Module 11 - RNA and Transcription
1/45
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
46 Terms
Review: Flow of Genetic Info, in Cells
DNA goes through transcription to produce mRNA, rRNA, and/or tRNA.
The RNA then travels to the ribosomes for translation.
The RNA is then translated into proteins.
Primary Structure of RNA:
contains uracil instead of thymine
hydroxyl group (-OH) on the 2’-carbon atom of its sugar component, whereas DNA has a hydrogen atom.
RNA is more reactive than DNA
Primary structure of a nucleic acid is…
the sequence of nucleotides
RNA forms secondary structures:
RNA molecule folds to form secondary structures due to the H-bonding between complementary bases on the same strand.
stem-loop (aka hairpin)
All cellular RNA types are generated by transcription: In All Cells
messenger RNA (mRNA)
ribosomal RNA (rRNA)
transfer RNA (tRNA)
All cellular RNA types are generated by transcription: Only in Eukaryotes
pre-messenger RNA (pre-mRNA)
small nuclear RNA (snRNA)
small nucleolar RNA (snoRNA)
microRNA (miRNA)
small interfering RNA (siRNA)
piwi-interacting RNA (piRNA)
All cellular RNA types are generated by transcription: Only in Prokaryotes
CRISPR RNA (cr RNA)
RNA replication only occurs with the genomes of some…
RNA viruses
Transcription
the transfer of genetic info. from DNA by the synthesis of a complementary RNA molecule copied from the DNA template.
Transcription Requirements:
template: DNA (a gene)
enzyme: RNA polymerase
free NTPs
no primers
RNA is transcribed from the…
template DNA strand
The template strand (transcribed strand) is known as…
antisense
The non-template strand is known as…
sense
Transcription Unit
a stretch of DNA that encodes an RNA molecule and the sequences necessary for its transcription.
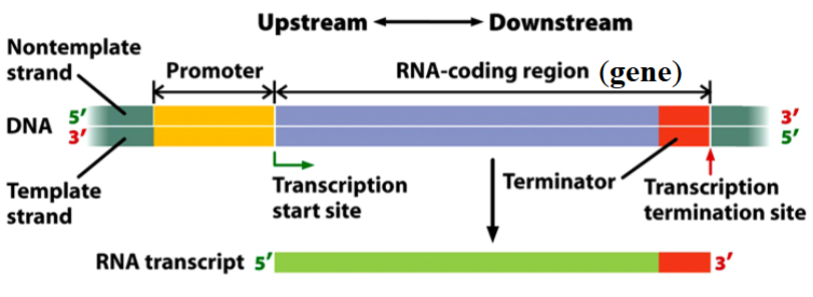
Transcription Unit Components:
promoter
RNA-coding region
terminator
Promoter
transcription unit component; the binding site for RNA polymerase and the transcription initiation apparatus.
RNA-coding region
transcription unit component; a sequence of DNA nucleotides that is copied into a RNA molecule (the gene).
Terminator
transcription unit component; a sequence of nucleotides that signals where transcription is to end (part of the gene).
The gene is only the ____ of the transcription unit.
transcribed portions
Final gene product must be a functional…
RNA molecule or protein,
Formation of the phosphodiester bonds during RNA synthesis:
NTP-NMPn+NTP → NTP-NMPn+1 + P-P (Growing chain + Precusor dNTP).
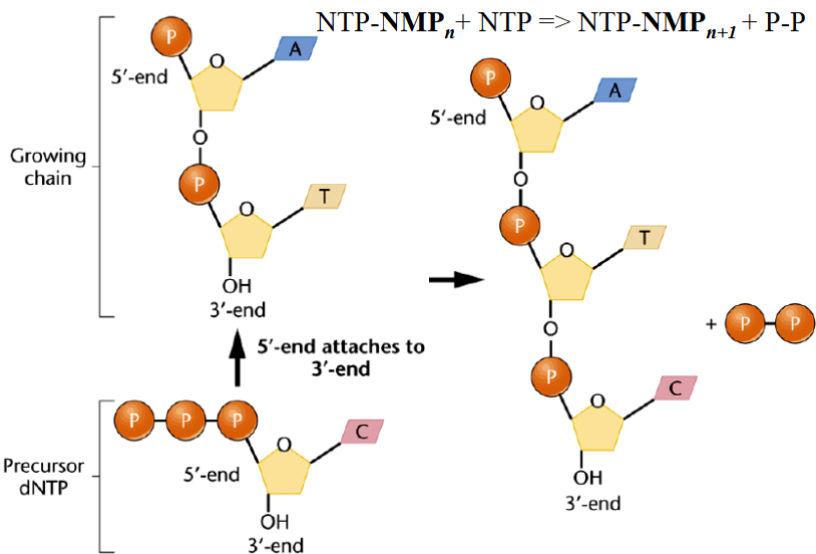
Initiation of RNA Synthesis in Transcription:
NTP + NTP → NTP-NMP + PPi
Elongation of RNA Synthesis in Transcription:
NTP - (NMP)n + NTP → NTP - (NMP)n+1 + PPi
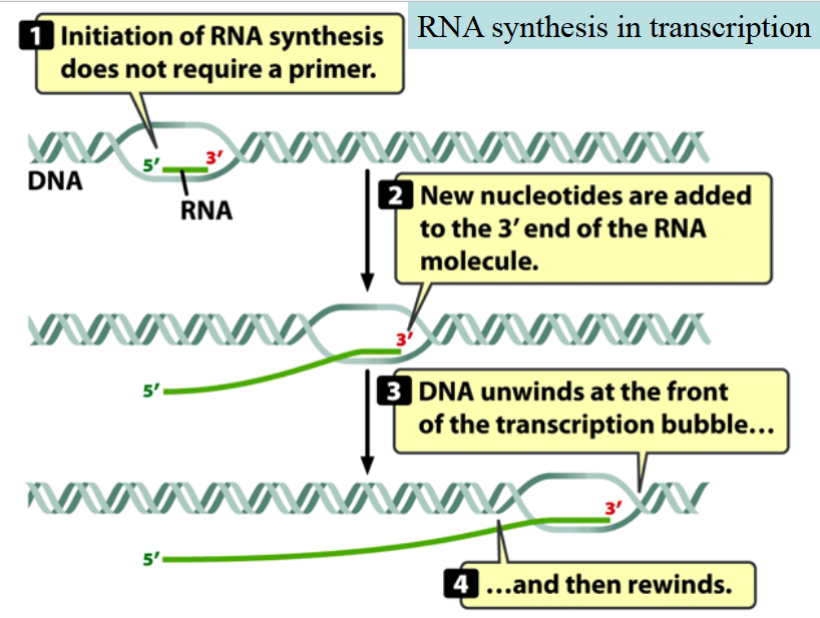
RNA Synthesis in Transcription:
initiation of RNA synthesis does not require a primer
new nucleotides are added to the 3’ end of the RNA molecule
DNA unwinds at the front of the transcription bubble
and then rewinds.
Consensus Sequences in Bacterial Promoters:
a consensus sequence comprises the most commonly found nucleotides at a specific DNA site; conserved sequence.
Pribnow Box
-10 consensus sequence → TATAAT
the bacterial pribnow box has similar function as the eukaryotic TATA box
Transcription is catalyzed by…
RNA polymerase
What is the binding site of RNA polymerase on the DNA molecule?
promoter; right before +1 and is not transcribed
What recognizes the promoter and directs RNA polymerase to it?
the sigma factor
Initiation of transcription begins at the…
transcription initiation site (+1, the first base to be transcribed)
The sigma factor dissociates after…
initiation
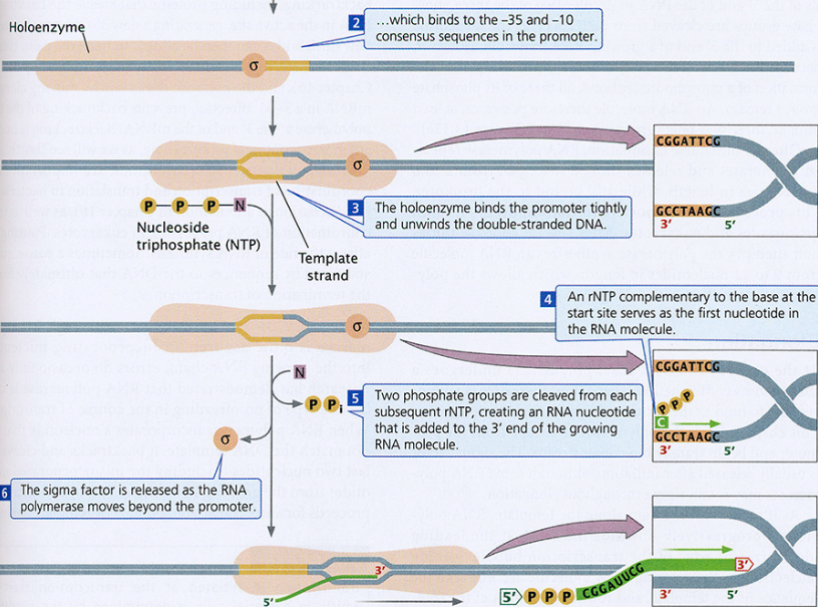
Transcription in Bacteria:
sigma factor associates with the core enzyme to form a holoenzyme…
…which binds to the -35 and -10 consensus sequences in the promoter.
holoenzyme binds the promoter tightly and unwinds the double-stranded DNA.
A rNTP complementary to the base at the site serves as the first nucleotide in the RNA molecule.
Two phosphate groups are cleaved from each subsequent rNTP, creating a RNA nucleotide that is added at the 3’ end of the growing RNA molecule.
Sigma factor is released as the RNA polymerase moves beyond the promoter.
Termination of Transcription in Bacteria:
Depends on the formation of the hairpin loop secondary structure at the terminator site.
Hairpin loop forms after inverted repeats in the gene are transcribed into the RNA molecule and form intramolecular base pairs
Formation of the hairpin loop destabilizes the DNA-RNA pairing, and transcription ends,
Termination of Transcription in Bacteria: Rho-dependent Termination
the Rho protein (a helicase) is needed to break the DNA-RNA pairing and terminate transcription.
Termination of Transcription in Bacteria: Rho-independent Termination
the weak DNA-RNA pairing at the terminator site is enough to destabilize the interaction and terminate transcription.
Termination in bacteria: Rho-dependent Mechanism:
Rho binds to an unstructured region of RNA and moves towards it 3’ end.
When RNA polymerase encounters a terminator sequence, it pauses and Rho catches up.
Using helicase activity, Rho unwinds the DNA-RNA hybrid and brings transcription to an end.
Termination in bacteria: Rho-independent Mechanism:
A Rho-independent terminator contains an inverted repeat followed by a string of approx. 6 adenine nucleotides.
The inverted repeats are transcribed into RNA.
The string of Us causes RNA polymerase to pause…
…and the inverted repeats in RNA fold into a haripin loop…
…which destabilizes the DNA-RNA pairing.
The RNA transcript separates from the DNA template, terminating transcription.
RNA polymerase II and transcription elongation in eukaryotes:
RNA polymerase maintains a transcription bubble during elongation, in which about 8 nucleotides of RNA remain base-paired with the DNA template strand.
RNA polymerase II and transcription elongation in eukaryotes Mechanism:
DNA double helix enters a cleft in the polymerase and is gripped by jaw-like extension of the enzyme.
The two strands of the DNA are unwound, and RNA nucleotides entering the enzyme through a pore are added to the 3’ end of the growing RNA molecule
As it funnels through the polymerase , the DNA-RNA hybrid hits a wall and bends at a right angle, keeping the bubble open and positioning the DNA-RNA hybrid at the active site of the enzyme.
The newly synthesized RNA separates from the DNA and runs through another groove before exiting the enzyme.
Termination of transcription of protein-coding genes in eukaryotes:
termination in eukaryotes does not occur by itself but requires the activity of the Rat1 exonuclease.
after the protein-coding region of the gene is transcribed, the RNA molecule is cleaved by a RNA endonuclease at a consensus cleavage site.
then, Rat1, a 5’ → 3’ RNA exonuclease, binds to the unprotected 5’ end of the trailing non-coding RNA fragment and begins to degrade it until it catches up with the RNA polymerase, which is still transcribing DNA
the complete degradation of the trailing fragment terminates transcription.
Termination of transcription of protein-coding genes in eukaryotes: Mechanism
RNA polymerase II transcribes well past the coding sequences of most genes.
Cleavage takes place at the 3’ coding region of the RNA…
…while RNA polymerase continues transcribing.
The Rat1 exonucleases attaches to the 5’ end of the trailing RNA.
…and moves towards the RNA polymerase , degrading the RNA as it goes.
When Rat1 reaches the polymerase, transcription is terminated.
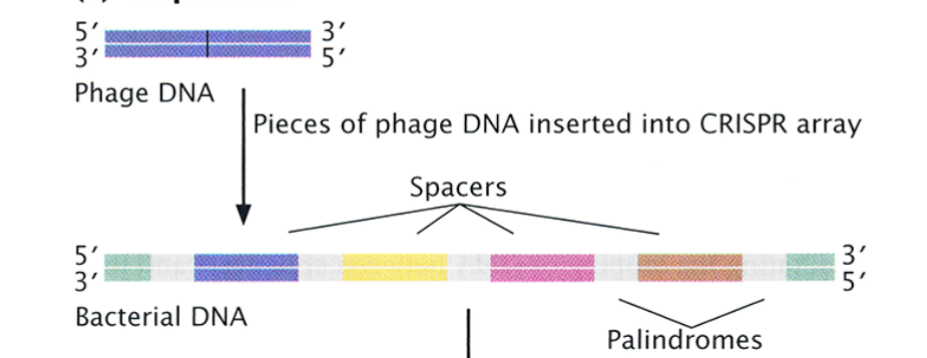
CRISPR
Clustered regularly interspaced short palindromic repeats.
DNA arrays consisting of a number of short palindromic sequences separated by spacer sequences. spacer sequences.
3 Stages of CRISPR:
Acquisition
Expression
Interference
CRISPR: Acquisition
foreign DNA enters enters the cell, and it is identified, processed, and inserted into the CRISPR array as a new spacer (a memory of the invading DNA).
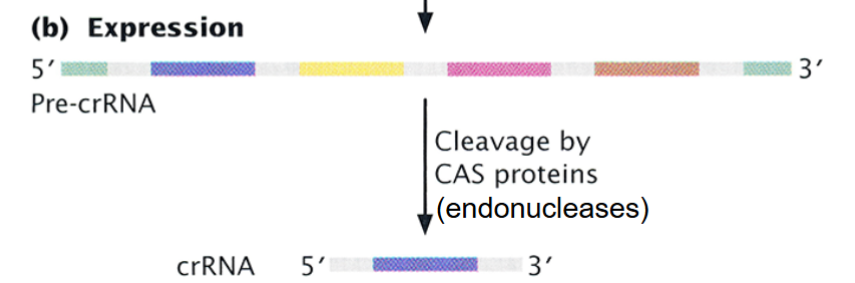
CRISPR: Expression
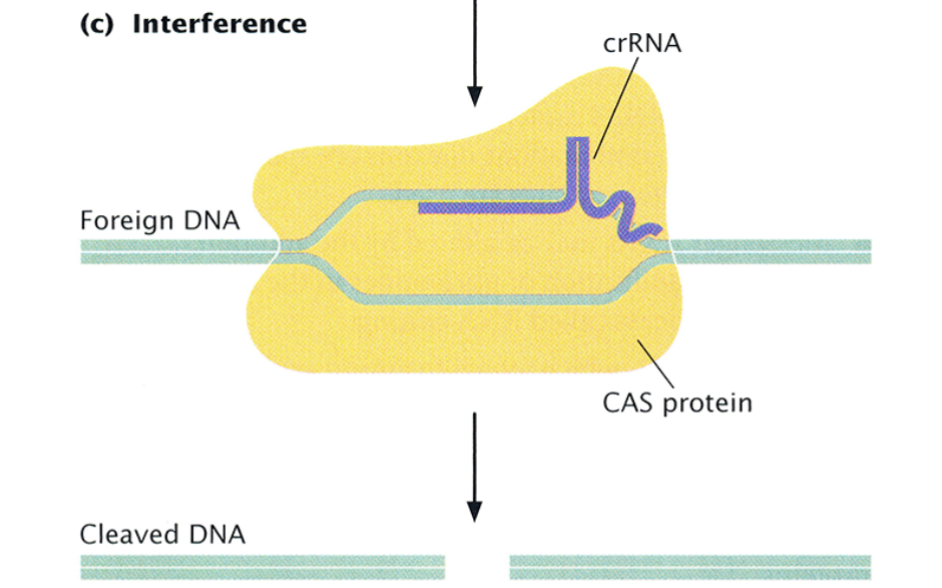
the entire array is transcribed into a long CRISPER precursor RNA and is the cleaved by a CAS protein into CRISPR RNAs, each one containing one spacer homologous to foreign DNA.
Each crRNA combines with a CAS protein to form an effector complex.
CRISPR: Interference
If the same foreign DNA enters the cell again, the effective complex recognizes it by its base-pair complementarity to the crRNA and the CAS protein cleaves it with its endonuclease activity.