PL5 : Translation 2 (elongation and termination)
1/13
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
14 Terms
What are ribosomes responsible of?
All protein synthesis.
Which is the most abundant RNA-protein complex in the cell?
The ribosomes
At what rate does elongation proceed?
3 -5 amino acids/second
What is peptidyltransferase activity?
Creation of peptide bonds in growing polypeptide chain without protein.→ enzymatic activity in RNA molecule called ribozyme
Which structure is responsible of peptide transferase activity?
In bacteria : 23s rRNA; in eukaryotes : 28s rRNA
Explain translation elongation in y steps:
Again flame:
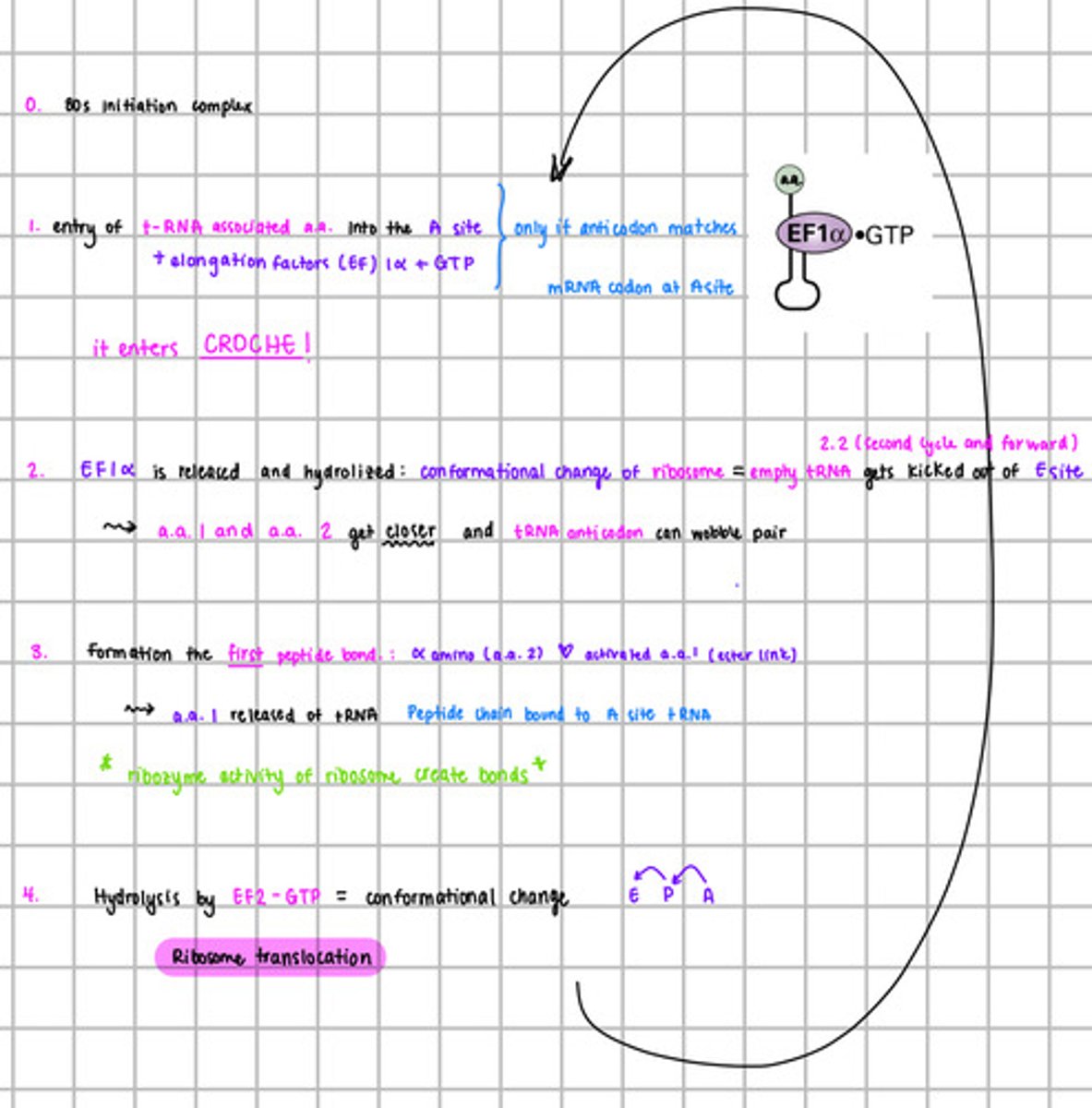
Explain translation termination
When stop codon at A site : eRF1 (termination release factor) associated with GTP-bound eRF3 bind to A site → cause GTP hydrolysis
Result : cleavage of polypeptide chain on P site tRNA
Disassociation of large and small ribosomal subunit so that ready to be used again.
Put these 3 events in order : peptide cleavage, release of tRNA at E site and release of polypeptide chain
1- peptide cleavage 2- release of the 2 other things
What favors the reinitiation of translation? How?
Interaction of the s' and 3' ends of the mRNA
They create a loop
mRNA stabilization = efficient transcription
What does ribosome profiling enable?
- Detailed measurement of translation
- instantaneous measurement while others measure are steady state.
What does ribosome profiling do?
It quantifies the levels of new protein synthesis and gives information on where the ribosomes are ( helped to discover new translation products)
How is ribosome profiling done?
1 - Isolation of mRNA ASSOCIATED with ribosomes
2 - cleaved with nuclease : not affected if associated with ribosomes = ribosome footprints
3 - Sequencing and mapping of ribosome footprints which should indicate reading frames ; the more they are present the more they should be actively translated
If gene A is sequenced 10x more than gene B, does it mean that gene A is translated 10x more efficiently than gene B?
NOOOOOOOOOO!
How can we correcly interpret result from ribosome profiling?
We need to compare the ratio of ribosome-bound mRNA to the ratio of total mRNA.
If they are the same, both genes are as efficiently translated.
If they are different, the one that has a bigger ribosome-bound ratio is translated more efficiently