Structural Bioinformatics
1/21
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
22 Terms
what is some information on the proteins of the extracellular matrix
fibrous proteins, crosslinking enabled by disulfide bridges
extensively glycosylated, contributes to cell adhesion
composed of distinct domains with specialised functions
cell adhesion
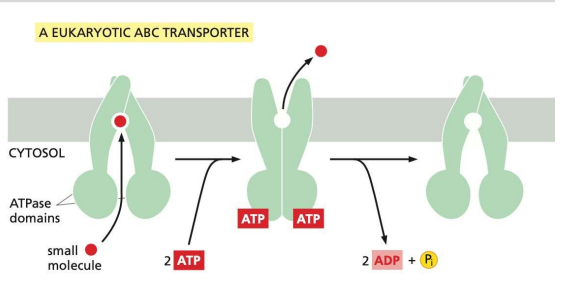
what is some information on membrane transport
hydrophobic belt of residues
ATPase activity
Dimer
conformational flexibility
channel specifically matching the chemical nature of small molecules
where is N & C terminus located
N = start (start Now)
C = end (Cut off)
what are the different levels of protein structure
primary - refers to sequence of amino acids
secondary - refers to initial shape formed by H-bonds
e.g. alpha helices / beta sheets
tertiary - how protein folds into a globular shape
quaternary - structure formed by multiple proteins interacting
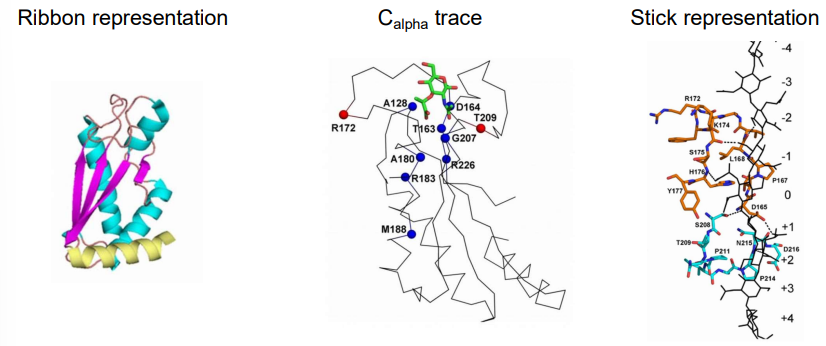
what are some different ways to display protein structure
ribbon representation
Calpha trace
stick representation
space filling representation
surface representation
what are some functions of oligomerisation
allows proteins to function
increases the stability of the protein
what does folding minimise in the system
minimises the free energy of the system
the configuration is optimized to minimize the free energy
partially folded intermediates have higher free energy
how is free energy minimized
through choosing a configuration that maximizes favorable interactions between amino acid residues, this includes maximizing:
favorable interactions between hydrophobic residues inside the hydrophobic core
interactions between polar residues on the surface and water
what forces drive the folding process
van der Waals forces - random dipole moments
3.5-4.0 A
hydrogen bonds - O, N, F
>=3.1 A
electrostatic interactions - ion pairs
>=3.1 A
hydrophobic interactions - polar vs non-polar interactions
what are domains
structural units that are part of a continuous protein chains, but that fold independently of each other
different domain often associated with different functions
what is sequence homology analysis
compare the amino acid sequence of a protein with sequences of other known proteins in databases
what information can be learnt from multiple sequence alignments
conserved residues can be identified
could be important for correct folding
could be important for function
2 proteins with >25% identity in amino acid sequences usually share the same structure.
sequences can diverge (evolution) while retaining the same stable structural fold
why might protein structure be challenging to study
cannot produce protein in high amounts
unstable or degrades fast
protein is too big/small for some techniques
what is resolution
the shortest distance between 2 objects where they can still be distinguished as 2 separate entities
what features are discernible at different resolutions
10-20 Å – blobs, overall shape of the molecule
Can dock/fit a homology model into the overall shape
5-10 Å - Can discern secondary structure elements, helices seen as sausages, beta-sheets as plates
<5 Å - Can perform backbone tracing
3.2-3.5 Å - Can see amino acid side chains
~1.5 Å - Can resolve individual atoms
what are some different levels of structural information
3D atomic structure
shape & symmetry
protein contact (interaction)
subunit proximity (distances)
what is the principle between X-ray crystallography
electrons within a single molecule will scatter X-rays, however we are unable to measure the scattering
crystal, containing a repeating pattern of molecules, provides a way of amplifying the signal of the scattered X-rays
what are some pros & cons of X-ray crystallography
PROS:
ultra-rare resolution
insight into dynamics
chemical reaction intermediates trapped in the crystal
CONS:
not all proteins crystallize
high protein concentration required
static picture
what is the principle behind nuclear magnetic resonance
Certain atoms (H, C, N) have a nuclear spin that gives rise to an NMR signal. The signals in the NMR spectra are affected by:
the chemical environment of the atom and
how flexible the region is where it is located
what are some limitations of NMR
high protein concentration required
requires efficient isotope labeling
outcome is multiple structure that must be interrupted
what are some advantages of cyro-electron microscopy
helps us visualize large proteins & protein complexes
only low concentration are needed
no need to crystallize
why is the structure of functionally homologous proteins more conserved than the amino acid sequence
Once a protein has evolved and folded into a stable conformation, natural selection will ensure that any subsequent random mutations do not change this fold.
Mutations emerging in the course of evolution may change the properties of the protein (e.g. reaction specificity or substrate specificity) but the stable fold will be retained