MD8 - Anti-metabolites
1/26
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
27 Terms
Main groups of antimetabolites
folate antagonists e.g. methotrexate, pemetrexed and non-classical lipophillic antifolates
pyrimidine antagonists e.g. 5-fluorouracil, fluorodeoxyuridine and azacytidine
purine antagonists e.g. 6-mercaptopurine, thioguanine, tiazofurin
sugar-modified nucleotides e.g. cytarabine, fludarabine and gemcitibine
Folate
Vitamine B9
cannot be synthesised in the body
Folic acid is converted into a folate by the body
used as a dietary substance and is required by the body for the synthesis of DNA and RNA and to metabolise AA
Aminopterin
folate antagonist
Farber realised a folate deficiency helped patients with cancer get better
provided TEMPORARY remission in trials
Dihydrofolate vs Tetrahydrofolate
Dihydrofolate - saturated only one of the double bonds in the aromatic ring
Tetrahydrofolate: saturated all the bonds in the aromatic ring
Folate Cycle
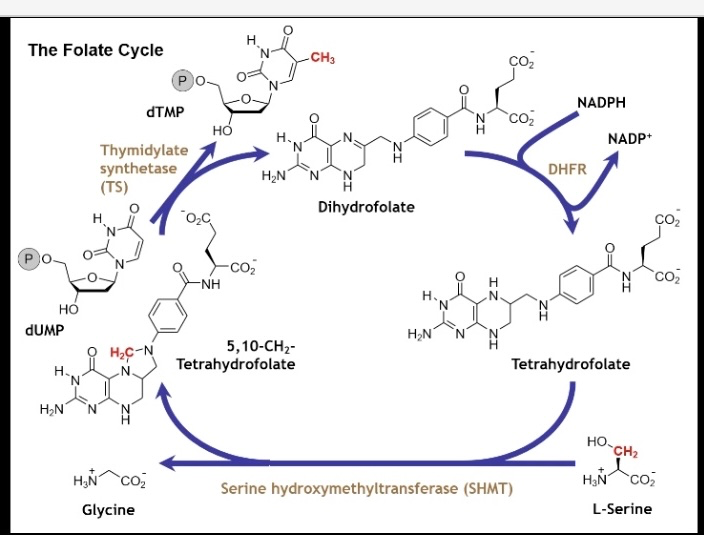
L-Serine is converted to Glycine by Serine hydroxymethyltransferase by removing a CH2 and adds it to tetrahydrofolate
this forms 5,10-CH2- tetrahydrofolate (5-membered ring in the molecule between the two nitrogens and the carbon unit) and a conformational change from this
then 5,10-CH2-tetrahydrofolate is converted to dihydrofolate by thymidylate synthase via conversion of dUMP to dTMP by adding the CH2 from the L-serine to it (5,10-CH2tetrahydrofolate acted as a carrier for it)
we are left with dihydrofolate
to create tetrahydrofolate again it is converted by DHFR (reductase) with NADPH giving a H+ to create this
What are the best targets for this cycle and why?
DHFR and TS because they are the rate-limiting enzymes
What is methotrexate and why is it’s structure important?
Analogue of folate
N in the middle of the molecule is methylated which is very important because in the folate process, the 5,10-CH2 tetrahydrofolate has an N in the middle and it is part of a 5 membered ring. Because of the Me in methotraxate it cannot form that ring so it cannot be converted to something similar to this intermediate
Structural differences between dihydrofolate and methotrexate
methylated N in the middle
unsaturated aromatic ring (only one carbon saturated)
amine replacing the carboxyl in the dihydrofolate structure
Methotrexate as a folate inhibitor
Inhibits DHFR at the folate binding site
very potent competitive inhibitor
too polar for passive diffusion into cell so taken up via reduced folate carrier (RFC)
must be polyglutaylated to be retained in cells
often used in high dose regimen for folate rescue in normal cells and widely used for cancer
Mechnisms of resistance to methotrexate
mutations to DHFR enzyme to modify the folate binding site
MDR (multi-drug resistant) phenotype causing active efflux of drug
mutations to RFC reducing drug uptake
Lipophillic antifolates
don’t need RFC to enter cells - can enter via diffusion
Pyrimethamine
Lipophillic antifolates
inhibits DHFR in many species
used atm as antibacterial
Methylbenzoprim
very potent experimental lipophilic inhibitor of DHFR
Piritrexim
Potent lipophillic inhibitor of DHFR and active in several tumour types
Nolatrexed
Inhibits DHFR and TS and is active against liver carcinoma
Pemetrexed and Raltitrexed
competitive inhibitors of TS and bind to 5,10-CH2-tetrahydrofolate binding site
MOA of TS
TS has a sulfur that attacks the dUMP in a 1,4 addition
TS is now covalently bound to dUMP
TS binds to the 5,10-CH2-THF complex to form a ternary complex (3 components)
TS is released via the loss of a proton on the uracil and a regeneration of a double bond
complex breaks down to give dTMP and DHFR
How does 5FU inhibit TS?
5FU is metabolised to FdURD
FdURD is metabolised to FdUMP
FdUMP binds to the TS and 5,10 CH2 complex but the F is in place of the proton
F is electronegative so cannot be released so TS is stuck and it acts as an inhibition
Azacytidine
weak inhibitor of TS
phosphorylated to form azacytidine TP the incorporated into RNA and mimics C in RNA but it’s unstable so it decomposes and causes damage to RNA
inhibits DNA methyltransferase
Metboli activation of purine analogues
parent compounds are hypoxanthine and guanine
hypoxanthine → 6-mercaptopurine (6-MP) → Thio-IMP (by HPRT) →Thio-GMP
guanine → 6-thioguanine (6-TG) → Thio-GMP (by HPRT) → Thio-GTP by TG-RNA → Thio-dGTP by TG-DNA
HPRT - hypoxanthine phosphoribosyltransferase
Tiazofurine
Experimental drug
converted to TAD
mimics NAD+
Inhibiting the biosynthesis of purine nucleotides
Thio-IMP nd Thio-GMP inhibit by binding at the purine binding site
Thio-IMP also inhibits IMP → AMP
Ara-C
Cytarabine - sugar modified nucleoside
can be converted to triphosphate which inhibits DNA polymerase as an analogue of dCTP
there is some incorporation into the DNA but all it does is make the DNA non-functional
Fludarabine
sugar modified nucleoside
converted to triphosphate
inhibits FNA polymerase as an analogue of dATP
Gemcitabine
sugar modified nucleoside
two fluorines on the sugar
converted to triphosphate very efficiently but also converted to diphosphate
inhibits DNA polymerase as analogue of dCTP but 100x more potent than Ara-C
Metabolism of Gemcitabine
Gemcitabine → F2dCMP by deoxycytidine kinase
f2dCMP → F2dCDP by UMP/CMP kinase
F2dCDP → F2dCTP by NDP kinase
How does Gemcitabine work?
blcoks CTP synthase and ribonucleotide reductase which are needed to convert UTP → CTP and CTP→ dCTP respectively
dCTP is a negative feedback loop inhibitor of dCK so less dCTP means mroe dCK which means more F2dCMP from gemcitabine