Protozoa Molecular Biology
1/33
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
34 Terms
Name the examples of eukaryotes needed for this lecture series.
Dinoflagellates
Diatoms
Trypanosomes
Define protists
Single celled eukaryotes that are not algae or fungi.
How are protists shown to be complex - example?
Nematodinium
Eye of a single celled organism, thought to be used for detecting shadows and predators. has a refracticle structure derived from a mitochondria, has a pigmented light sensitive region from the chloroplast.
How did eukarya evolve? What is the evidence for this?
rRNA sequences analysis by Woese showed that eukaryotes embedded within archaea. First eukaryote can be described as the first eukarya to acquire a mitochondrion.
Acquisition first shown in Lokiarchaeota part of the Asgard superphylum from environmental samples which were shown to eukaryote type sequences.
What features do eukaryotes share with archaea?
RNA polymerase. specfically RNA pol II composition of subunits.
for example, whilst RBP 1,2,3,6,11 homolgous in bacteria and archaea.
only RPB 4,5,7,8,10,12 in archaea.
RPB 9 no homologues in bacteria or archaea.
What is the issue with the model organisms used presently?
Claim diversity eg from yeast to humans, however, across the evolutionary tree very small part sampled. little work done on protists.
What is different about the nuclei in ciliates?
They have two types of nuclei:
Micronucleus (small, diploid, transcriptionally silent during vegetative growth) Macronucleus (highly polyploid [800n], transcriptionally active) (They may have multiple macronuclei.)
Shown through staining with DAPI
How is ciliate sexual and asexual cell cycle different to mammalian?
Cells (e.g. Paramecium) have a sexual cycle, followed by asexual propagation Ciliate sexual cycle – conjugation between two cells .
Describe the ciliate sexual cycle.
1) Micronucleus of two cells undergoes meiosis; macronuclei destroyed
2) Haploid nuclei from each partner fuse to generate new diploid micronucleus
3) Macronucleus develops from the new micronucleus (mitosis/binary fission)
Describe how the macronucleus develops from the micronucleus.
Micro genome eliminates repeated sequences, genome fragmented with telomeres.
IES - single copied sequences, no recognisable sequence motifs to this (IES flanked by TA on each side of it). excised from the sequence.
Elimination events involve: (i) Elimination of tandem repeats and transposons, fragmentation of genome and addition of telomeres (ii) Excision of approx. 60,000 internal eliminated sequences (IES).
Determined by RNA in the macronucleus which somehow allows for retention of the sequence in the next macro nucleus.
What is the experimental evidence for RNA driving the inclusion of genes in subsequent macronuclei?
Evidence comes from looking at d48 strain, which fails to express a surface antigen.
Gene is present in the micronucleus, but gets eliminated from the macronucleus.
Restore it to the macronucleus by transformation – it then gets retained by the micronucleus in future rounds of sexual reproduction.
that therefore shows there is a signal from mac to mic for sequence retention in the form of RNA.
What is the evidence using RNAi about the ciliate sexual cycle?
Get dsRNA from Ecoli that is specific to the sequence of RNA of interest.
Feed into the ciliate - will have in the new macronucleus.
However if also supplement with rest of RNAi machinery, RNA is destroyed and the macronucleus new will not have the DNA to the RNA sequence in it.
How does elimination of genes happen?
RNA binding porteins have been implicated.
Genetic data indicates chromatin remodelling proteins in the micronucleus are important .
therefore RNA is the determinant of inheritance
knockdown of RNA binding poroteins shows decrease in excisiion.
How is Oxytricha’s elimination different?
Some ciliates have different rearrangements in the macronucleus. Oxytricha eliminates sequences and also ‘unscrambles’ them – changing the order in which they are located. Again, seems to depend on RNA.
eg. Telomere and binding protein, sequence changes from MIC to MAC. Hit with dsRNA to destroy = aberrant macronucelus.
The macronucleus has a huge number of chromosomes, each of which has telomeres, so this (Tetrahymena) was important in the early studies by Blackburn on telomeres. Many cloning vectors for eukaryotes also include telomeres from Tetrahymena. Makes sense to use since so polyploid.
How are dinoflagellate nuclei unusual?
Dinoflagellate nuclei are highly unusual, often referred to as a ‘dinokaryon’ and described as ‘mesokaryotic’:
• chromosomes remain condensed
• have a fibrillar appearance (‘liquid crystalline’)
• have a high content of 5-hydroxymethyl uracil
• very high DNA content – up to 200 pg per nucleus, c.f. 3 pg per haploid nucleus in humans
• high concentration of cations associated with DNA
• largely dispensed with conventional histones
What is different about nucleosome packagaing in haematodinium?
does not show nucleosome patterning on nuclease digestion and histones not seen in westerns.
What do protists have instead of histones - 3 examples?
1) Proteins similar in sequence to bacterial histone-like proteins (HU proteins) that coat DNA in prokaryotes – Histone Like Basic Proteins (HLPs).
• Absent from early-branching dinoflagellates, but present in core species (e.g. Symbiodinium). Maybe acquired by lateral transfer from bacterium
2) Dinoflagellate/viral nucleoprotein (DVNP)
• Similar in sequence to a DNA binding protein from viruses with very large genomes (hundreds of kbp) that infect algae - Phycodnaviridae show interaction using fluorescence.
• Expression of DVNP in eg Saccharomyces impairs growth. (serial dilution and nuclear localisation signal shows reduced viability.)
• Also leads to reduction in histone levels (compare right hand column with left)
• Mutations that reduce histone levels help to restore viability when DVNP expressed
dinoflagellate infection of DVNP virus → survived, lateral gene transfer = expressed, cell respond by decreasing histone and DVNP replaced the histone
• Suggests a sequence of events leading to displacement of histones by DVNP
And yet there are also: 3) Histone genes (no idea what they are used for maybe in some stage of the cell cycle or in some region of DNA.
• divergent, yet retain recognizable features
• transcribed, but transcript levels low (1/25 what is ‘typical’)
• polyA tail (unusual for histone mRNAs)
How are some bacterial histone like proteins similar to eukaryotic?
Structural similarity to histone, convergent evolution, decorate the outside not around it like a histone.
Archaea: have histone fold domain with non specific DNA binding and they don’t have tails like mammals, so there is no modification overall.
It is difficult to distinguish real histones from examples of convergent evolution.
Summary: not all eukaryotes have histones but histones may appear outside of eukaryotes.
Why are most eukaryotic RNAs monocistronic?
consequence of translation initiation. ORFs downstream not translated since only monocistronic and IRES can be done before stop signal.
How are trypansomes not monocistronic?
Mature transcripts monocistronic but transplicing occurs of polycistronic primary transcripts. less transcription start sites and open reading frames so transcribes as polycistronic.
What disease can trypanosomes cause?
Chronic disease Euglena causes Chagas Disease
How does transplicing work in trypanosomes?
So splice leader RNAs of around 140bp from a different part of the genome single locus and full of tandem repeats gets spliced to one open reading frame and a poly A tail gets added.
What are other unique examples of transplicing?
Chlamydomonas has a reaction centre in the chloroplast which has three separate exons which are fused together by transplicing. Not the normal pattern though this is the exception to the rule in this case.
How do hydrogenosomes and mitosomes break the paradigm that chloroplasts are for photosynthesis and mitochondria are for aerobic respiration?
Hydrogenosomes - No complex III, IV, ATP synthase, but still have bioenergetic role: Nyctotherus – anaerobic ciliate – has remnant mt genome, encoding some components of complex I. Organelle has complex I and II; possibly running complex II backwards Blastocystis – stramenopile - has complex I and II (also an alternative oxidase) mt genome encodes complex I subunits and several ribosomal proteins Trichomonas – Parabasalid - has complex I (probably no mt genome)
Mitosomes – No bioenergetic role, but still function in Fe-S cluster biogenesis e.g. Giardia for other proteins
What is unusual about dinoflagellate chloroplast genome?
Dinoflagellates have a highly unusual chloroplast genome: • Highly reduced – just encodes key proteins of photosynthetic electron transfer chain (and some open reading frames) • All other genes (including most tRNAs) moved to nucleus • Fragmented into plasmid-like ‘minicircles’ with ‘core’ believed to be involved in replication, expression • Movement of genes to nucleus may reduce burden of ensuring a full set of genes transferred to daughter cells on division • Transcripts given polyU tails – function not clear
each gene is on a different mini circle - genome organisation is similar.
Essentially, some event happened stimulating fragmentation and strong selective advantage to move to nucleus after this otherwise risk of losing genes and function.
What other organisms is this pattern of fragmentations visible in?
Otherwise, the pattern with inverted repeats etc is recognisable even in Apicomplexa such as Plasmodium, Theileria, and Toxoplasma, which have lost photosynthesis.
they have a photosynthetic ancestory and remnant chloroplast which the only function is for biosynthesis FeS. possible exploitation for antimalarial agents.
What are two things that can be used for antmalarial agents?
Protein synthesis inhibitors (e.g. doxycycline) Isoprenoid biosynthesis inhibitors (e.g. fosmidomycin)
How were the mitochondria and chlopolast gained?
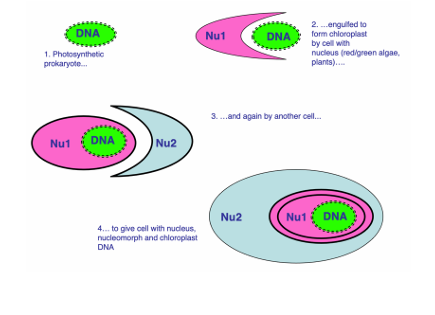
Mitochondria (or mitochondrion-related organelles – MROs) are present throughout eukaryotes. Chloroplasts are widely distributed, but result from lateral transfer from the original lineage that contains red/green algae and plants. These are ‘secondary’ endosymbioses. The exact number is unclear.
Acquisition of mitochondria seen in archaea - marking the beginning of eukaryotes. Chloroplasts acquired from photosynthetic bacteria
What do we mean by secondary endosymbiosis and why are there 4 genomes in a cryptophyte?
Secondary endosymbionts - acquired by another lateral gene transfer event. multiple of these happened seeing as there are both red and green protists.
Secondary endosymbiosis explains the mysterious fourth genome, the nucleomorph, seen in chlorarachniophytes and cryptophytes. 4 membrane chloroplast and 2 membranes = nucleomorph which has happened twice in evolution (at least).
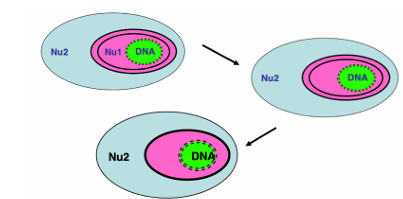
How was the nucleomorph 4 genomes simplified in some lineages?
This got simplified in some lineages • Loss of nucleomorph • Reduction of membranes Organisms with 4 or 3 membranes and no nucleomorph (eg dinoflagellates)
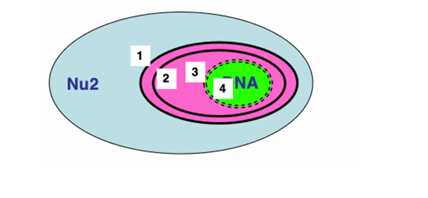
What are the usual systems for transport of proteins into the chloroplast (one with 2 membranes)?
Toc and Tic complexes (Translocon outer/inner membrane of the chloroplast) a bit like the mitochondrion
What are the systems for protein trafficking from nucleus to the thylakoid lumen in 4 membrane system - from the cytoplasm? How does this differ in three membrane systems like dinoflagellates?
Membranes 3 and 4 are the easy ones, equivalent to the green chloroplast envelope, and a similar system is system used.
Likewise for entry into thylakoid lumen – equivalent to entry into the thylakoid lumen for green chloroplasts (and cyanobacteria).
In many algae with 4 membranes round the outside, Membrane 1 is continuous with ER. Getting across it into compartment between 1 and 2 is analogous to getting into ER lumen. So ER targeting takes care of that.
Getting across Membrane 2 is equivalent to getting something out of the ER. There is a pathway for that in other organisms – the ERAD system (ER-Associated Degradation).
An equivalent of that is used here - SELMA (Symbiont specific ER-Like Machinery).Where Membrane 1 doesn’t exist (eg dinoflagellates), proteins are probably delivered to Membrane 2 by vesicles. No need for SELMA.
What is an example of an independent primary endosymbiosis?
Paulinella chromatophora is an example of an independent primary endosymbiosis separate from green and red algae/plants, in the Rhizaria.
There are two membranes round the chromatophore. Hundreds of proteins get imported, and there are two novel (ie seem to be different from what happens in the green plant lineage) import pathways for these:
Proteins < 90 amino acids don’t have an N-terminal targeting sequence Proteins > 270 amino acids do.
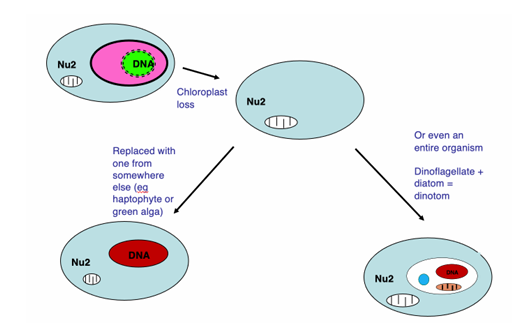
How have some dinoflagelllates regained a chloroplast? (2 methods)
Some dinoflagellates have lost their chloroplast and replaced it with one they acquired from elsewhere (haptophyte or a green alga).
Others have replaced it with an entire organism – a diatom – to form a ‘dinotom’.
So in the dinotoms we have two different nuclei, two different mitochondria and one chloroplast. There are also two different mitochondria. That makes five genomes in one cell in total!
More than enough for anyone to cope with, you might think – but not the ciliate Mesodinium rubrum. It eats cryptophytes and maintain the cryptophyte nucleus (1), nucleomorph (2), chloroplast (3), and mitochondrion (4).
It has its own mitochondrion (5) and, being a ciliate, has a micronucleus (6) and macronucleus (7). That’s seven genomes in one cell.