Biochemistry- Chapter 2 & 3 Mississippi State University
0.0(0)
Card Sorting
1/87
Earn XP
Description and Tags
Study Analytics
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
88 Terms
1
New cards
Water has a higher melting point, boiling point, and heat of vaporization than
most other common solvents
2
New cards
Hydrogen bond
electrostatic attraction between the oxygen atom of one water molecule and the hydrogen of another.
3
New cards
Hydrogen bonds are
relatively weak.
- bond dissociation energy = ~23 kJ/mol in liquid H2O
- 10% covalent, 90% electrostatic.
- bond dissociation energy = ~23 kJ/mol in liquid H2O
- 10% covalent, 90% electrostatic.
4
New cards
Hydrogen bonds are fleeting
- lifetime of each hydrogen bond is just 1 to 20 picoseconds in liquid
- when one hydrogen bond breaks, another forms.
- Flickering Clusters
- when one hydrogen bond breaks, another forms.
- Flickering Clusters
5
New cards
Hydrogen bonds readily form between
an electronegative atom and a hydrogen atom covalently bonded to another electronegative atom
6
New cards
electronegative atom
the hydrogen acceptor.
7
New cards
hydrogen atom covalently bonded to another electronegative atom
the hydrogen donor
8
New cards
Hydrogen atoms covalently bonded to carbon atoms
DO NOT do hydrogen bond
9
New cards
Alcohols, aldehydes, ketones, and compounds containing N—H bonds all form
hydrogen bonds with water.
10
New cards
directionality of the Hydrogen Bond
strongest when the acceptor atom is in line with the covalent bond between the donor atom and H
- maximizes electrostatic interaction
- maximizes electrostatic interaction
11
New cards
Hydrophilic
describes compounds that dissolve easily in H2O; generally charged or polar compounds.
12
New cards
Hydrophobic
nonpolar molecules such as lipids and waxes.
13
New cards
Amphipathic
contain regions that are polar (or charged) and regions that are nonpolar.
14
New cards
H2O dissolves
salts and charged biomolecules by screening electrostatic interactions.
15
New cards
The increase in entropy of the system is largely responsible for
the ease of dissolving salts in water.
16
New cards
water has a high
dielectric constant
17
New cards
Dielectric constant (ε)
a dimensionless physical property that reflects the number of dipoles in a solvent - for water at 25 °C, ε = 78.5 - for nonpolar benzene at 25 °C, ε = 4.6
18
New cards
the force (F) of ionic interactions in solution depends on:
- the magnitude of the charges (Q)
- the distance between the charged groups (r)
- the dielectric constant of the solvent (ε)
- the distance between the charged groups (r)
- the dielectric constant of the solvent (ε)
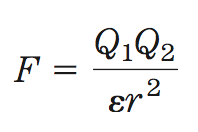
19
New cards
ionic attractions or repulsions operate
over 10 to 40 nm.
20
New cards
CO2, O2, N2
are nonpolar.
21
New cards
Why is CO2, O2, N2 non polar?
Their movement into aqueous solution decreases entropy by constraining their motion.
22
New cards
Nonpolar compounds interfere with the hydrogen bonding among H2O molecules by
increases enthalpy (∆H) and decreases entropy (∆S)
23
New cards
the free-energy change (∆G = ∆H −T∆S) for dissolving a nonpolar solute in water is unfavorable because
- ∆H has a positive value
- ∆S has a negative value
- ∆G has a positive value
- ∆S has a negative value
- ∆G has a positive value
24
New cards
H2O molecules form a highly ordered, cage like shell around each solute molecule
- maximizes solvent-solvent hydrogen bonding
- H2O molecules are not as highly oriented as those in clathrates (crystalline compounds of nonpolar solutes and water)
- H2O molecules are not as highly oriented as those in clathrates (crystalline compounds of nonpolar solutes and water)
25
New cards
Hydrophobic effect
- nonpolar regions cluster together
- polar regions arrange to maximize interactions with each other and with the solvent
- polar regions arrange to maximize interactions with each other and with the solvent
26
New cards
micelles
thermodynamically stable structures of amphipathic compounds in water.
27
New cards
van der Waals interactions (London dispersion forces)
distance-dependent weak attractions and repulsions between transient dipoles.
28
New cards
van der Waals radius
measure of how close an atom will allow another to approach.
29
New cards
Noncovalent interactions are
much weaker than covalent bonds, Continually forming and breaking.
30
New cards
For macromolecules, the most stable structure
usually maximizes weak interactions.
31
New cards
In every living organism
proteins are constructed from a common set of 20 amino acids.
32
New cards
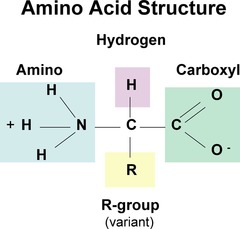
amino acids share common structural features
-α carbon and four substituents - α
carbon is the chiral center
• Tetrahedral
• Exception: Glycine has a second hydrogen atom instead of an R group
carbon is the chiral center
• Tetrahedral
• Exception: Glycine has a second hydrogen atom instead of an R group
33
New cards
Two possible stereoisomers
enantiomers
34
New cards
D and L system specifies
absolute configuration.
35
New cards
L-amino acids are
used by the cells to make proteins. (NH3 on left.)
36
New cards
amino acids form
zwitterions
37
New cards
In neutral pH (pH=7)
both alpha amino and carboxyl group are ionized.
38
New cards
The charged form of the amino acid is called a
zwitterion.
39
New cards
Amino groups, Carboxyl groups, and ionizable R groups =
weak acids and bases.
40
New cards
Zwitterion
occurs at neutral pH.
41
New cards
pKa
analogous to pH and defined by the equation:
pKa = log 1/Ka= −log Ka
pKa = log 1/Ka= −log Ka
42
New cards
The stronger the tendency to dissociate a proton
the stronger the acid and the lower its pKa.
43
New cards
All amino acids have
at least two dissociation constants.
44
New cards
Several Amino acids have an ionizable side chain with a
third pKa
45
New cards
The pH at which the net electric charge is zero is the
isoelectric point (pI)
46
New cards
For amino acids without ionizable side chains, the Isoelectric Point(equivalence point, pI) is:
At this pint, the net charge is zero.
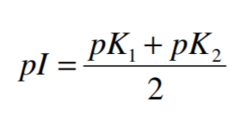
47
New cards
Henderson-Hasselbalch equation
describes the shape of the titration curve of any weak acid.
-Can be used to determine the fraction of ionizable groups found in each of the possible ionization states at a given pH.
-Can be used to determine the fraction of ionizable groups found in each of the possible ionization states at a given pH.
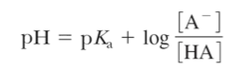
48
New cards
Amino acids can be sorted into four groups on the basis of the general characteristics of their R groups:
- Hydrophobic amino acids
- Polar amino acids
- Positively charged amino acids
- Negatively charged amino acids
- Polar amino acids
- Positively charged amino acids
- Negatively charged amino acids
49
New cards
pKa of amino acids depends on
the neighboring molecules in a protein.
50
New cards
essential amino acids
histidine, isoleucine, leucine, lysine, methionine, phenylalanine, threonine, tryptophan, valine
51
New cards
non-essential amino acids
alanine, arginine, asparagine, aspartic acid, cysteine, glutamic acid, glutamine, glycine, proline, serine, tyrosine
52
New cards
Polar Amino Acids Have Side Chains That Contain an
electronegative atom
53
New cards
Histidine is found at the active sites of many enzymes that require a
proton donor or proton acceptor.
54
New cards
Alpha-amino acid pKa
9.30
55
New cards
Arg pKa
12.48
56
New cards
Lys pKa
10.53
57
New cards
Tyr pKa
10.07
58
New cards
Cys pKa
8.18
59
New cards
His pKa
6.00
60
New cards
Glu
4.25
61
New cards
Asp
3.65
62
New cards
Alpha-Carboxyl
2.10
63
New cards
Amino acid side chains play a pivotal role in
protein folding and protein-protein interaction.
- These side chains may become ionized at different pKa values.
-This has an effect on overall protein structure.
- These side chains may become ionized at different pKa values.
-This has an effect on overall protein structure.
64
New cards
If the side chain does not have an ionizable group
then the pI is simply the average of the α-NH3 and α-COOH pKa values.
65
New cards
If the side chain has an ionizable group then
all three pKa values must be considered.
66
New cards
If the side chain is acidic (asp and glu), then
average the sidechain pKa with the α-COOH pKa
67
New cards
If the side chain is basic (his, arg, and lys), then
average the sidechain pKa with the α-NH3 pKa.
68
New cards
For other ionizable groups (tyr and cys), determine which is the middle pKa
average it with the α-COOH pKa.
69
New cards
When the pH > pI
a protein has a net negative charge.
70
New cards
When the pH < pI,
a protein has a net positive charge.
71
New cards
Proteins can be separated and purified based on
size
- charge
- binding properties
- protein solubility
- charge
- binding properties
- protein solubility
72
New cards
Methods for Purifying Proteins
1. first step = break open tissue or microbial cells. (crude extract= releases proteins in solution.)
2. second step = fractionation = separate proteins into fractions based on size or charge- "salting out" = lower solubility of proteins in salt to selectively precipitate proteins
3.third step = dialysis = use semipermeable membrane to separate proteins from small solutes
2. second step = fractionation = separate proteins into fractions based on size or charge- "salting out" = lower solubility of proteins in salt to selectively precipitate proteins
3.third step = dialysis = use semipermeable membrane to separate proteins from small solutes
73
New cards
column chromatography steps:
first step = buffered solution (mobile phase) migrates through porous solid material (solid phase)
2. second step = buffered solution containing protein migrates through solid phase• protein properties affect migration rates
2. second step = buffered solution containing protein migrates through solid phase• protein properties affect migration rates
74
New cards
Ion-Exchange Chromatography
separates based on sign and magnitude of the net electric charge.
-pH and concentration of free salt ions affect protein affinity
-Recall the relation between pH and pI
-uses bound charged groups:
-- cation exchangers
-- anion exchangers
-pH and concentration of free salt ions affect protein affinity
-Recall the relation between pH and pI
-uses bound charged groups:
-- cation exchangers
-- anion exchangers
75
New cards
pH=pI
no net charge
76
New cards
pH>pI
negative charge
77
New cards
pH
net positive charge
78
New cards
Size-Exclusion Chromatography
--also called gel filtration chromatography --separates based on size
--large proteins emerge from the column before small proteins do
--large proteins emerge from the column before small proteins do
79
New cards
Affinity Chromatography
--separates based on binding affinity
--eluted by high concentration of salt or ligand
--eluted by high concentration of salt or ligand
80
New cards
Amino acid sequence can inform:
- 3D structure
- function
- cellular location
-evolution
- function
- cellular location
-evolution
81
New cards
consensus sequence
reflects most common amino acid at each position
82
New cards
Bioinformatics:
- identifies functional segments in new proteins.
--establishes sequence and structural relationships to known proteins
--establishes sequence and structural relationships to known proteins
83
New cards
Essential amino acid residues =
conserved over evolutionary time.
84
New cards
Less important amino acid residues =
vary over evolutionary time.
85
New cards
Horizontal gene transfer =
transfer of a gene or group of genes from one organism to another.
-- proteins derived from transferred genes are not good candidates for bacterial evolution studies.
-- for example, rapid spread of antibiotic-resistance genes in bacterial populations
-- proteins derived from transferred genes are not good candidates for bacterial evolution studies.
-- for example, rapid spread of antibiotic-resistance genes in bacterial populations
86
New cards
Homologs = homologous proteins =
members of protein families.
--identified by comparing protein sequences to a database of protein sequences.
--identified by comparing protein sequences to a database of protein sequences.
87
New cards
paralogs
homologs in same species
88
New cards
orthologs
homologs in different species.