experiment 7 week 1 - introduction to protein column chromatography
1/61
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
62 Terms
chromatography
the name given to a broad selection of processes that are designed to separate different molecules from each other for purposes of identification, isolation and analysis.
stationary phase
-the stationary phase can reside on the outer edges of some kind of rectangular or cylindrical chamber, but can also simply fill the entirety of the chamber wherein separation occurs.
role of stationary phase
it will interact to varying degrees with different types of molecules in the mobile phase that moving past it.
-if a molecule in the mobile phase interacts strongly with the stationary phase, it will spend a certain portion of its time immobilized on the stationary phase material.
-conversely, different molecules, which interact weakly or not at all with the stationary phase will spend more of their time moving.
the cumulative effect of this variable interaction between molecules in the mobile phase with the stationary phase
-molecules will exit the chamber in which the separation is taking place at different times depending upon the particular molecule.
generic chromatography set up
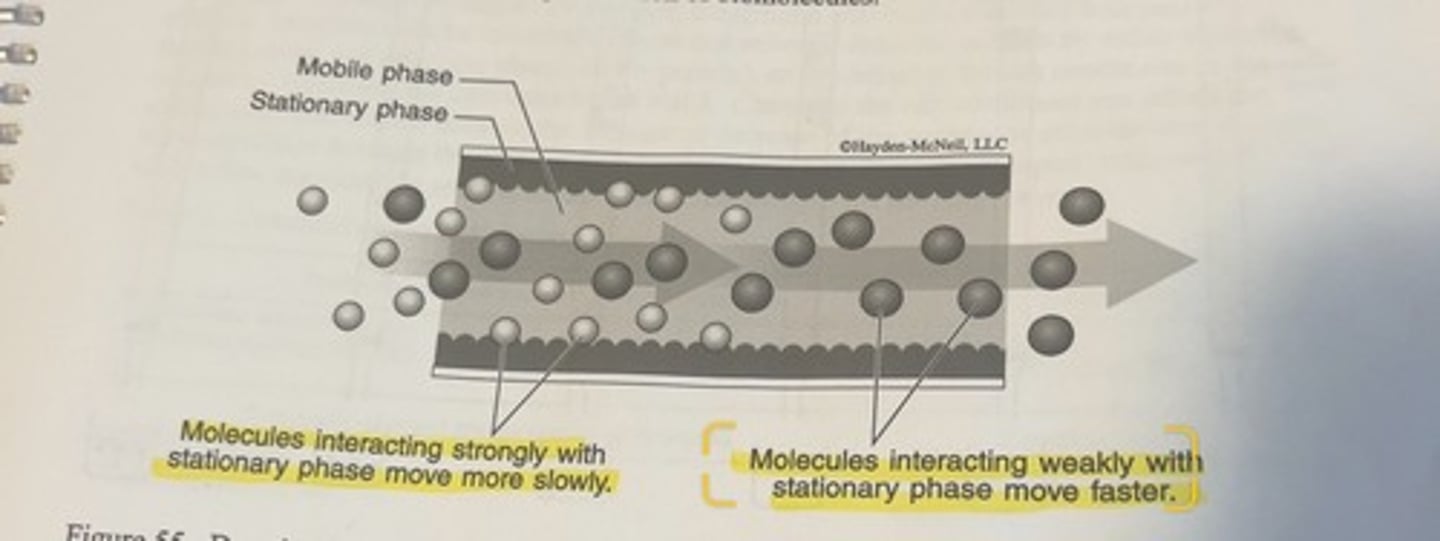
column set up for the purification of proteins
the standard arrangement is for a stationary phase packed inside the column, and then a protein mixture is added to the top of the column and the separated proteins are collected out of the bottom of the column after they have passed through the stationary phase.
the stationary phases for protein purification columns
are almost always an inert matrix to which functional groups are covalently attached.
the inert matrix
it is frequently spherical beads made from some kind of polymer such as polystyrene or an agarose-based polysaccharide such as Sepharose.
-the functional groups give this the specificity to the column, and can be either charged molecules, or molecules modelled after biological molecules to which proteins can interact.
how is stationary phase material selected
selected based upon the specific type of protein purification method being used.
separation of 2 different protein molecules by generic protein purification column
-the faint yellow interior region of the column represents the stationary phase which can be any one of the vast variety of different types of stationary phase materials which are selected based upon the specific type of protein purification method being used.
-a mixture of different proteins is added to the top of the column and allowed to flow through the column assisted by the continuous addition of an appropriate buffered solvent.
-the proteins will separate depending upon the specific properties of the proteins and the stationary phase, and the flow through is collected continuously from the bottom of the column.
-the first 3 fractions collected in this hypothetical situation only contain solvent
-the fourth fraction contains the faster moving green molecules.
-the final fraction contains the slower moving pink molecules.
-in this example, the pink molecules would have a greater interaction with the stationary phase.
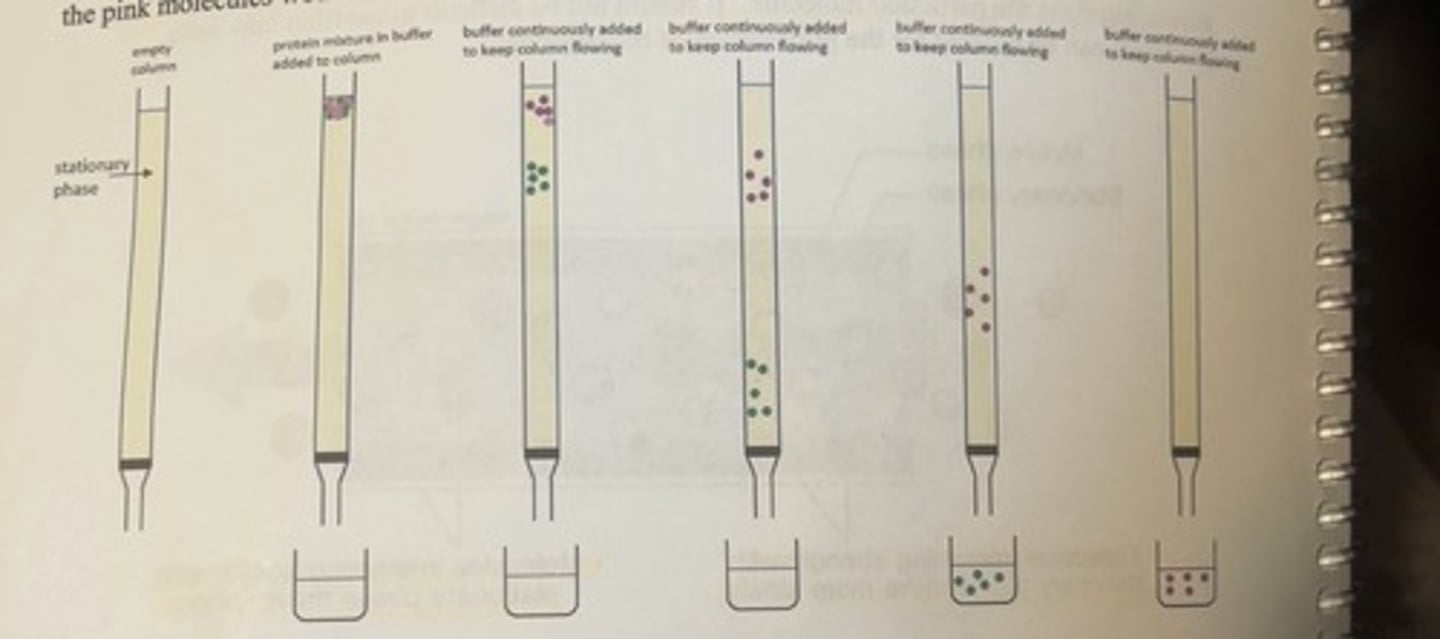
buffer/protein mixture
this can be regarded as the mobile phase.
fractions
different collected samples.
what are the 3 types of protein column chromatography that we considered in this course
ion exchange (IEX), affinity chromatography (AC) and size exclusion chromatography (SEC).
size exclusion chromatography is sometimes referred to as
gel filtration
ion exchange chromatography
based upon the reversible electrostatic interactions between charged molecules and charged functional groups bound to inert resin beads, such as polystyrene or a polysaccharide, packed in a column.
why is ion exchange chromatography an effective protein purification method
because different proteins have different amino acid compositions and therefore, different proteins will have different numbers of positively or negatively charged amino acid side chains at any particular pH.
the overall net charge on any particular type of protein
will be determined by the overall sum of their positively and negatively charged residues.
-at any particular pH, some proteins will have a net positive charge, and some proteins will have a net negative charge, and it is this overall charge which is used for the separation.
if functional groups on the IEX resin are positively charged
the column will bind negatively charged proteins
if the functional groups are negatively charged
the column will bind molecules with positive charges.
how can molecules be bound/unbound
molecules can be selectively bound and unbound from the column by either changing the pH (which will affect the charge on the protein), or by changing the salt conditions in the column by increasing the concentration of NaCl.
changing the salt concentration
can affect the electrostatic interactions because the increase or decrease of the sodium or chloride ions changes the competition between the charged proteins for binding spots on the charged resin surface.
the most commonly used charged groups for IEX resins

affinity chromatography
it is a specific and reversible high affinity interaction between a protein and a ligand molecule that is covalently linked to an insoluble solid support packed into a column.
what is the biggest challenge of affinity chromatography
finding a ligand that will bind to your protein, but not other proteins.
-for proteins that have known binding partners, like enzymes or receptor proteins, these binding partners can serve as the ligand if there is a way to conveniently attach it to the column.
-unfortunately, many proteins will not have such unique binding partners.
so what is done about this challenge?
because of revolutionary techniques introduced by molecular biology, proteins can be synthesized to include sequences of amino acids which are known to bind ligands that can be easily attached to the column.
tags
additional sequences added to proteins.
most commonly used tag
His-tag
his-tag
typically a sequence of 6 consecutive histidine residues that is attached to the beginning or the end of the protein sequence.
-proteins with his-tags will bind to columns that have a nickel ion immobilized on the column.
why are his-tags useful
-since consecutive histidine residues are rare in natural proteins, this ability to bind the immobolized nickel makes a protein with the tag unique compared to other proteins in the mixture and allows for easy purification using the appropriate column.
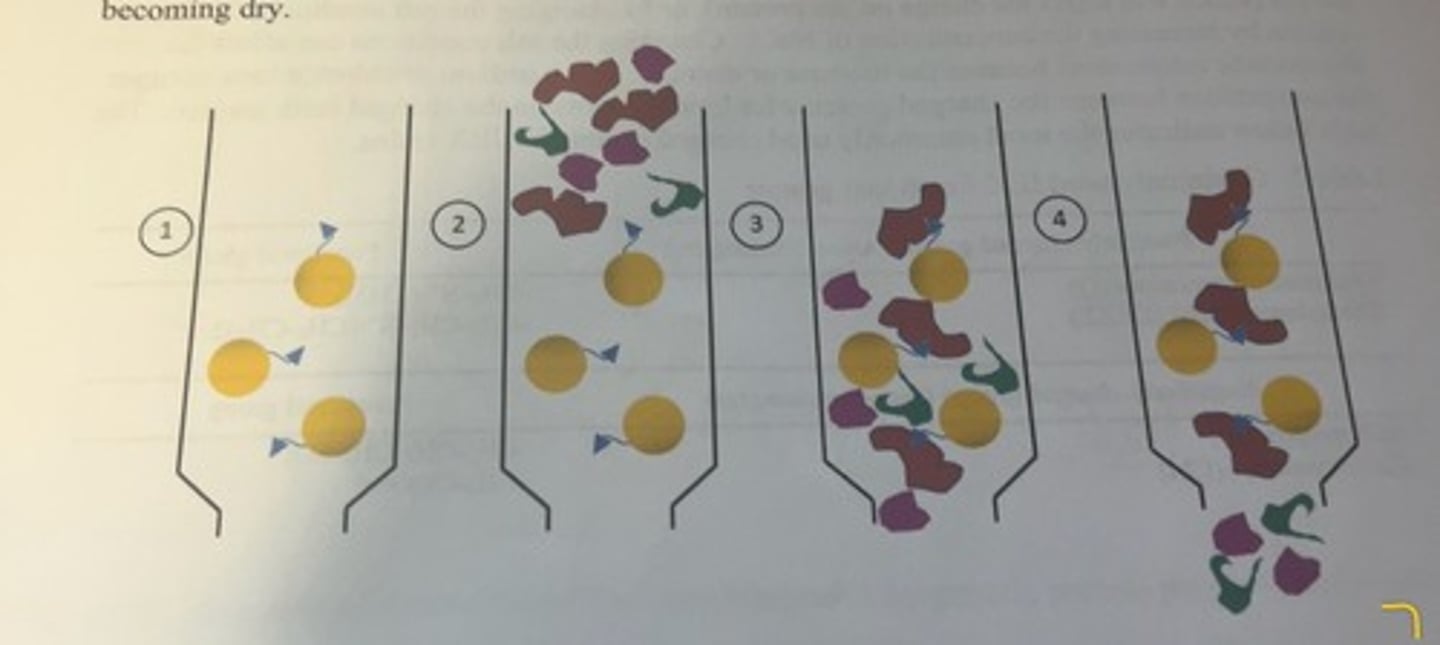
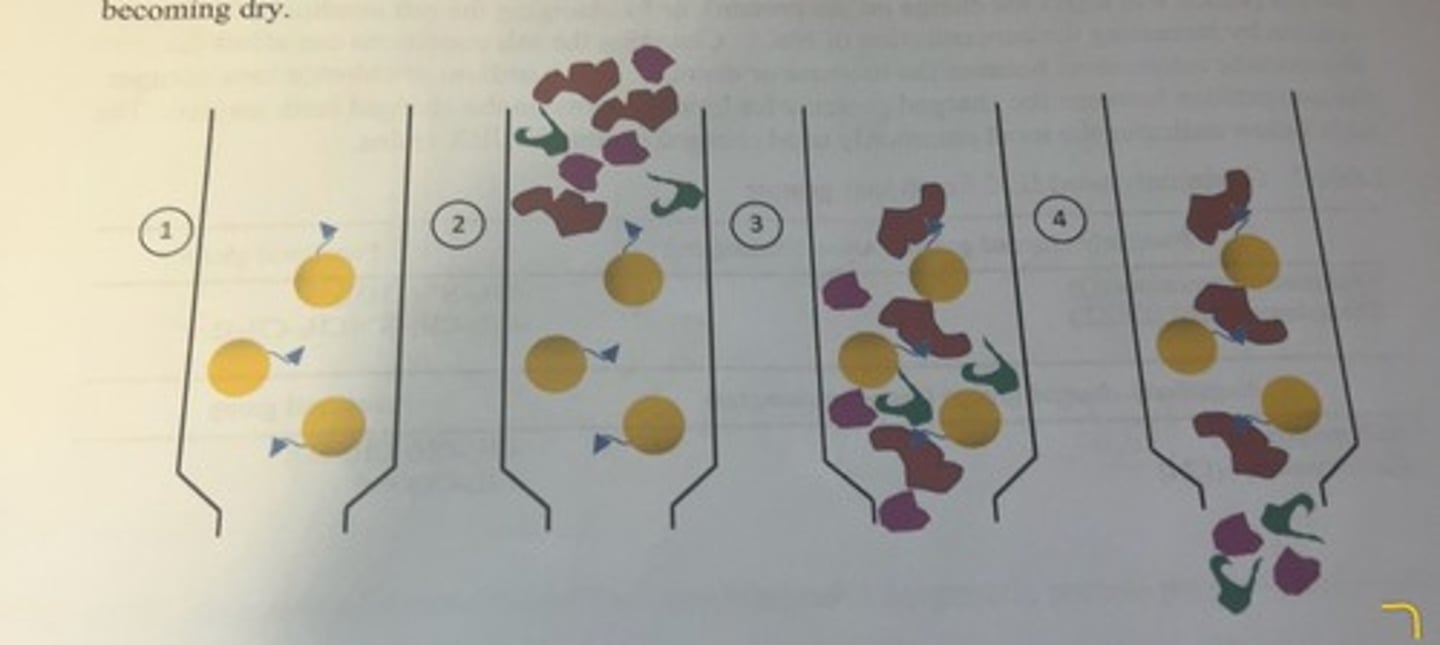
sequence of steps for separating desired protein from protein mixture by affinity chromatography
-the solid support is the yellow bead and the blue triangle attached to the bead is the ligand.
-the column is then equilibrated with a buffer similar to the buffer that was used to extract protein from the cells or tissues.
-following the extraction of proteins from the cells of the tissue into solution, a small volume of the crude lysate is applied to the top of the column and allowed to flow down into the column (2).
-once the sample has entered the column, a wash buffer is continuously added to the top of the column to promote the flow of protein solution within the column, and to prevent the column from becoming dry.
-the protein that we want to purify (indicated by the red structure) has the ability to bind to the functional group on the beads, whereas other proteins (purple and green structures) do not and these proteins flow right through the column (3 and 4).
-the column will be washed with several column volumes of wash buffer to ensure nonbinding protein has been removed from the column.
-ideally at this point, only buffer and the protein of interest (bound to the resin) remain in the column.
steps to remove the bound protein of interest
-there are multiple ways to do this.
-the most common way is to add a high concentration of another molecule (usually very similar in structure to the ligand that is covalently bound to the resin) that also has the ability to bind to the protein of interest.
-since the protein bound to the column also has the ability to bind to the ligand that has been added to the column, the high concentration of the added free ligand out competes the column for binding to the protein with the result that the protein elutes from the column and can be collected in a fraction relatively pure for the protein
elution buffer
the buffer that is used to remove the protein from the beads
elution of bound protein using ligands that bind to column
-another method to elute the protein is add a high concentration of molecule that can also bind to the ligand attached to the column which will outcompete the bound protein for binding spots on the column.
-for example, for proteins with a his-tag that are bound to the nickel ions on the column, a high concentration of free histidine can be added in the elution buffer with the result that some of these free histidine will bind to the column instead of the protein which results in protein elution form the column.
composition of the elution factor
it will be comprised of buffer, the protein of interest and the ligand in the elution buffer that was used to remove the protein from the column.
high concentration of the ligand in the elution buffer
it is highly likely that it is undesirable for further analysis of the protein so now you have to separate this ligand from the protein.
how to separate ligand from protein
-in most cases, the ligand is quite small relative to the size of the protein, so size can be used as the basis for separating the protein and the ligand from each other.
Size-exlusion Chromatography
method that separates molecules on the basis of their size.
beads of size exlusion chromatography
the beads do not have functional groups attached to them.
-instead, the beads are porous and filled with channels of varying sizes.
-the beads are relatively inert and do not have any chemical interaction with proteins or other molecules in a buffer.
how do the beads work
-molecules of varying sizes can be in solution around the bead and within the bead itself
-when a mixture is applied to a column filled with size exclusion beads and allowed to sink into the column, molecules may or may not be able to enter into the pores and channels within the beads depending upon how big the molecules are.
small molecules
can enter the bead, move around in the bead before exiting the bead and returning to the general flow of the mobile phase
larger molecules
may not be able to enter the bead because they are too big to fit which means they remain in the mobile phase moving throughout the column.
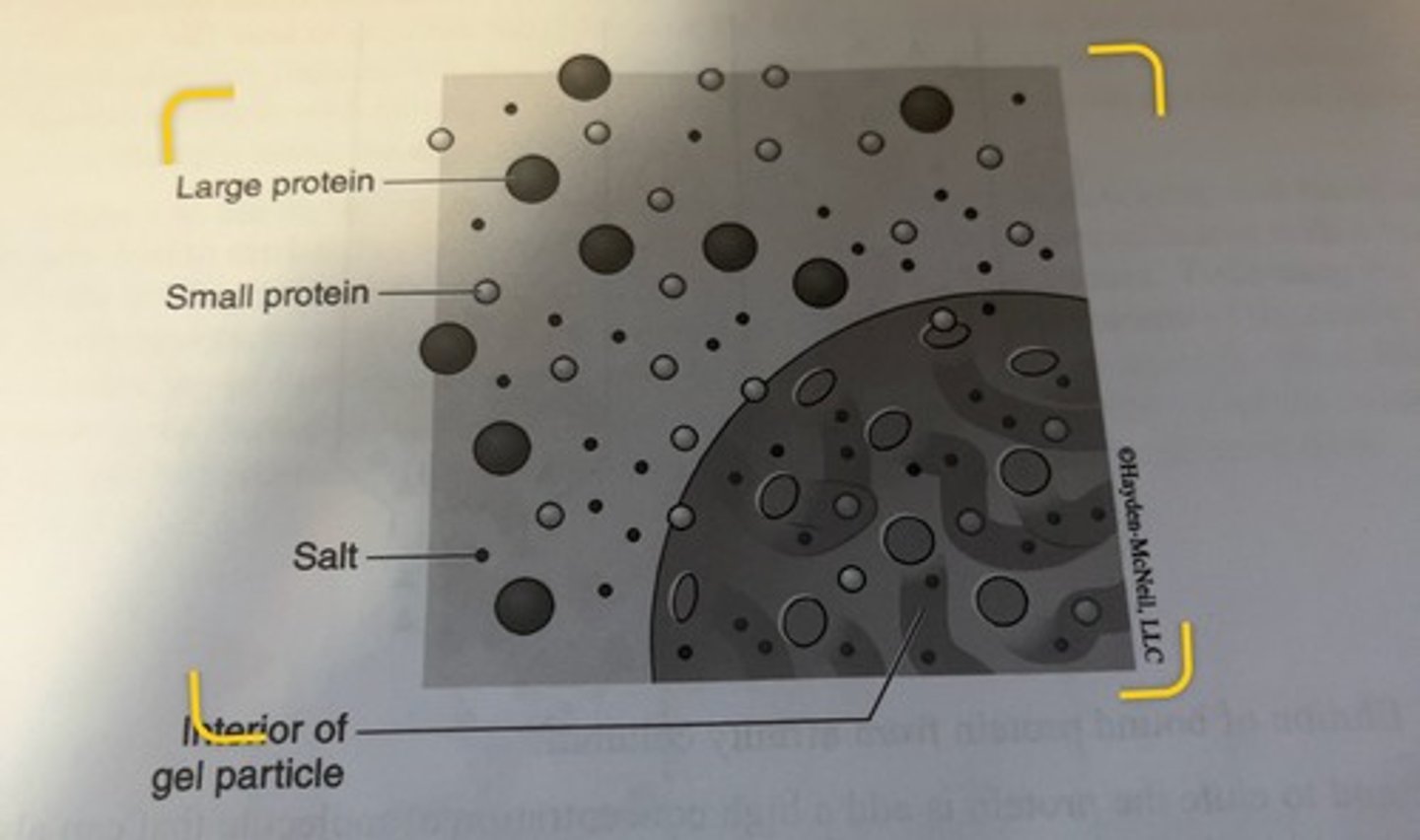
molecules that cannot enter beads
will elute first
-smaller molecules will elute later according to their size, with the smallest molecules eluting last
channels within beads
this ability of the column to segregate molecules by size arises from the fact that the channels within the beads will be of varying sizes, and some molecules will only be able to enter some of the channels, but not all of the channels.
-the overall effect of a varying ability to enter the bead by different sized molecules is that the larger molecules will have a shorter path down the length of the column as compared to smaller molecules.
all molecules that are too big to fit within a bead
they will elute together even if they are different sizes.
all molecules that can fit within all of the pores
will elute together even if they are different sizes.
size exclusion beads
can be manufactured with a variety of pore and channel sizes
-this allows for a tuning of the range of molecules that can be separated.
limits
there are limits to how effectively you can separate different sized molecules within the separation range of the particular type of bead, so the most common use of gel filtration is to clean up a partially purified protein from small molecules, in particular, salts.
separation of the protein
when the protein is mixed with a small ligand, we can effectively separate the protein from the ligand because the protein will be large and will elute from a size exclusion column long before smaller ligand.
size exclusion chromatography as a analytical technique
can be used as an analytical technique for relatively pure protein mixtures.
-if you were trying to determine whether or not a protein is comprised of monomers, dimers or whether it complexes with other proteins, you could determine this because the different structures would likely be sufficiently different in size that you could separate them on a size exclusion column.
in experiment 7, week 1, what did you purify
lysozyme
what did you purify lysozyme from
chicken egg white
what column did you use
Sulfopropyl (SP)-Sephadex ion exchange column
what happened in this lab
a mixture of egg white proteins will be solubilized in acetate buffer (pH 5.3) which will be applied to the top of the column. different mixtures of proteins will be eluted from the column by washing the column with buffers of different pH. the column will be initially washed with acetate buffer (5.3), followed by addition of TRIS buffer (pH 8.1) and finally by the addition of carbonate buffer (pH 10.9).
what will the 3 fractions contain
you will collect 3 mL fractions and will quantify lysozyme activity and protein in various fractions.
anatomy of a column
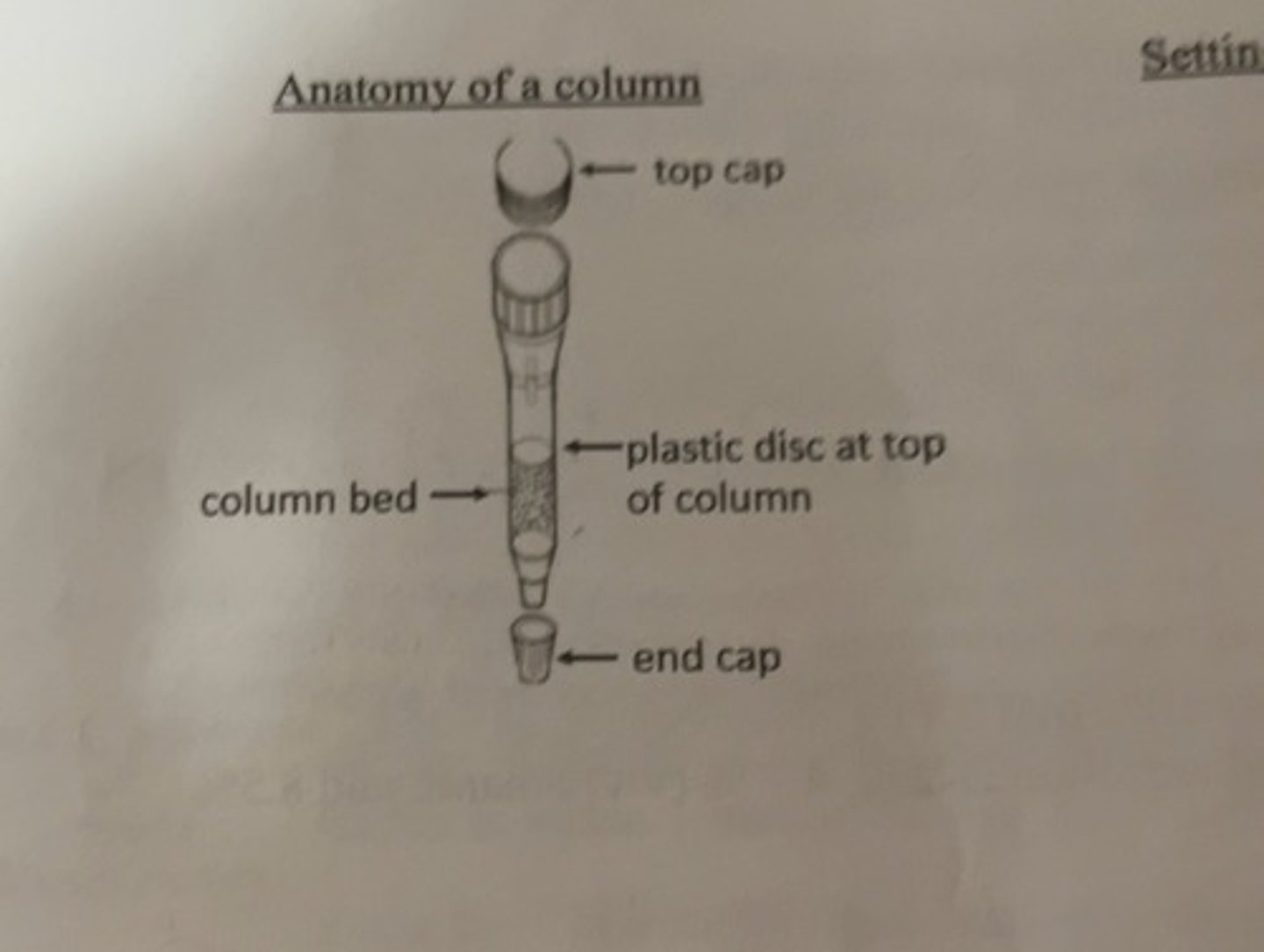
setting up column

running the column
-every time you switch from 1 step to the next step, always allow buffer atop column to fall to within 1 mm of top of column before adding the next required volume.

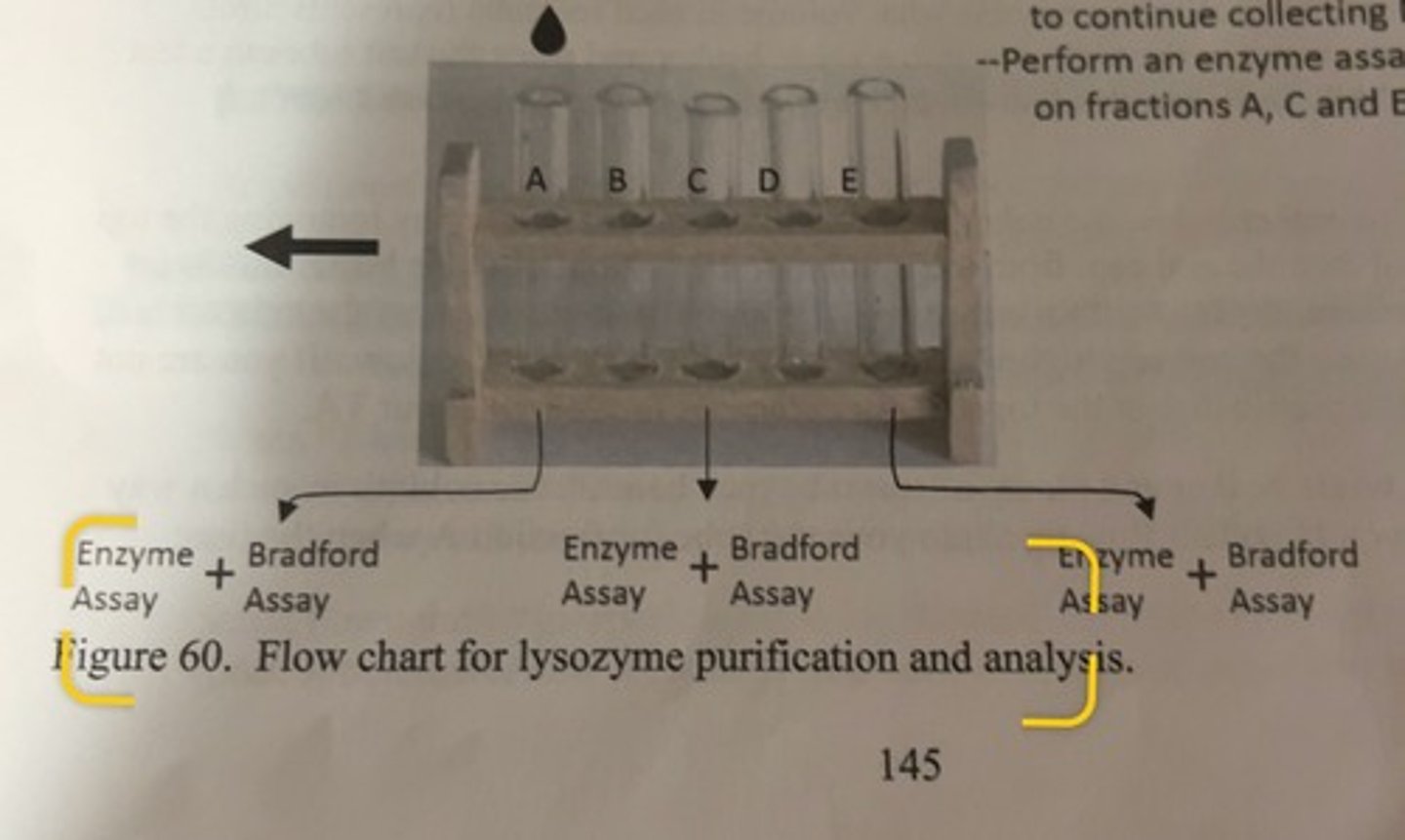
flow chart for lysozyme purification and analysis
-collect flow through in test tubes
-once you have collected 3mL in a test tube, shift the rack and continue collecting in the next tube until you have collected 6 fractions (A thru F).
-once fraction F is collected, return waste beaker to continue collecting NaCl and NaOH
-perform an enzyme assay and Bradford assay on fractions A, C and E.
what species did substrate come from
Micrococcus lysodeikticus
performing lysozyme assays
the cell walls of the substrate are opaque to light at 450 nm, so at the start of the assay, the spectrophotometer will show an absorbance reading well above zero, but if there is any lysozyme present, the bonds between the monomers of the cells walls will be hydrolyzed by the enzyme and the absorbance will go down because more light is now reaching the detector.
-since the presence of lysozyme causes the absorbance to fall with time, the slope of the line will be negative if there is lysozyme in that fraction.
when the assay is completed, the rate of cell wall hydrolysis will be displayed as
absorbance per minute (abs/min)
why did we dilute cuvette A
we diluted it 10 fold because there is too much protein in the undiluted fraction to accurately determine its protein content.