Lecture #9 | DNA Repair, Mutagenesis, and Human Disease
1/14
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
15 Terms
Which is more dangerous UV A, B or C
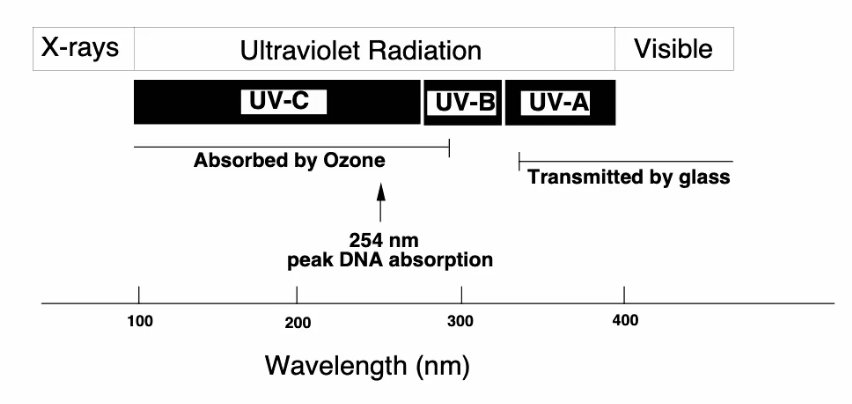
UV C shorter wavelengths are more dangerous
in practicality: UV C is absorbed by Ozone
UV B responsible for sun burns
We are much more exposed to UV A so its more dangerous
What do different types of mutations suggest?
Different mutations are characterizes by different types of lesions and hence have different types of agents
these are signatures in the DNA
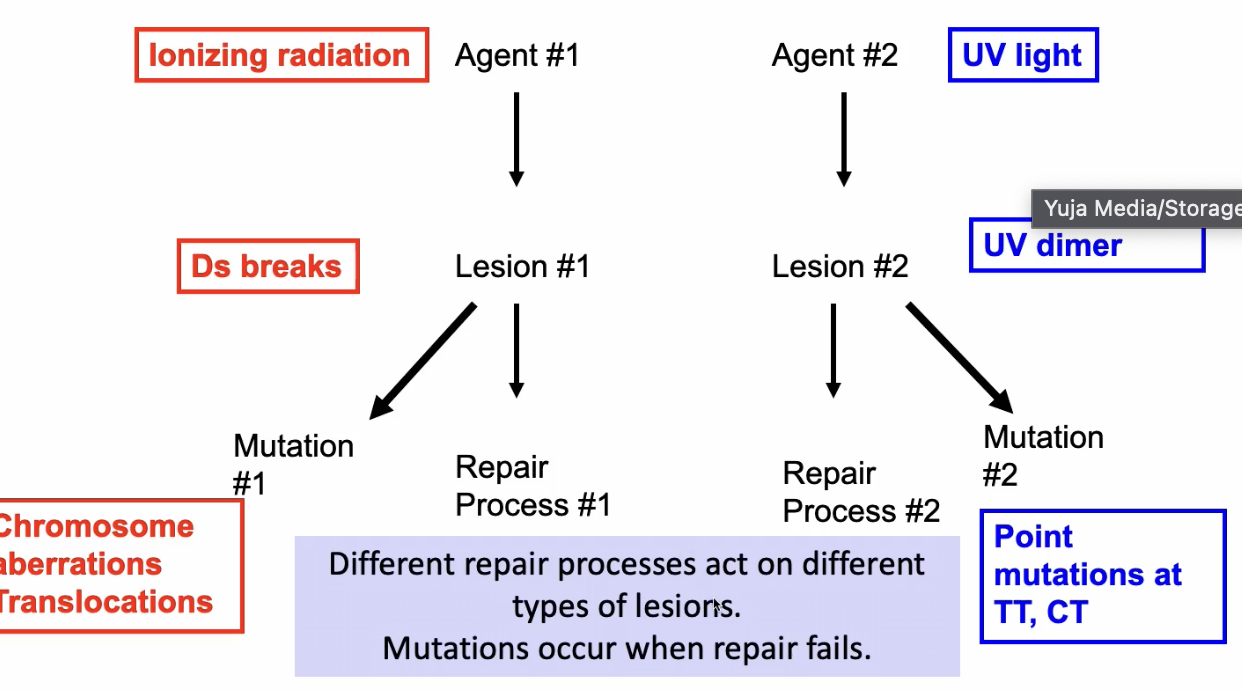
DNA damaging agent determined the lesion, the lesion determined the repair process
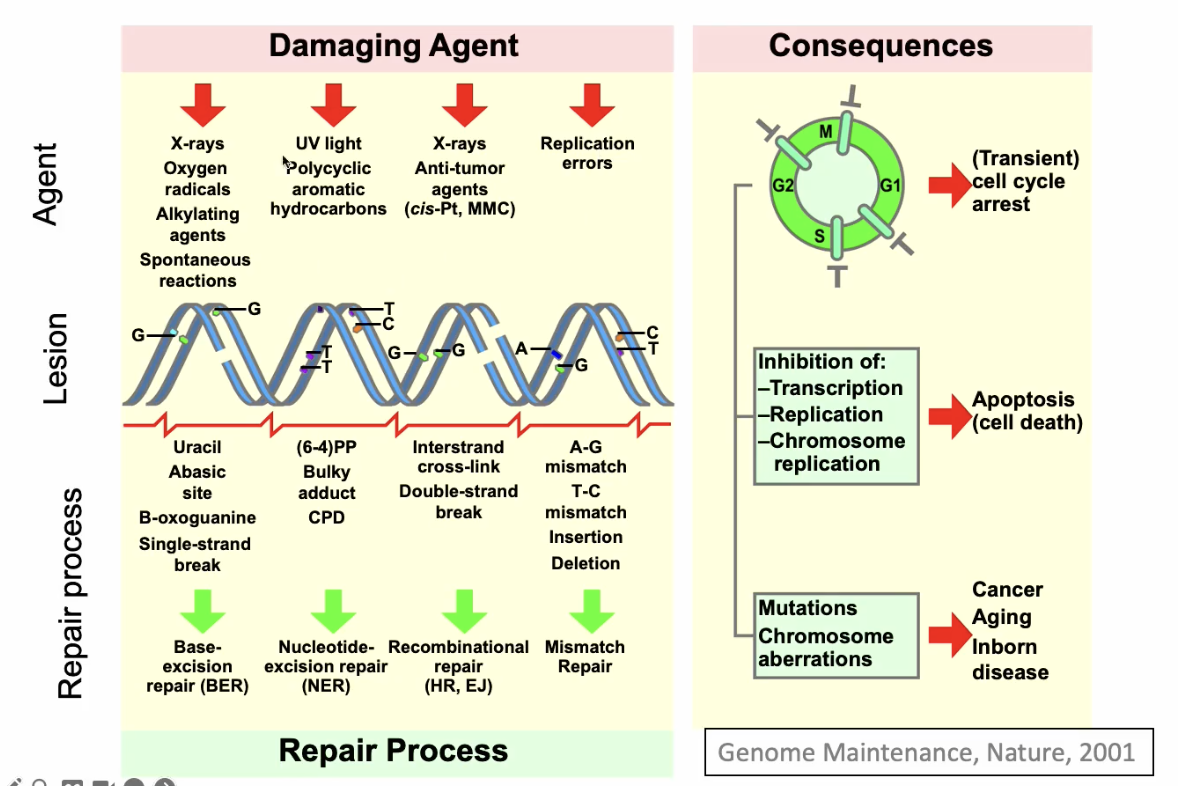
Common features of DNA repair systems
Highly conserved
Often inducible (always present and go into action when needed)
Repair systems are linked to other processes
replication, cell cycle, transcription
Common features of mechanisms
recognition of damage
excision
resynthesis
ligation
Enzymes: polymerase and lygase
#1 Mismatch repairs (MMR)
Lesion: DNA polymerase
Repair enzyme: MutS, MutL
Cancer: colon cancer
DNA Poly is highly efficient in proof reading but still makes some mistakes
if the organism is deficient in proof reading, it will die early from many different types of cancer
Repair: Mismatches, small insertions, deletions
MutS homologs (6 types) bind to site of mismatch through forming heterodimers based on the mismatch
Recruits MutL enzyme which acts as a bridge between the MutS and DNA helicase as it keeps track of the parental strand and they will act together to cut
DNA helicase and exonuclease expand the incision forming a very large gap
Gap repaired by a polymerase and a ligase
#2 Base excision repair (BER)
Lesion: Damaged based through oxidation or alkylation
Repair: Glycosylases, AP endonuclease, lyase
There are many glycosylases which depends on the type of lesions
Cancer: Not clear what cancers are created due to a deficiency in this system
Mechanism:
Improper base if there is deamination
DNA glycosylase removes modified base by cleaving glycosylic bond
And endonuclease classes the phosphodiester backbone
AP lyase removes the sugar phosphate
DNA polymerase fills the 1 base gap and ligase seals the nick
Types of lesions repaired by BER
Spontaneous damage: deamination and depurinaton
Alkylation
Oxidative damage
8-oxo-G
thymine glycol
#3 Nucleotide excision repair (NER)
Lesion: Bulky adducts
Repair enzymes: XP protein including helicase
Cancer: Skin cancer
Note: caffein can cause this distortion
Mechanism:
Bulk adducts forms (UV dimers, BPDE, AFB1 adducts, crosslinks) causes a distortion in the DNA helix
XP-C recognizes the distortion in the double helix
A precision complex is formed
TFIIH complex containing helicases XP- and XP-D melts 30 bp around the damage
XPA confirms the presence of damage by probing for abnormal backbone structure
RPA (replication protein A) stabilizes the open intermediate by binding to the undamaged strand
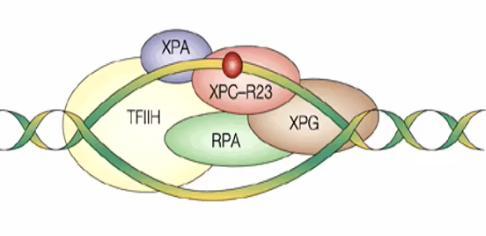
XP-F and XP-G are structure specific endonucleases that make dual incisions in the DNA
With the aid of proliferating cell nuclear antigen (PCNA), 24 to 32 new fragment containing the damage is released
#4 Transcription coupled repair
Lesions: modified bases, bulky adducts
Repair enzymes: CSA, CSB, XP proteins
Disease: Cockayne Syndrome
Basic features of TCR
Preferential repair of actively transcribed regions
Preferential repair of actively transcribed strand
Coupling factors that recognize stalled RNA polymerase
Proteins involved in Bothe DNA repair and transcription
TFIIH
Mechanism of TCR
RNA polymerase II stalls at a lesion in the transcribed strand
Stalled Pol 11 recruits CSB and UVSSA
CSB recruits TFIIH, CSA, XPG (endonuclease) and the repair proteins
#5 Recombination repair: post replication | Double-strand break repair
Homologous recombination (few errors)
Lesion: repair of ds breaks (radiation damage)
Repair enzymes: BRCA 1/2, Rad51, ATM
Cancer/Disease: Breast cancer
Non-homologous end joining (error prone)
Repair enzymes: Ku proteins, DNA-PK
Cancer: various
Recombination repair mechanism for ds break
Many proteins are involved in the initial steps: ATM, BRCA, RPA
Exonucleases chew back the ends of the DNA leaving ss overhangs
Rad51 forms nucleoprotein filaments starting in the ss regions and extending into ds regions
Rad proteins search for homology between the damaged strand and the other chromosomal allele
Strand invasion and elongation by DNA polymerase creating a new homologous strand
The newly synthesized strand crosses back over the original duplex and base pairs with the ds
DNA polymerase uses the newly synthesized strand as template and fills in the gap. Ligase seals the nicks
Non-homologous end-joining
Does not involve recombination
Kinase and KU80 and KU70 recognize ends of DNA and also play a role in VDJ doing in the immune system