasm and near plasma membrane
composed of proteins and RNA molecules
Nucleoid
region with chromosomes and proteins
chromosomes in bacteria
most circular and double stranded
how are the chromosomes longer than the length of the cell?
supercoiling
Nucleoid-associated proteins (HU and condensins)
Plasmids
small extrachromosomal self replicating DNA
circular (common) or linear (rare)
single or multi copy
episomes
plasmids integrated into and replicated with the chromosomes
curing
loss of a plasmid
spontaneously or by treatment
conjugative plasmids
transport themselves to other bacteria
F factor: fertility factor
R factor: resistance factor
Bacteriocin
encoding plasmids
virulence plasmids
more pathogenic plasmids
metabolic plasmids
genes for enzymes that degrade substances
TOL plasmids
degradation of aromatic compounds
why are gas vacuoles bound by proteins rather than lipid membrane?
gas vacuoles are more permeable
benefits plasmids give a bacterial cell
antibiotic resistance genes in plasmids
virulence plasmids can make bacteria more pathogenic
code to metabolize certain components
what would happen if the creS gene was removed
it would alter the shape of the bacteria
what would happen if the ftsZ was removed
would not be able to form the septum to divide
inclusions containing carbon are commonly found in the form of __________
poly-beta-hydroxybutyrate
certain bacteria accumulate magnetite in _______ that can be used to sense _______
magnetosomes; earths magnetic field
glycocalyx
outside cell wall
capsules and slime layers
capsules
usually made of polysaccharides
visible in light microscope
resistant to phagocytosis
protective to dessication
exclude virus and detergent
slime layers
similar to capsules but less organized
used for motility
S layers
Layers of protein or glycoprotein
S Layer in G-
outside outer membrane
S layer in G+
peptidoglycan surface
S layer function
protect from ion and pH fluctuation
promote adhesion and protect from host
self assemble
pili/fimbriae
short appendages, thinner than flagella
slender tubes of helical protein subunits
attach to solid surface
type IV in G- bacteria
motility
uptake of DNA during transformation
sex pili
larger, coded by conjugative plasmids
flagella
motility and attachment to surfaces
virulence factor
Distribution of Flagella
monotrichous- one
amphitrichous- one at each pole
lophotrichous- cluster at pole(s)
petritrichous- all over
flagella filament
cell surface to tip
hollow cylinder of flagellin subunits
capping protein at end
flagella basal body
embedded in the cell
rings drive flagellar motor
flagella hook
flexible link of filaments to basal body
flagella synthesis
complex (20 to 30 genes)
type III like secretion system
flagellin subunits transported through hollow core
spontaneously aggregate using filament cup
grows at the tip, not the base
swimming motility
flagella rotates like propellar
powered by motor in basal body (Proton Motive Force)
swimming counterclockwise
cell moves forward (run)
swimming clockwise
cell moves randomly (tumble)
Biased random walk
bacteria move randomly without chemical gradient
Attractant→ longer runs toward attractant
Repellant→ longer runs away from repellant
swarming motility
on moist surface as bacteria group behavior
produce molecules to aid movement
spirochete motility
corkscrew shape flexes and spins
endoflagella
multiple flagella form axial fibril in periplasm that wraps around cell
twitching
type IV pili
short jerky motions when cells in contact
pili extend (contact surface) and retract (oull cell forward)
gliding
smooth motions
endospore
dormant cell formed when nutrients are low
resistant to heat, radiation, chemicals, and desiccation
endospore structure
many layers
thin exosporium covers spores
thick layer of proteins form spore coat
core has nucleoid and ribosomes
what makes endospore so tough?
Ca2+and dipicolinic acid in the core
dehydrated core
protection from spore coat and exosporium
function of Ca2+ and dipicolinic in endospore
interact with small acid-soluble DNA-binding proteins (SASPs) to protect DNA
sporulation process
10 hours
axial filament formation
septum formation and forespore development
engulfment of forespore
cortex formation
coat synthesis
completion of coat synthesis, increase in refractility and heat resistance
lysis of sporangium, endospore liberation
germination activation
spore prepare for germination
germination
environmental nutrients detected
breakdown of peptidoglycan
release of Ca2+ and dipicolinic acid
increased metabolic activity
water uptake
germination outgrowth
emergence of vegetative cell