Microbiology Midterm #1
1/96
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
97 Terms
Microrganisms
Anything microscopic, except for large bacteria, algae, and microscopic worms. Includes bacteria, archaea, and eukaryotes. (such as fungi, algae, and protozoa)
What are smaller organisms good for?
Energy efficiency and there is a larger surface area to volume ratio
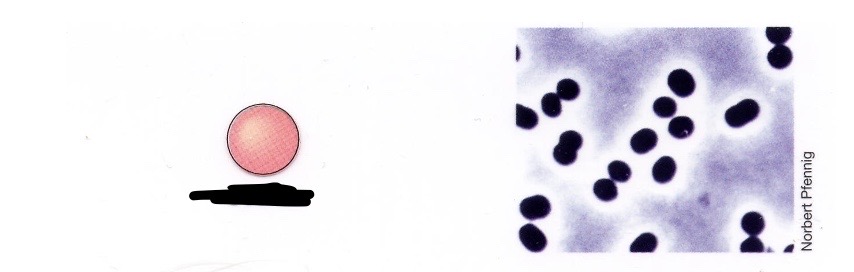
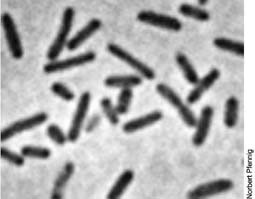
Coccus
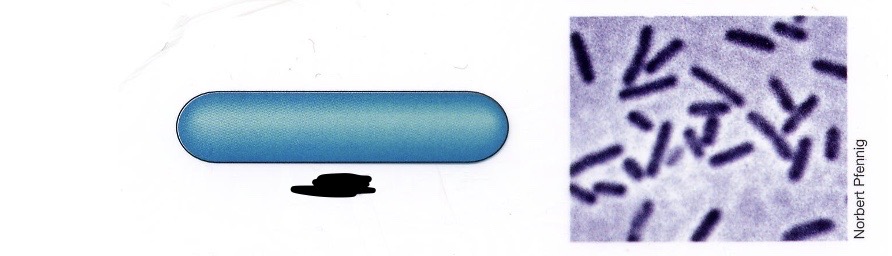
Rod
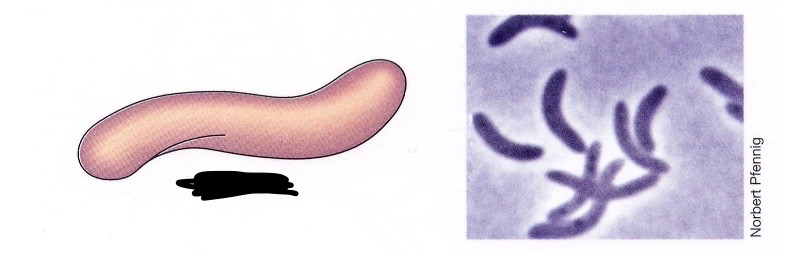
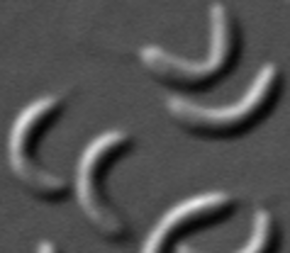
Spirillum
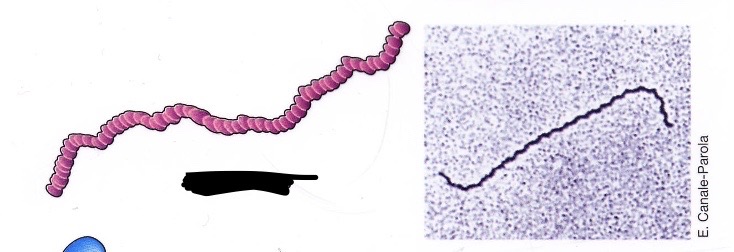
Spirochete
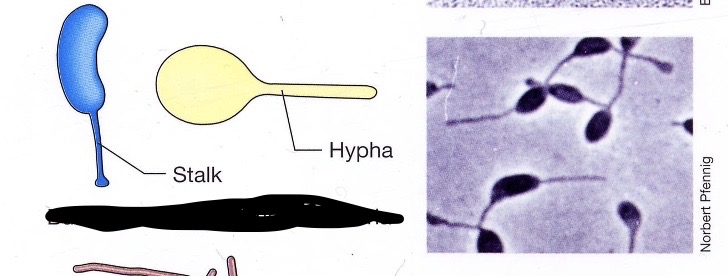
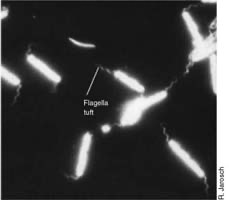
Budding and appendaged bacteria
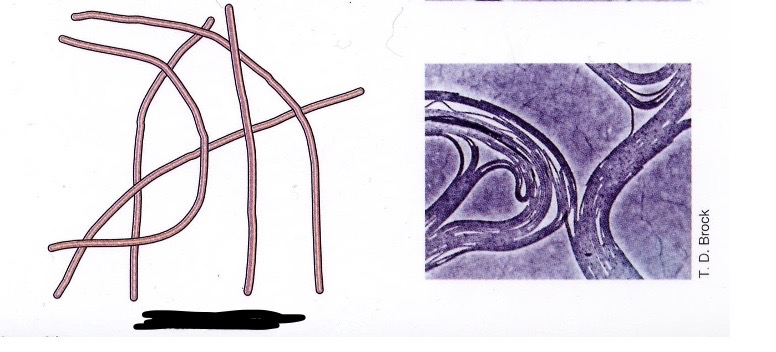
Filamentous
Light Microscopy
Using a compound microscope, light is the source of protons. You use an objective lens and an ocular lens. The light interacts with the objective to create a contrast. (100x magnification)
Absorption- Light Microscopy
The object blocks part of the light.
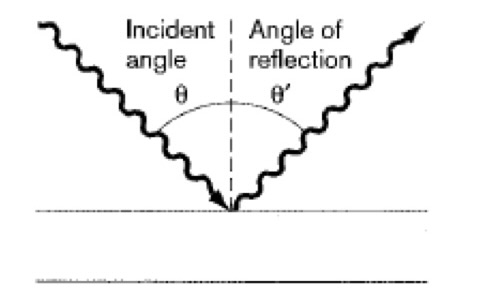
Reflection- Light Microscopy
The wavefront of the light bounces off the surface.
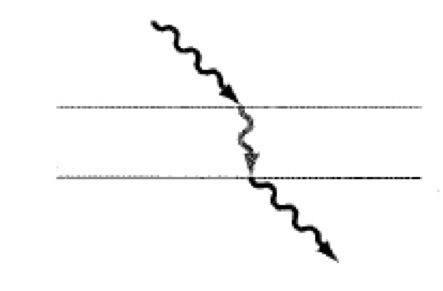
Refraction- Light Microscopy
The light bends when it enters a substance that changes its speed.
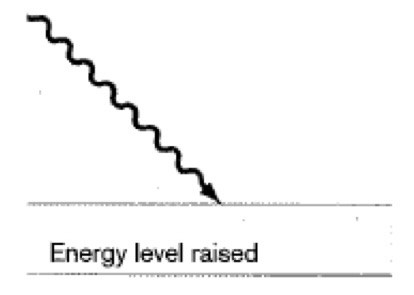
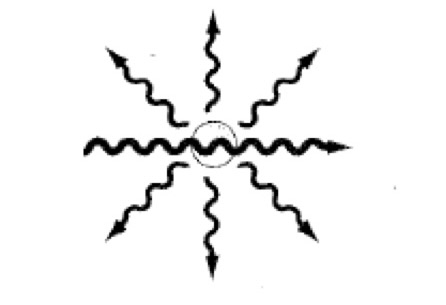
Scattering- Light Microscopy
A small fraction of the incident light is scattered in all directions. Occurs when the dimensions of the object are close to the wavelength of incident light. When you shine light, a little of it will get dispersed. The shorter the wavelength, the better the resolution.
Brightfield
Sample illumination is transmitted (i.e., illuminated from below and observed from above) with white light, and contrast in the image is caused by attenuation of the transmitted light in dense areas of the sample.
With this, people often use dyes that are normally positively charged. Sometimes the dyes are detrimental to bacteria.
Phase Contrast
Accentuates the contrast of bacteria compared to the background. Converts phase shifts caused by light passing through different densities into brightness changes, using a phase plate to shift the background light. Best For: Thin, transparent specimens, such as live cell cultures (bacteria, yeast).
Dark field
The environment is black because there is no scattering. Light is caught.
Light enters the microscope for illumination of the sample.
2. A specially sized disc, the patch stop (see figure), blocks some light from the light source, leaving an outer ring of illumination.
3. The condenser lens focuses the light towards the sample.
4. The light enters the sample. Most is directly transmitted, while some is scattered from the sample.
5. The scattered light enters the objective lens, while the directly transmitted light
simply misses the lens and is not collected due to a direct-illumination block
(see figure).
6. Only the scattered light goes on to produce the image, while the directly transmitted light is omitted.
Differential interference contrast
Creates a shadow of sides, looks 3D (like an illusion). Uses Nomarski prisms to split light, creating two slightly offset beams that interfere, revealing optical path differences (gradients). It is best for thick specimens like tissues and embryos.
Fluorescence
Have to have flor. Emit light at high fluorescence after activation with a laser. Can use dyes

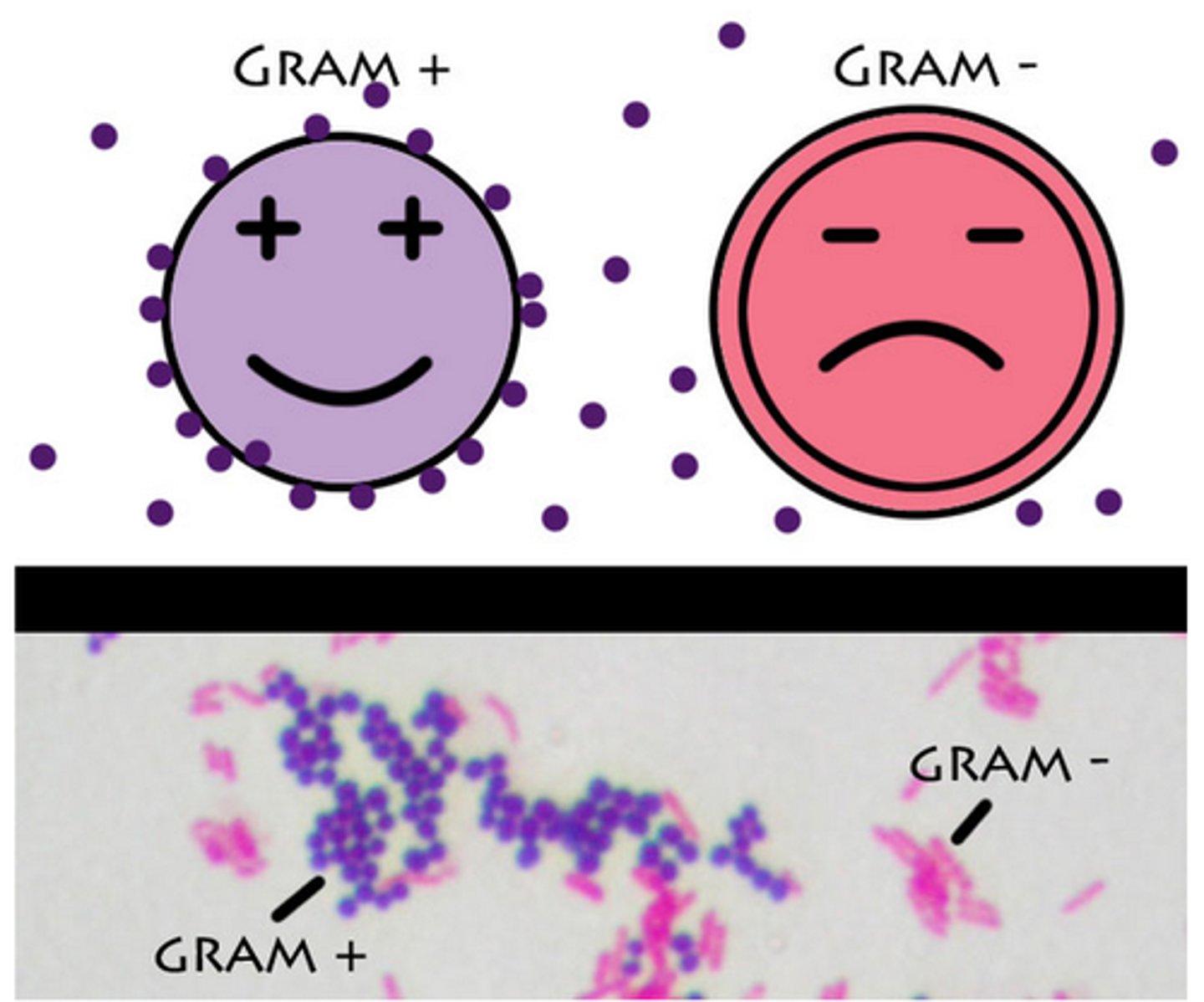
Gram Stain
Gram negative bacteria stains negative which is pink and gram positive bacteria stains positive which is purple. The difference is in the cell walls.
Electron Microscopy
Electron microscopes that use electrons instead of photons and electromagnets instead of lenses. You need to work under a vacuum because you do not want electron interference with the air.
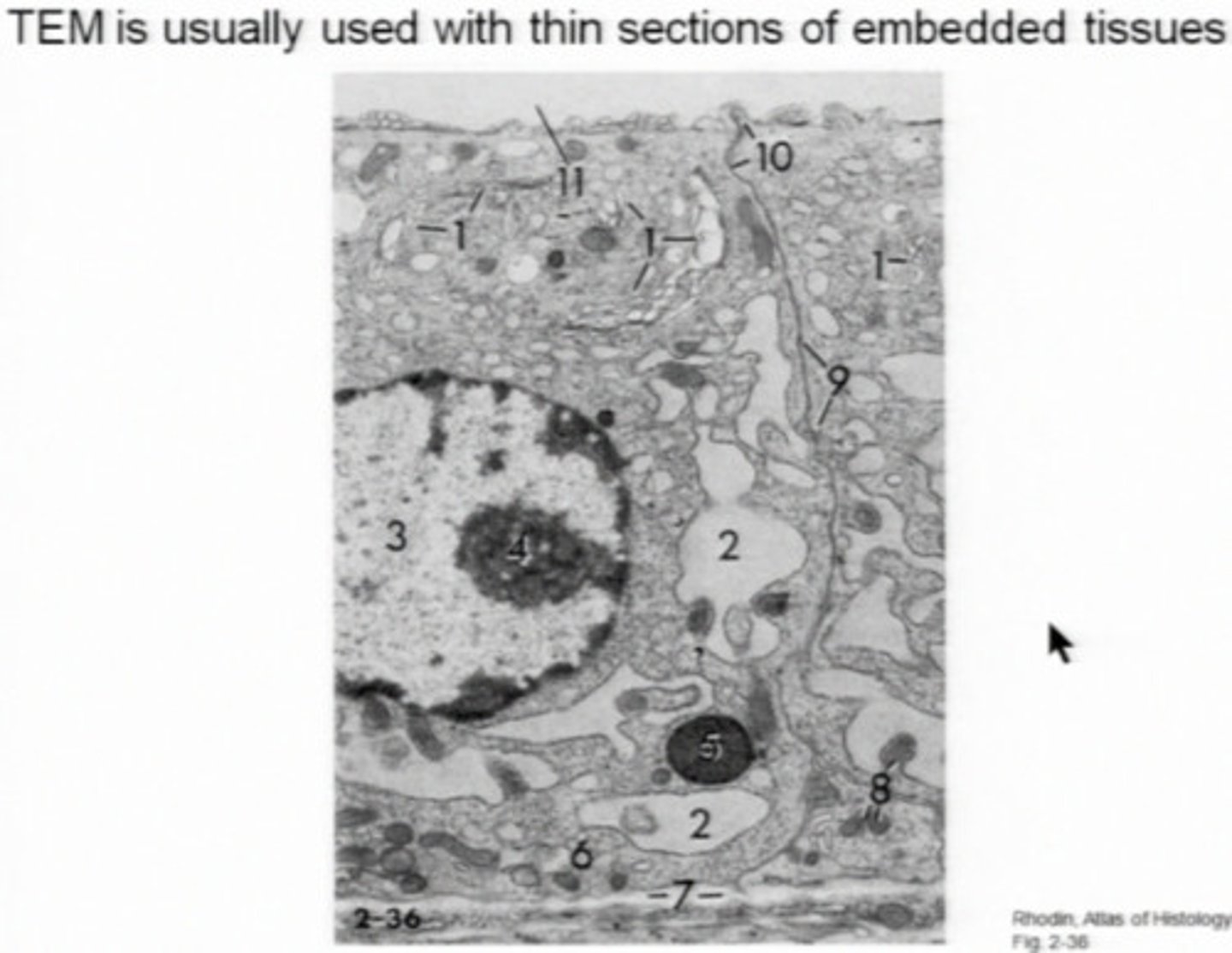
Transmission Electron Microscopy
Uses stains made of heavy metals like uranium. The highest resolution is 0.2 nm. You need to make thin sections (20-60nm) for visualization of internal structures. It can magnify up to one million. You can only see the surface, see what is inside, and make slices.
Cryo electro tomography
Uses a sample that is flash-frozen, thus in a near-native (hydrated) state. No staining is involved. The imaging occurs under cryogenic conditions to maintain the sample hydrated and reduce damage from the electron beam.
Tomography
Acquisition of projected images from different angles; no thin sectioning required if the sample is thin (< 0.5 μm)
Scanning Electron Microscopy
It uses a thin film of heavy metals like gold. it has magnification from 15X to 100,000X and uses an electron beam to scan back and forth, creating a 3d rendering. It is good for looking at the surface of bacteria.
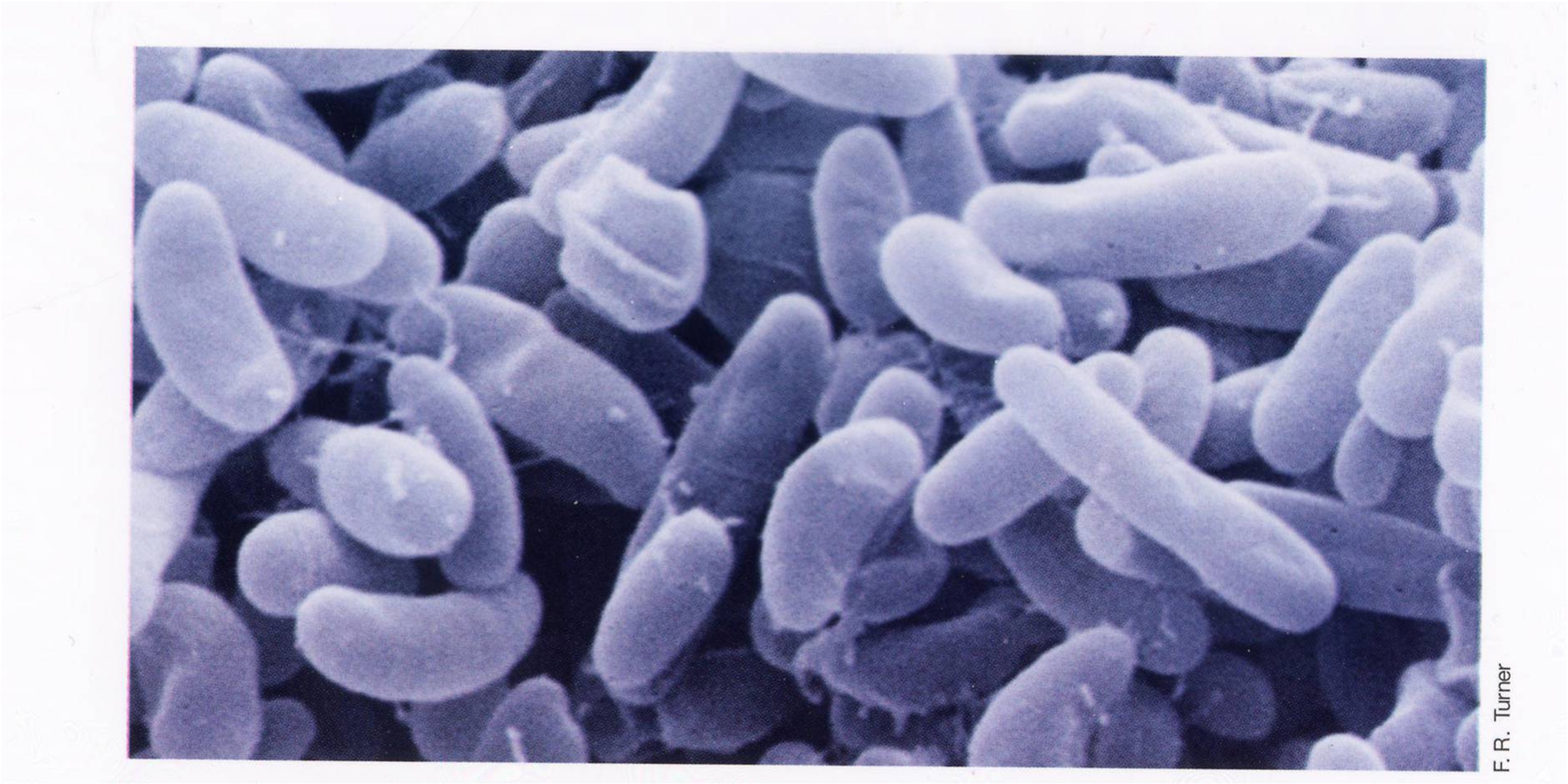
Scanning Electron Microscopy vs Cryo Electron tomography
SEM uses one image, and Cryo Electron scans with many different images to get the 3D aspect.
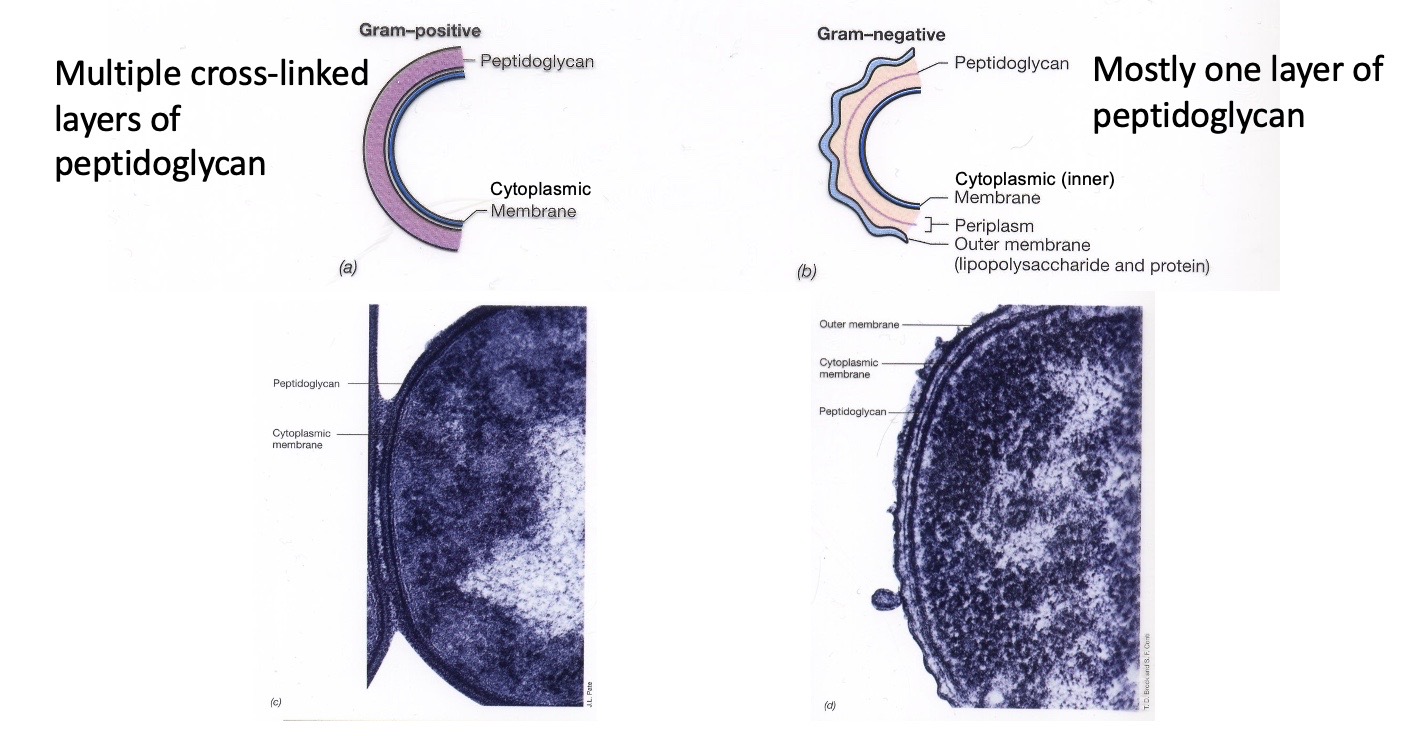
Bacterial Cell Envelope
The cell envelope interacts with the environment and adds protection. All Cell envelopes have cytroplasmic membranes, and the bacterial cell envelope also has a cell wall. The cell wall includes all of the external layers outside the cytoplasmic membrane. Peptidoglycan is one of the main components and gram negative bacteria also have an outer membrane. In the TEM image, the black line is the peptidoglycan.
The cytoplasmic membrane- A phospholipid bilayer
The bilayer is very fluid and has the viscosity of light-grade oil. It is missing sterols. It has a highly selective permeability barrier that allows the cell to concentrate specific metabolites and excrete waste materials. Water and dissolved gases (O2, CO2) and lipid-soluble molecules readily cross the phospholipid bilayer, unlike ions and charged molecules. The bilayer likes sterols such as cholesterol.
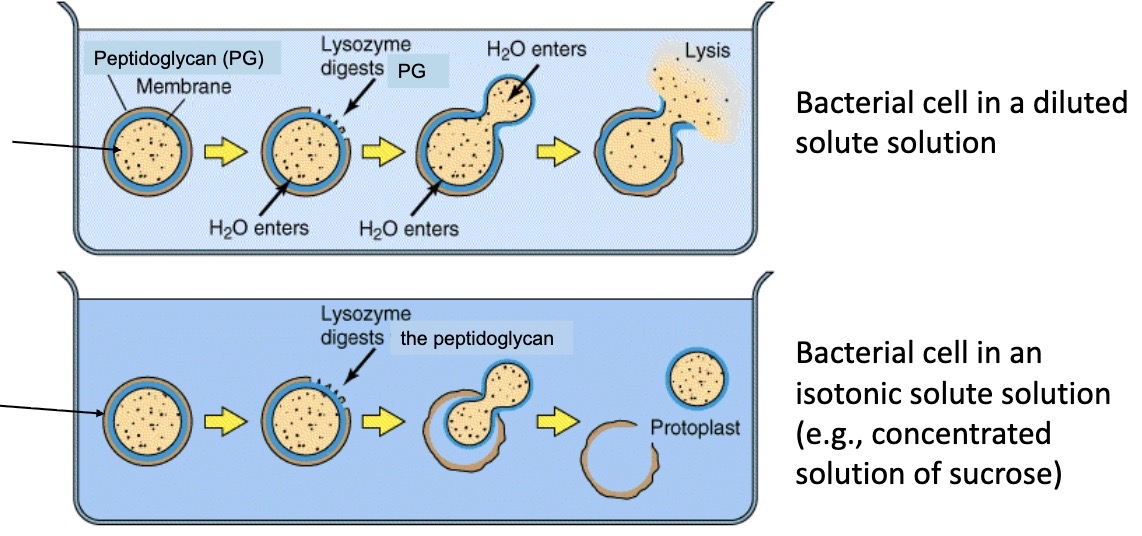
Peptidoglycan provides mechanical support and resistance to the intracellular osmotic pressure in hypotonic environments
When this occurs, lysozyme is produced as an enzyme for the first line of defense. The cell ends up breaking because of too much pressure, and water moves via osmosis. Peptidoglycan is like a tire that keeps the pressure in. With rod shapes, peptidoglycan keeps it in the rod shape, but when the shape is lost, protoplast becomes spherical.
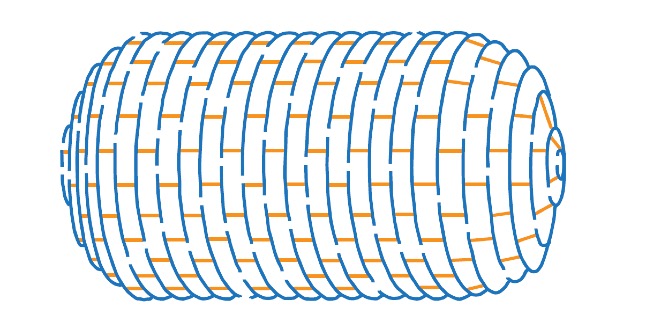
Peptidoglycan
A meshwork permeable to solutes and small proteins. It is a strong, elastic fabric made of Glycan strands connected by peptide bridges to form one giant molecule that surrounds the cytoplasmic membrane. There are multiple layers in Gram–positive bacteria; mostly only 1 layer in Gram-negative bacteria.
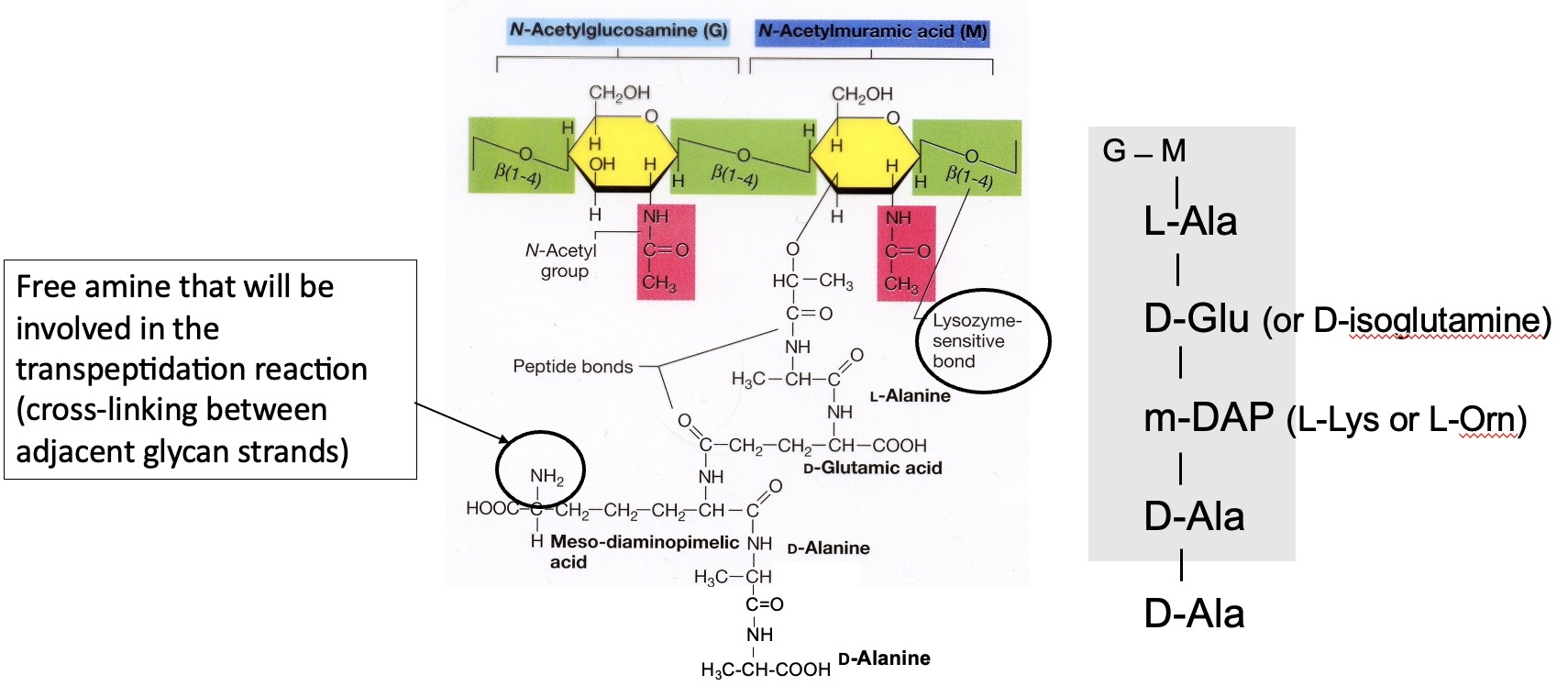
Basic Unit of Peptidoglycan
There is a N-acetyl-muramic acid, meso-diaminopimelic acid (DAP), and amino acids in the D configuration. It will always have two DAlas next to each other. The 3rd position of peptide stem will be an amino acid with a free amine because the amine is used for cross linking via a peptide bond.
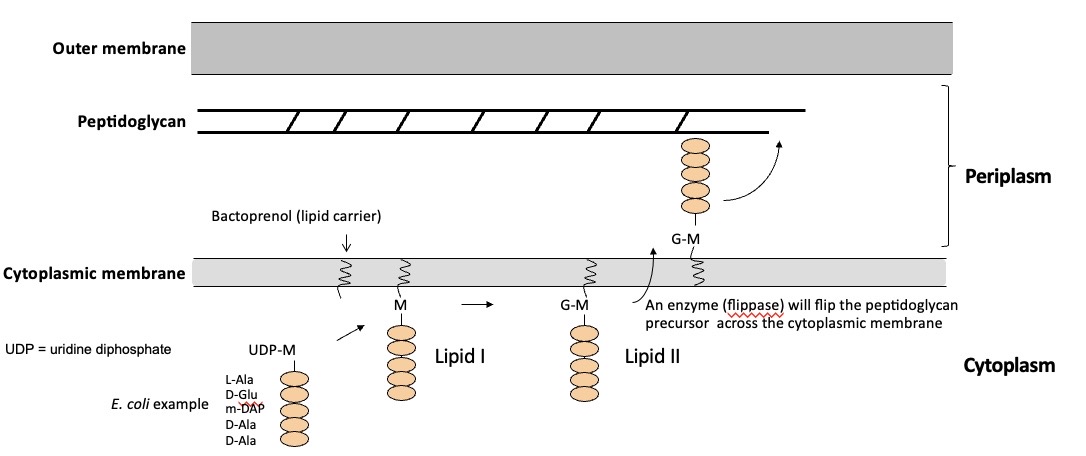
The peptidoglycan biosyntehtic pathway
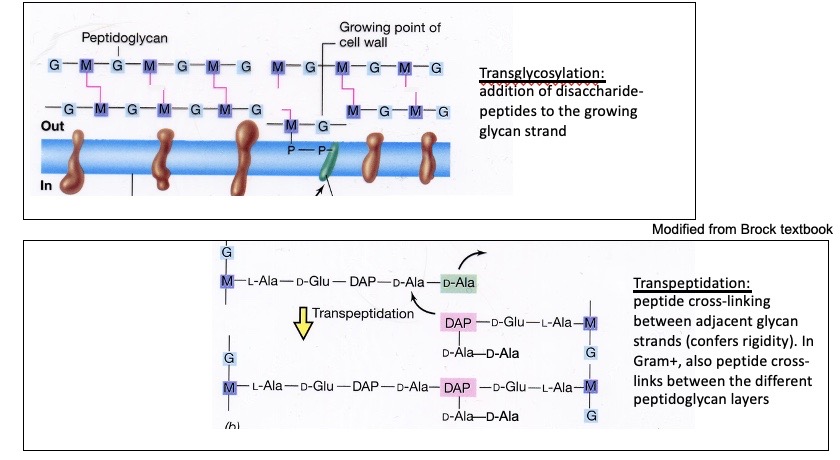
Peptidoglycan synthesis (Pink is the peptides and purple and blue are the glycan strands)
Transglycosylation occurs, which is the addition of disaccharide-peptides to the growing glycan strand. This happens with high molecular weight PBPs. Transglycosylation is the linking. Then Transpeptidation occurs, when a peptide cross-links adjacent glycan strands (which confers rigidity). Transpeptidation uses a free amine to bond to the fourth amino acid. Low molecular PBPs facilitate breaking of linkages so that the cell can grow.
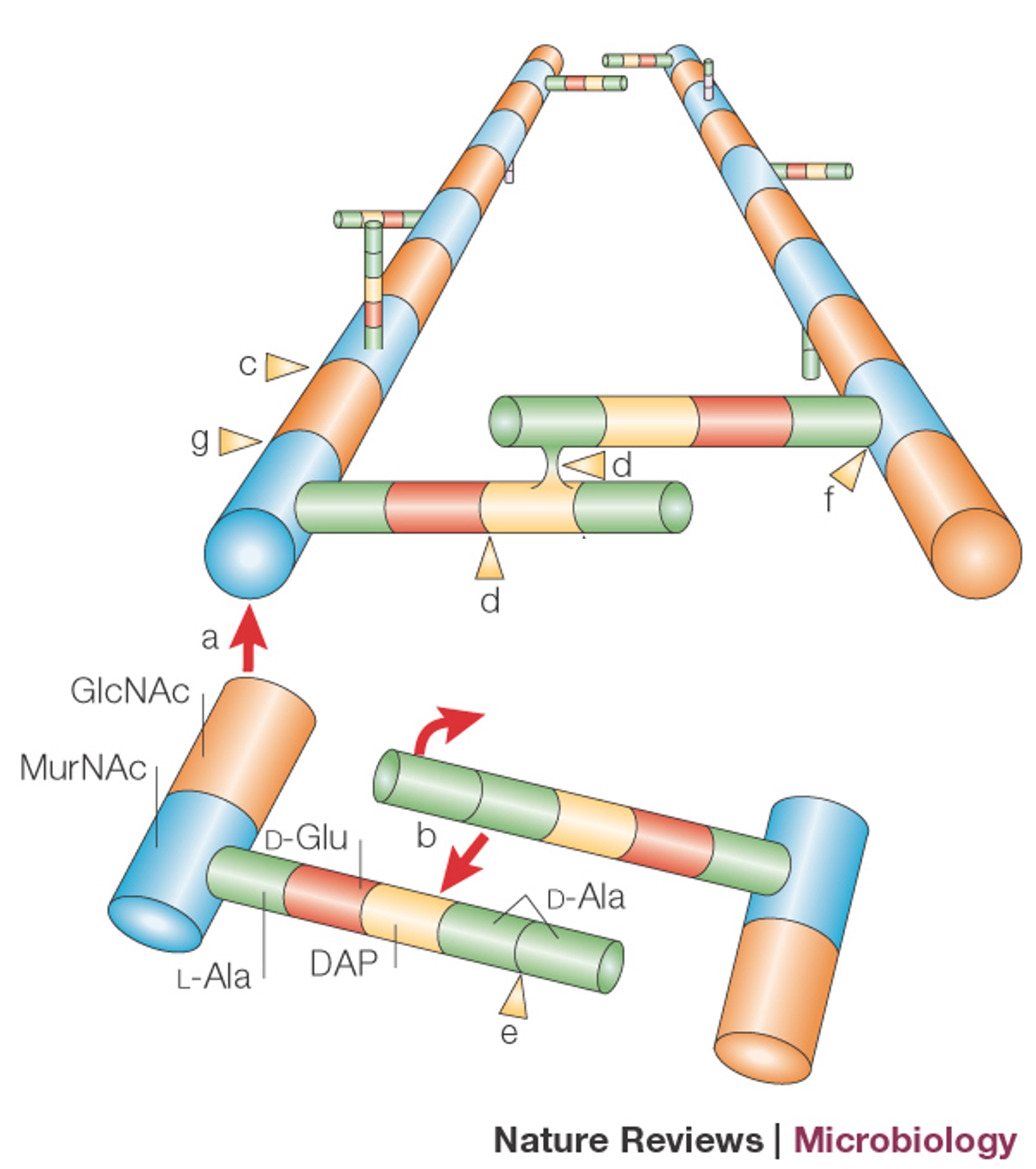
Enzymes invovled in Peptidoglycan synthesis
Penicillin-binding proteins (PBPs):
High-molecular-weight PBPs are involved in this: transglycosylase (a) and transpeptidase (b) activities, or just transpeptidase activity (b). Low-molecular-weight PBPs involved in peptidoglycan processing: carboxypeptidases (e) or endopeptidases (d).
Other processing enzymes: lytic transglycosylases (c); amidases (f), N-acetylglucosaminidase activity (g).
In order to expand, bonds must be broken by enzymes.
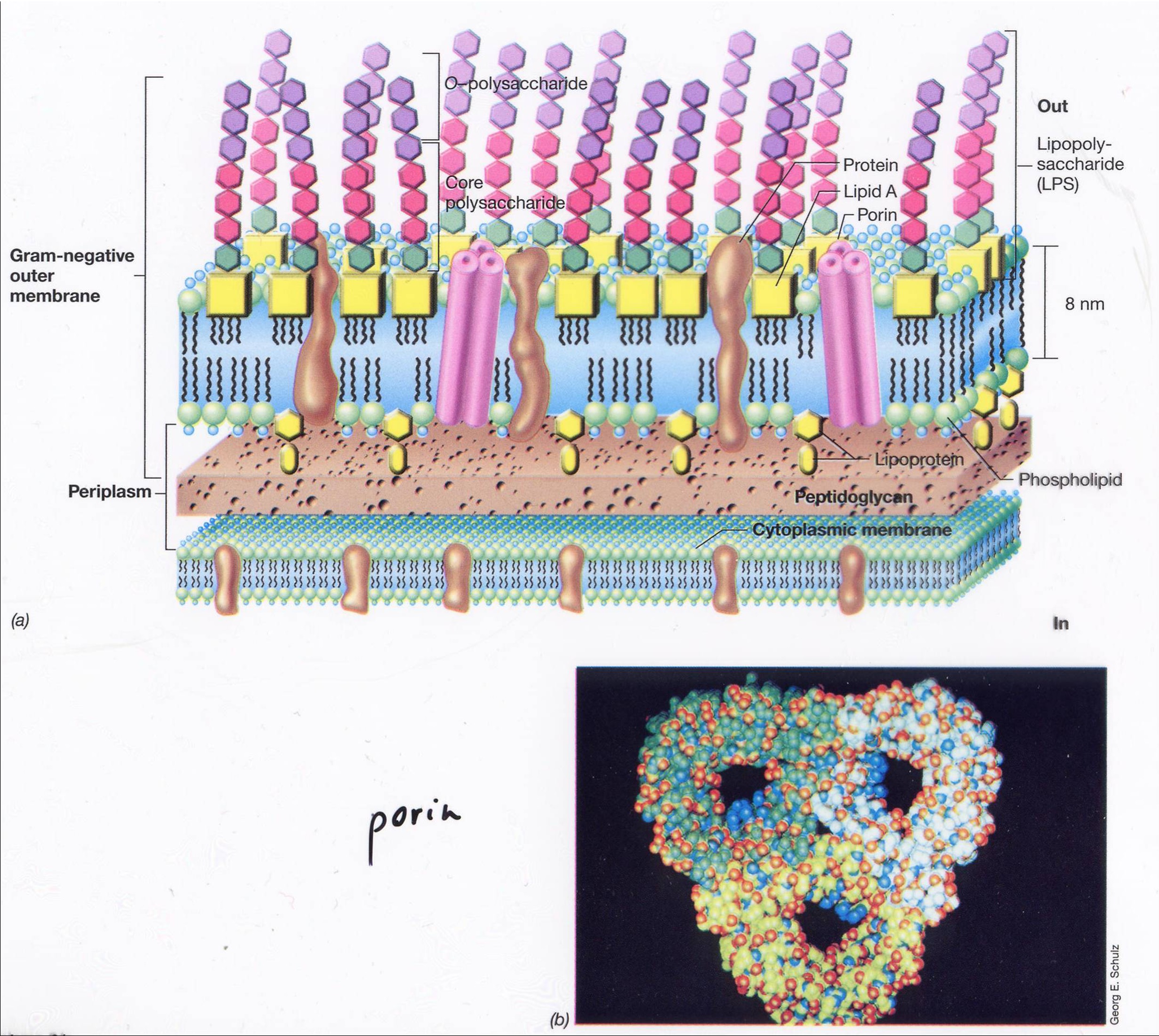
The Gram negative outer membrane
The outer membrane is rigid relative to the cytoplasmic membrane and, together with the monolayered peptidoglycan, it provides mechanical resistance to the intracellular osmotic pressure.The outer membrane is a semi-permeable barrier because of the presence of porins. The pink things are porins, and are less selective. Nutirients can go through the porins but lysosyme can’t. Lipopolysaccharide (LPS) interacts with the environment and it is not fluid. LPS does provide resistence to turbo pressure.
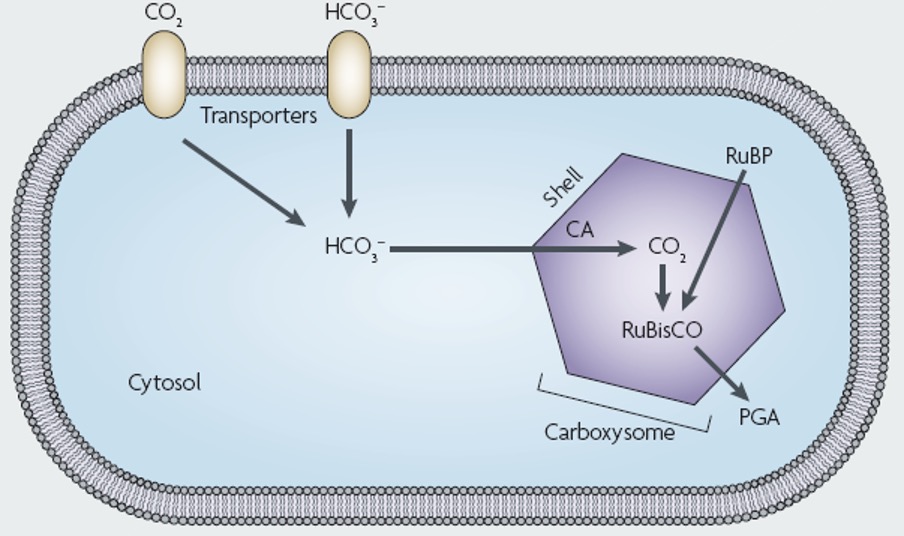
Bacteria can form microcompartments with Protein shells to sequester specific metabolic activities
A polyhedral protein shell encapsulates enzymes involved in carbon fixation, including Rubisco, which converts carbon dioxide into energy-rich molecules. Rubisco is a very poor enzyme, so in order to increase its activity, it must be condensed. CO2 gets concentrated, and CO2 goes into the shells and turns into bicarbonate. Bicarbonate goes into carboxysome, and then turns into CO2.
Major cytoskeletal systems- In Humans
Microtubules (tubulin)
Microfilaments (actin)
Intermediate filaments (intermediate filament proteins)
FtsZ assembles into a ring-like structure at the site of division
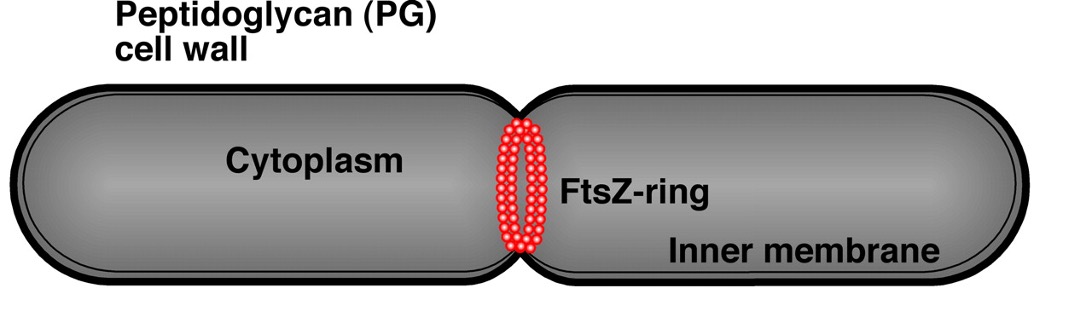
FtsZ ring constricts to create new cell poles
FtsZ ring directs peptidoglycan synthesis across the cell width, resulting in cell pole formation. The ring is right at the site of division.
FtsZ is required for cell division
Without FtsZ the cells will continue to grow but won’t divide. Removal of xylose (inducer) from the environment results in repression of ftsZ expression and a block in cell division (cell filamentation).
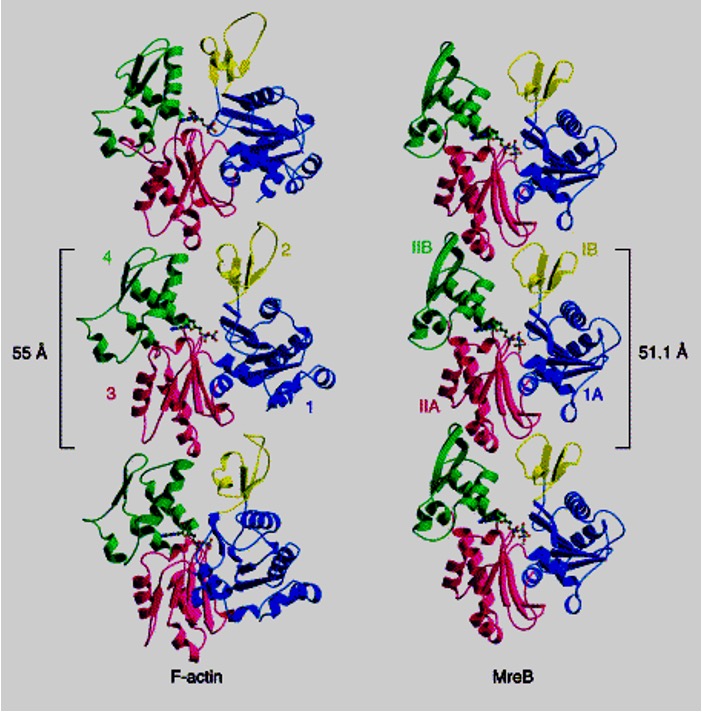
MreB is an actin homolog found in many rod-shaped bacteria
In live cells, short filaments rotate around the cell circumference. The current hypothesis is that they direct circumferential peptidoglycan synthesis. Glycans move in one direction and peptides move in another. This is mainly found in rod-shaped bacteria.
MreB maintains the cell width constant during cell elongation
Since MreB is a rod shape determinant, spherical species lack MreB which is why:
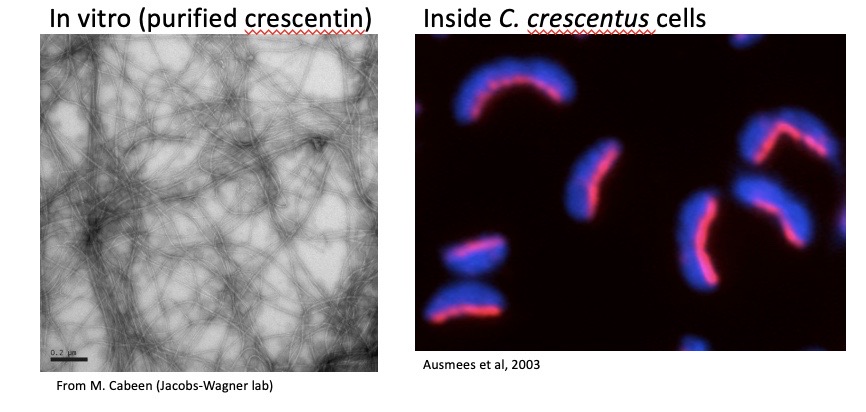
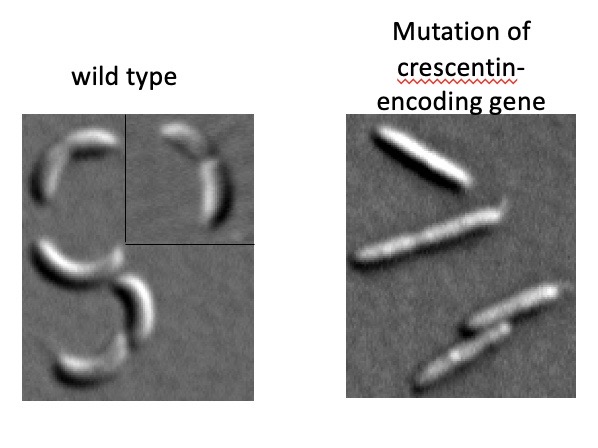
Crescentin is an intermediate filament-like protein in Caulobacter crescentus
Spontaneous polymerization occurs in the absence of nucleotides or cofactors. Crescentin assembles into a filamentous structure beneath the cytoplasmic membrane on one side (inner curvature) of the cell. Crescentin self-assembles inside the cell and is structurally similar to animal intermediate filament proteins.
Crescentin mediates the cell curvature of Caulobacter crescentus
Major cytoskeletal systems- Analogous Systems in Bacteria
Tubulin-like (FtsZ)
Actin-like (MreB, others)
Intermediate filament-like (crescentin, others)
Bacterial cell replication
Growth of an individual cell continues until it divides into two cells, typically after doubling its genome, ribosomes, macromolecules, metabolites and inorganic ions.
Generation
Interval for the formation of two cells from one during exponential growth
Growth rate
The change in cell number or cell mass per unit of time.
Generation time
The time required for the cell population to double during exponential growth. Also called doubling time.
Growth parameters (for exponential growth)

Microbes can grow into colonies visible to the naked eye
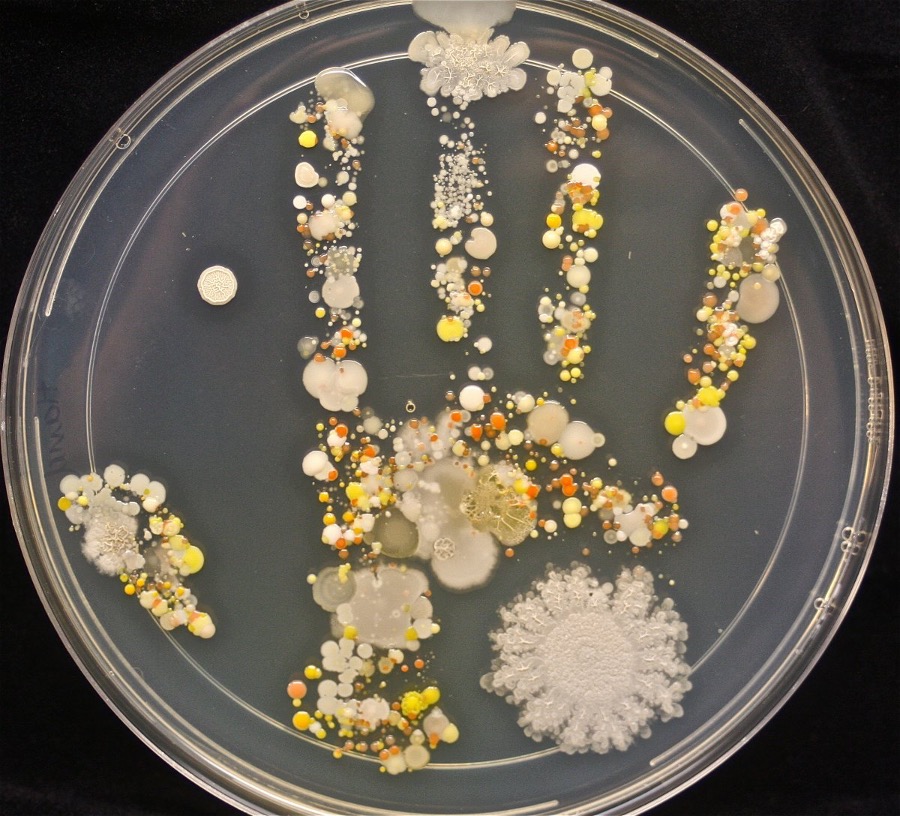
Viable counts using dilution and plating method
You perform a serial dilution then plate it. Then you can count. It tells you the number of fiable cells in the tube and the density of the cells.
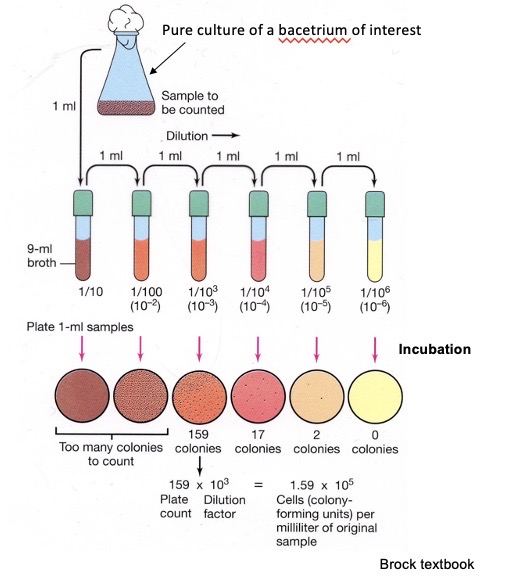
Counting chamber for direct cell count using microscopy
The number of cells under the big square is counted (in practice, several squares are counted and the numbers are averaged). Because the volume under the grid is known (e.g., 10-4 mL), this value can be converted to the number of cells per mL. The chamber holds a specific volume under square grids. The counting chamber provides Instant measurements; it counts live and dead cells. It is not a good method for cell suspensions of low density (<106 cells/mL), and motile cells must be immobilized. If the cells are lysed, you can distinguish live from dead cells. The number of cells per square decreases at low density.
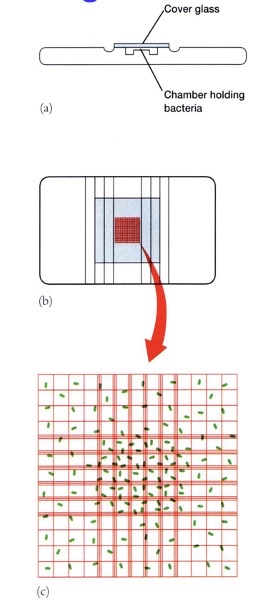
Turbidity Growth Measurements
Using a spectrophotometer, you can measure light that didn’t hit anything. The more bacteria there are, the more the light will be scattered. However, you need to use other techniques in addition to this for calibration. When the cell density is too high, light is rescattered, so you will need to dilute to obtain a correct measurement. With this, you get instant measurements, can count live and dead cells, and need a standard curve to convert optical density (OD) measurements into cell numbers per mL.
Calibration of turbidity measurements
Optical density (OD), or absorbance, measures a material's ability to block or absorb light (𝑂𝐷=1 means 90% of light is blocked), proportional to cell numbers, so you can use a standard curve for conversion. However, it is less reliable at high OD.
Microbe growth is dependent on many environmental factors
•Nutrients
•Temperature
•Water availability
•pH
•Oxygen
•Saline/osmolarity
•Pressure
•Radiation
•Others
Microbes are not “culturable”
We can’t meet all conditions needed to grow in pure culture (e.g., needs other co-existing organism to grow)
Extremophiles
Microbes that live in environments considered as extreme. e.g. psychrotolerant organisms: grow near 0º C but have optima of 20-40º C, found in temperate climates (psychrophiles are pickier and die outside ideal range)
The benefits are exploitability/applicability in lab experiments.
Temperature classes of microorganisms
The higher the temperature, the faster the enzymatic activity that facilitates growth. at a certain threshold temperature = denaturation. Microbes cannot regulate their temperature.
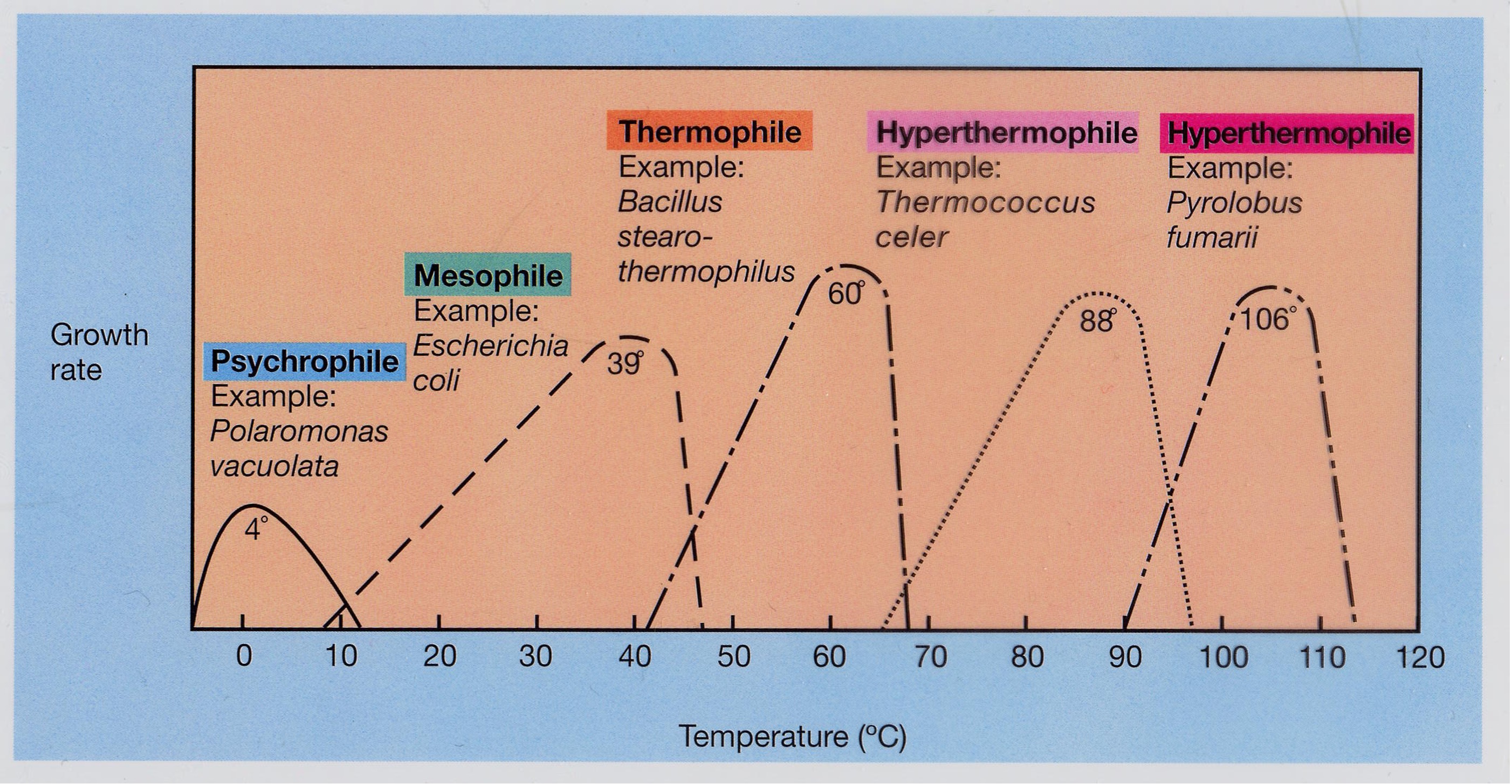
What happens if you put a thermophile in lower temperature (outside its ideal range)?
It becomes more stiff and rigid, therefore has lower activity
Why sterilization (e.g., autoclave) is needed
If boiling a solution, some bacteria are heat-resistant because they have SPORES so the fix is to autoclave (increases pressure to increase temperature)
Pasteurization (doesn’t kill all organisms and is for heat sensitive foods)
Used when we cannot sterilize (e.g., lots of protein denaturation in milk) but want to reduce amount of organisms, which will still grow overtime hence fast expiration)
The longer the time of heat exposure = the greater the reduction in bacteria
Irradiation
Kills bacteria by exposing food to ionizing radiation (gamma rays, X-rays, or electron beams), which damages microorganisms' DNA, preventing them from reproducing or causing illness. It causes irreparable DNA damage. (UV, x-rays, 𝜸-rays, electrons)
Deinococcus radiodurans
A radiation-resistant microorganism originally isolated from an irradiated can of meat. They have evolved to this degree of resistance via desiccation and have a very efficient DNA repair mechanism
Methods for Microbial growth control
•Heat
Sterilization (e.g., autoclave)
Pasteurization (doesn’t kill all organisms; for heat-sensitive foods)
•Irradiation
(UV, X-rays, γ-rays, electrons)
•Filter sterilization
for heat-sensitive liquids or gases
•Chemical growth control
antimicrobial agents
Filter sterilization
This removes microorganisms from heat-sensitive liquids or gases using membrane filters, typically with pores, without heat or radiation. Common methods include syringe filters for small volumes, vacuum filters for intermediate, and cartridge/capsule filters for large-scale production. This is meant for heat-sensitive liquids or gases. Most bacteria 0.5-5 μm in size and the typical pore sizes are: 0.3-0.5 μm = small enough to trap most bacteria but large enough to let liquid/gas pass through
Chemical - antimicrobial agents
Substances that kill or inhibit the growth of microorganisms (bacteria, viruses, fungi, parasites), acting through mechanisms like disrupting cell membranes, DNA inhibition, or protein denaturation. Common types include alcohol (70% ethanol/isopropanol), phenolic compounds (triclosan, hexachlorophene), halogen-releasing agents (chlorine bleach, iodine), heavy metals (silver, copper), and soaps/quaternary ammonium compounds. Arrows show when this was added during the exponential growth of the bacterial culture.
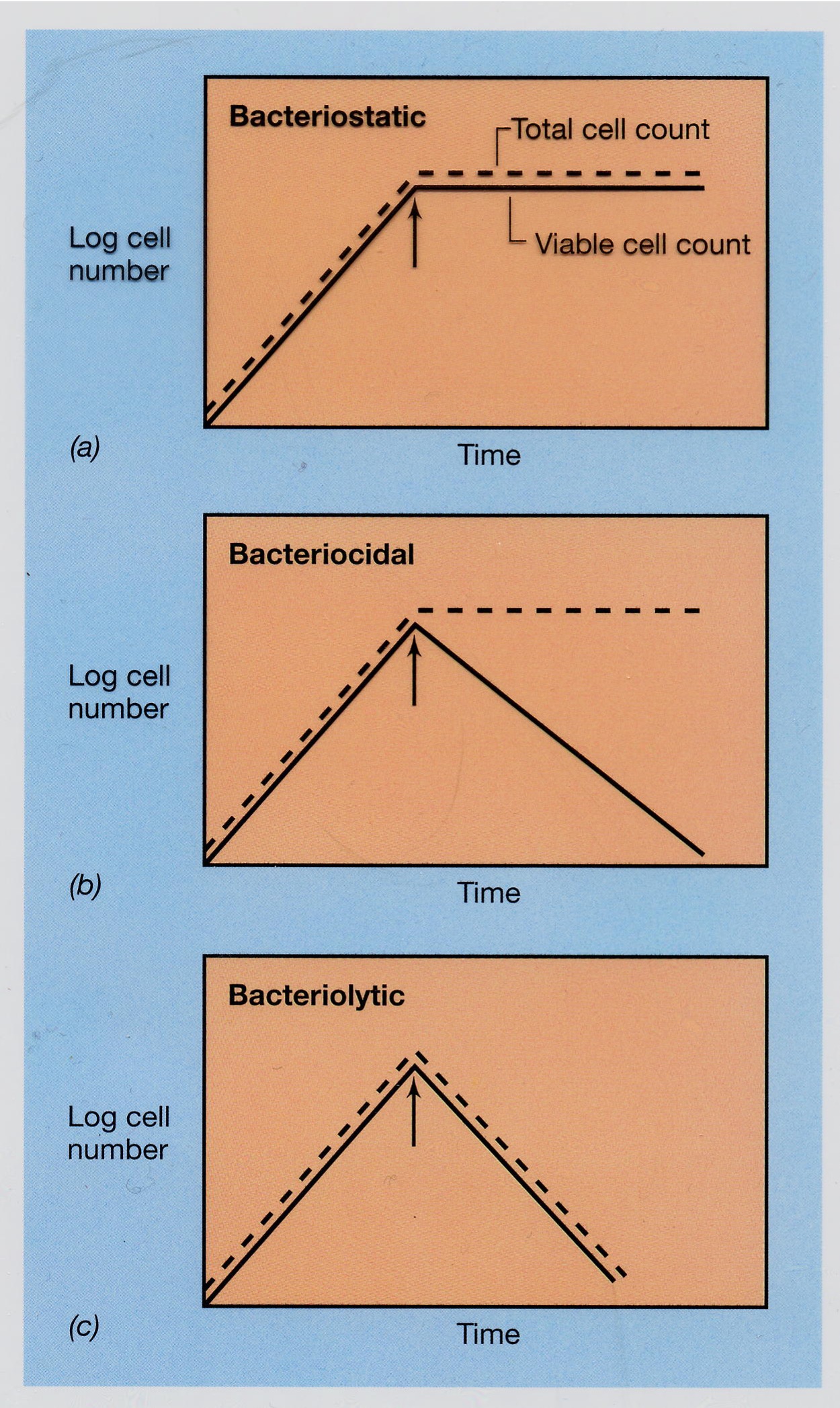
Bacteriostatic
Arrests bacterial growth (# of viable plateaus, not increases) If the antimicrobial agent is removed, bacterial growth would recover (reversible)
Bacteriocidal
Substances that actively kill bacteria by disrupting cell walls, membranes, or essential enzymes, If the antimicrobial agent is removed, bacteria cannot recover (irreversible), count continues declining
Bacteriolytic
Bacteriocidal but ALSO cells lyse completely = non-visible, unable to count
Antibiotics
Drugs that selectively inhibit bacterial growth or kill bacteria (and not the host). Growth inhibition can be enough for an antibiotic to be effective because the reduced population size is now enough for our immune system to combat.
Importance of bacterial susceptibility vs. host sensitivity (e.g., toxicity, allergic reaction)
An antibiotic might be good at killing bacteria (high BS) but still be unusable if the patient has severe side effects (high HS). A good antibiotic ensures both high BS AND low HS!!
Minimal inhibitory concentration (MIC)
Lowest concentration of an antimicrobial drug (e.g., antifungal, antibiotic) that will inhibit the visible growth of a given microorganism after incubation in a growth medium.
start at high concentration, then gradually increase dilution to lower concentration
**does not tell use anything about bacterial killing just inhibition
example to the right: MIC = 1.0 i.e. the tube with 1.0 μg/mL is the lowest concentration where you see no visible bacterial growth (clear tube) i.e. growth successfully inhibited
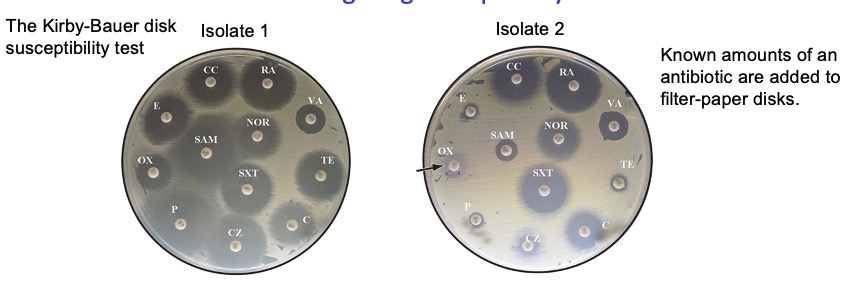
Kirby-Bauer disk susceptibility test
Known amounts of an antibiotic are added to filter-paper disks, then they diffuse through the agar to establish a concentration gradient where the lowest point of the gradient is where the bacteria is resistant enough.
the larger the gray circles = the farther the antibiotic has diffused (greater radius) = the more lethal it is against the bacteria → clinical significance? indicates that this is the antibiotic to be using for patients
performed under standardized conditions & in standard-sized zones of inhibition for each antibiotic
results usually reported as sensitive, intermediate, or resistant, based on zone of inhibition’s size:
if observed zone of inhibition size >= standard zone size → microorganism deemed susceptible to antibiotic
if observed ZOI size < standard zone size → microorganism deemed resistant
The size of a zone of inhibition in a KB test is inversely related to the minimum inhibitory concentration (MIC), which is the amount of antibiotic required to prevent bacterial growth in an overnight culture. The MIC (in µg/ml) can be calculated from known standard-curve graphs based on the diameter of the observed inhibition zone diameter (in millimeters).
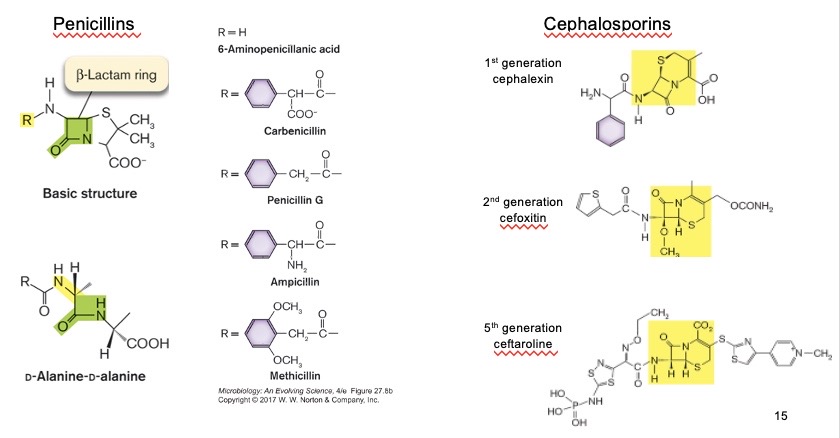
β-lactam antibiotics
Can inhibit penicillin-binding protein (PBP) activity:
For example: (penicillins (5-member ring), cephalosporins (6-member ring)) structurally mimic D-Ala-D-Ala and make a covalent bond with PBPs to block transpeptidation. Using the active site serine in PBP attacks the carbonyl carbon of the β-lactam ring → ring opens → forms a stable acyl-enzyme complex (covalent bond) → PBP is permanently inactivated
(Enz = example of a PBP)
High-molecular-weight PBPs
Involved in peptidoglycan synthesis: transglycosylase (a) and transpeptidase (b) activities, OR just transpeptidase activity (b).
Low-molecular-weight PBPs
Involved in peptidoglycan processing: carboxypeptidases (e) or endopeptidases (d)
Site of antibiotic action
Many bactericidal antibiotics also generate reactive oxygen species (ROS), superoxide (O2•—), hydrogen peroxide (H2O2), & hydroxyl radical (OH•) by affecting metabolism
ROS damage DNA, proteins, & lipids
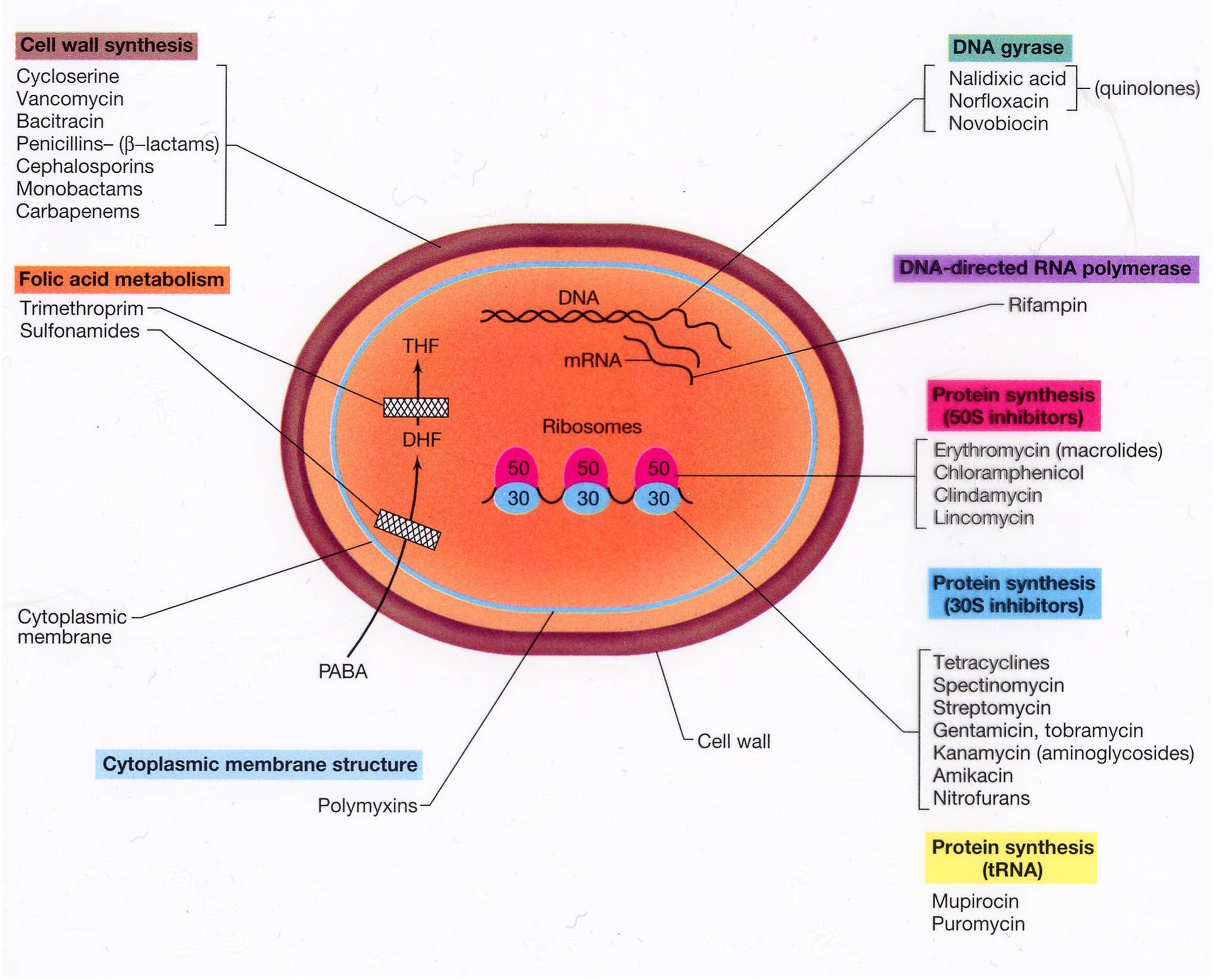
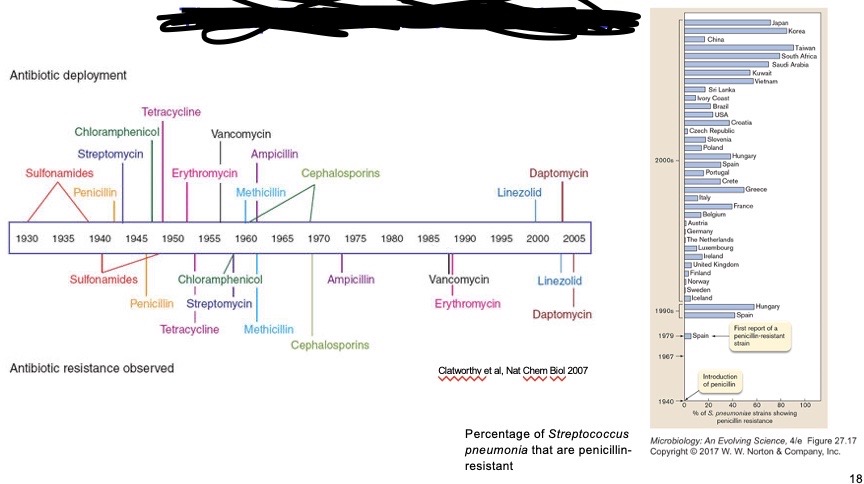
Rapid rise of antibiotic resistance & decline of antibiotic development
Daptomycin affects the function of bacterial cell membranes. Linezolid is a protein synthesis inhibitor
Antibiotic resistance mechanisms:
prevent access to target
target site protection
pump the antibiotic out of the cell
ex. multi-drug efflux pumps in gram– and gram+ bacteria
a single pump can often export diff kinds of antibiotics
↑ expression of efflux pumps → multi-drug resistance
either chromosomally- or plasmid-encoded
alter the target
ex. mutation
inactivate the antibiotic (cleavage or modification)
ex. hydrolysis
β-lactamases
ex. steric hindrance
Our gut ecosystem is losing diversity due to these factors:
• Diet
• Sanitization
• Antibiotics
• Baby Formula
• C-sections
Narrow nutrient categories
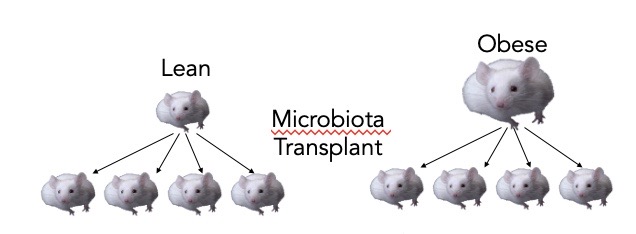
Microbiota transplantation can cause weight gain
If you put an obese microbiome in germ free mice, they will gain weight. The microbiota of obese mice shows an increased capacity for energy harvest.
Prebiotics
Complex carbohydrates that promote gut residents
To manipulate biology using the microbiota you can use:
Live microbes:
•Microbiota transplants, bacterial cocktails
Synthetic biology applications:
• Therapeutic delivery
• Diagnostics
Diet
Probiotic
Live culture of bacteria derived from fermented foods but don’t take residence in the gut
The microbiota is connected to human biology via:
•Metabolism/Obesity
•Immune System
• Central Nervous System ex. the brain gut axis and inflamation
The human body as a microbial habitat
The microbial cells that inhabit our bodies are estimated to outnumber our human cells by 10-fold.
The collective number of genes in one person’s combined microbial genomes is >100 times the number of genes in the human genome.
We are elaborate culturing flasks that have been optimized by our microbial inhabitants for their growth and spread.
We are 10X more microbial than human…
When comparing the number of bacteria in the human body (3.9 × 1013) to the number of nucleated human cells (≈0.3 × 1013) we do get a ratio of about 1 to 10.
The B/H ratio is actually close enough to one, so that each defecation event, which excretes about 1/3 of the colonic bacterial content, may flip the ratio to favor human cells over bacteria.
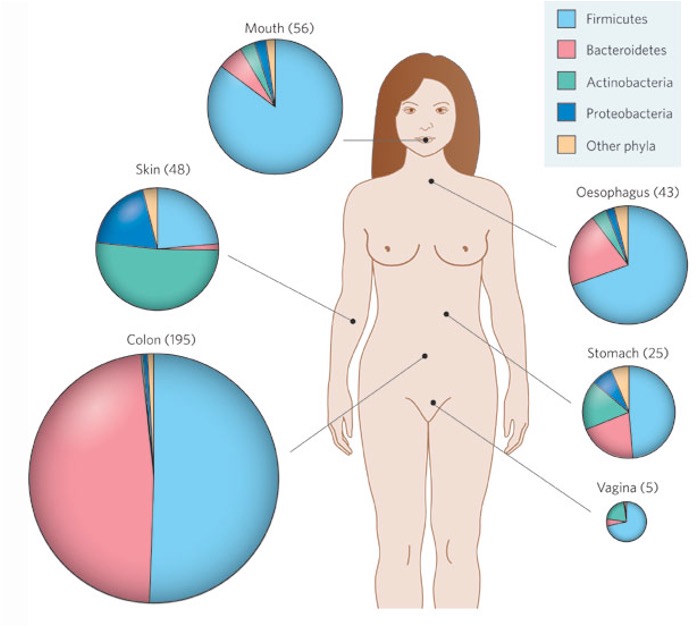
The human body as a composite organism: microbial and human parts
Microbiota is composed of bacteria, eukaryotes, viruses, and archaea
Two bacterial phyla dominate the intestinal microbiota of humans: >90% of bacteria are Bacteroidetes and Firmicutes.
Largest collection of microbes resides in the intestine (~10-100 trillion organisms): 100-1000 species in an individual
Methods for Analysis of the gut microbiome
Pure culture- Culturomics
Sequencing: Shotgun sequencing (where you purify all of the DNA and sequence all of it), 165 rRNA Amplicon sequencing (which takes specific primers). Shotgun makes Metagenome and 165 rRNA Amplicon makes Amplicons
Metabolomics which is used to study small molecules
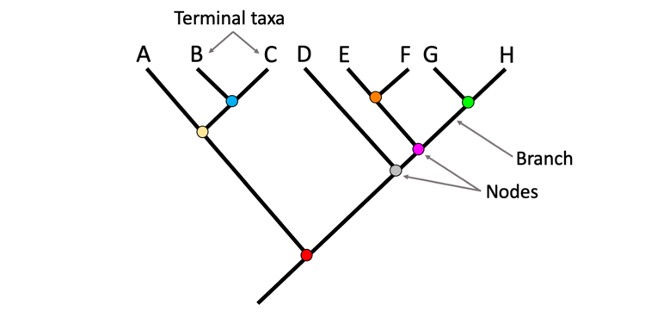
Phylogenetic trees
It is a way of understanding relationships. We get to see how close different microbes are related which is indicated by how close the branches are.
Gut microbiota have diversity at the level of species/strain
All of our microbiomes are similar to each other in some way, however there is lots of diversity at the tipds of the trees
3 key features that make the gut microbiota well suited for precision health (personalized medicine)
•Plasticity/malleability
•Impacts to most aspects of our biology
•Individualized
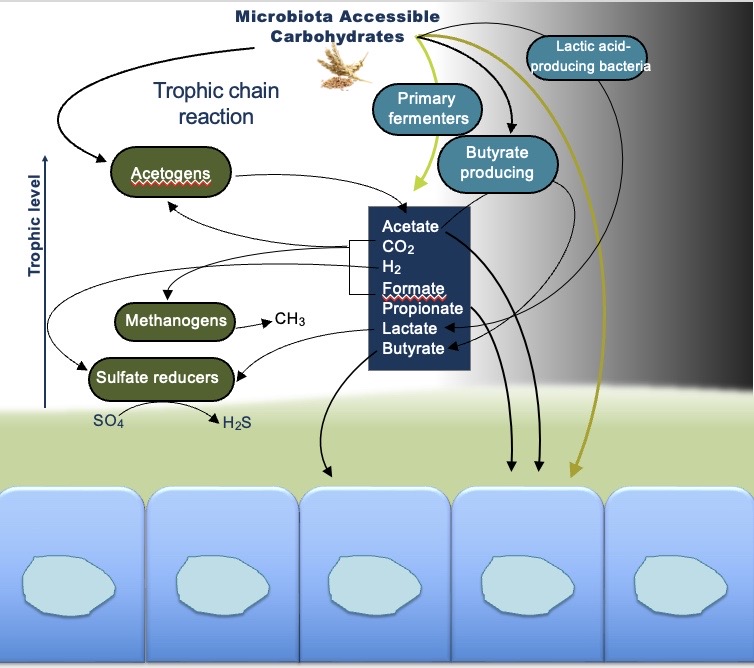
Microbiota Accesible Carbohydrates
Types of -omics
DNA- genomics
RNA- transcriptomics
Protein- proteomics
Metabolomics
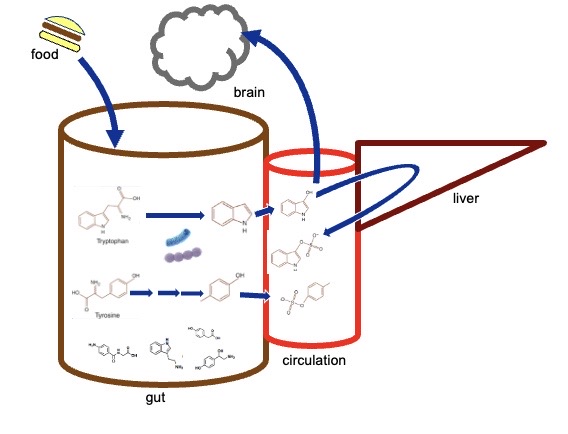
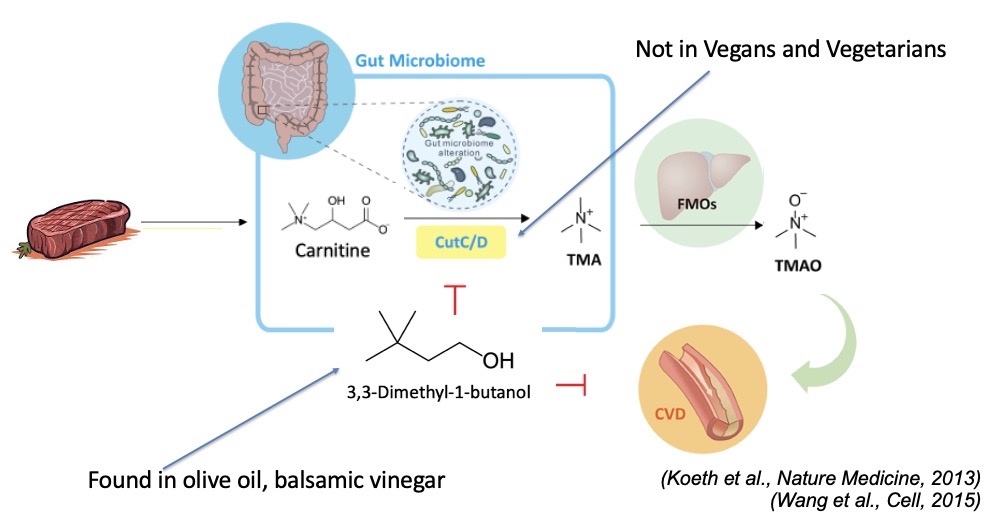
An unsupervised pharmacy in our gut?
The molecules that microbiota produces is like a pharmacy. They are absorbed into our blood stream.
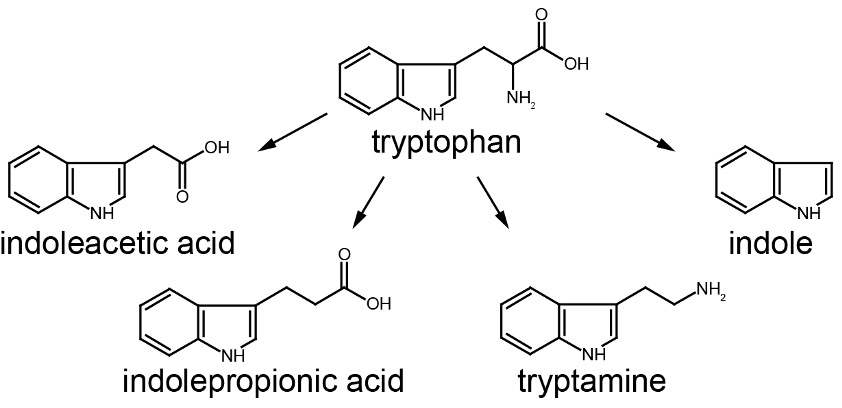
Same precursor can be metabolized through different pathways by gut microbes
Intestinal microbial metabolites of carnitine promotes heart disease
Carnitine is cut by Cut C/D which turns it into TMA, which is then oxidized in the liver and turned into TMAO.