Protein Translation in Eukaryotes
1/31
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
32 Terms
What are the five ways by which eukaryotic protein synthesis is more complex than prokaryotic protein synthesis?
The ribosomes are larger (40S+60S=80S)
Protein synthesis begins with methionine. A special initiator tRNA called Met-tRNAi is required.
The initiator codon is always the first AUG from the 5’ end of the mRNA. More protein initiation factors (eIFs) are required.
The structure of mRNA is more complex. There is a step that circularizes the mRNA.
Protein synthesis occurs in the cytosol while RNA synthesis occurs in the nucleus.
What are the two mechanisms of initiation in eukaryotes?
Cap-dependent and cap-independent.
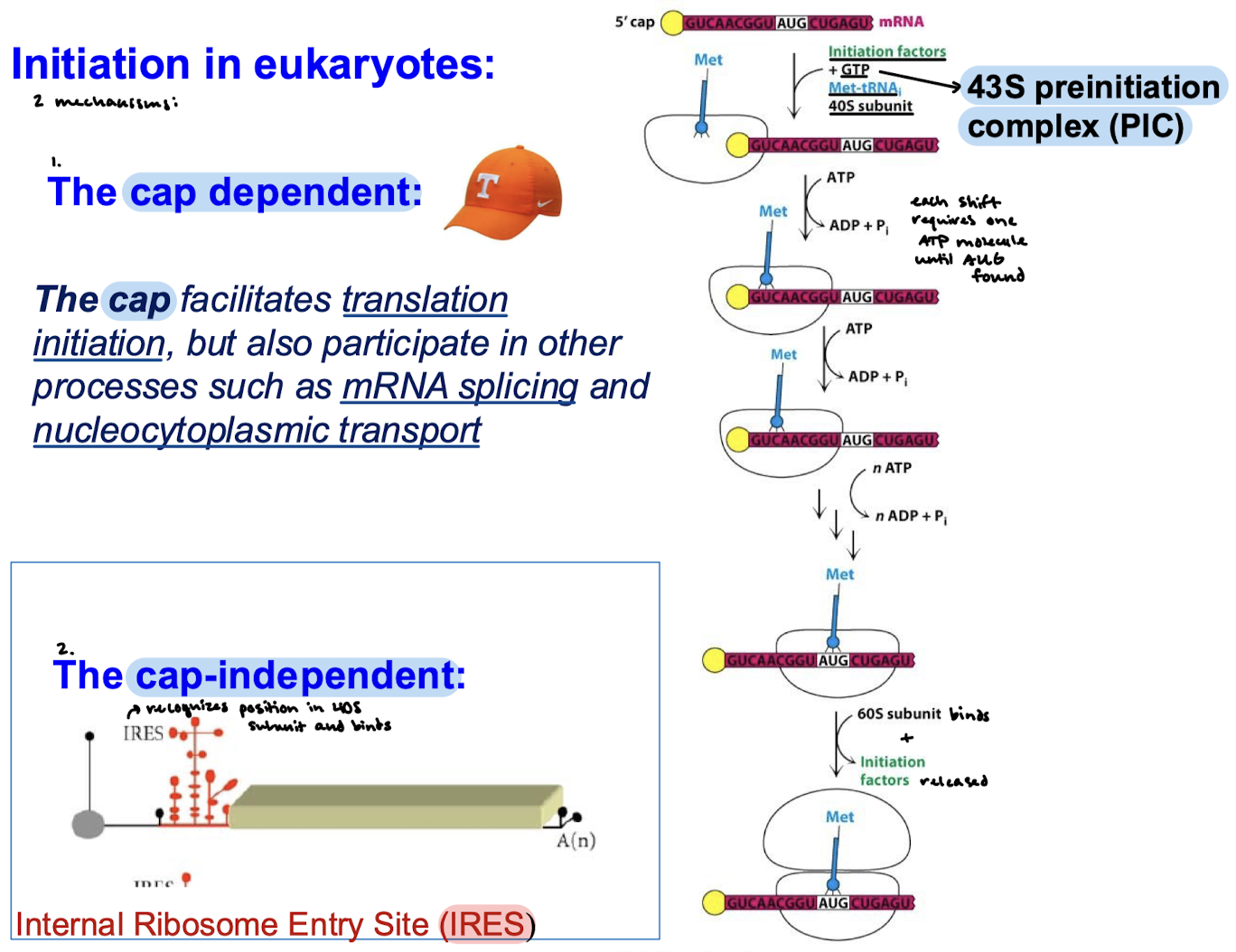
What does cap facilitate (in more than just protein synthesis)?
Translation initiation, mRNA splicing, and nucleocytoplasmic transport.
How does cap-independent initiation work?
Internal ribosome entry sites (IRESs) on mRNA recognize a position in the 40S subunit and bind.
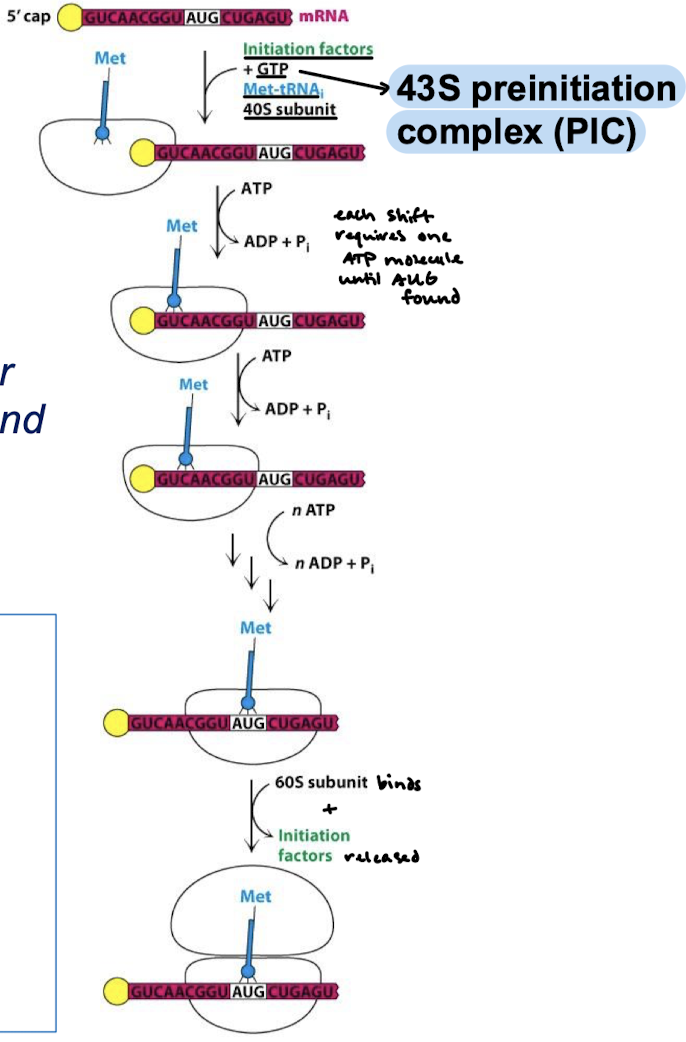
When the small subunit bound with Met, eIFs, and GTP (43S preinitiation complex) is scanning for the start codon, how much ATP is used?
When shift requires one molecule of ATP until AUG is found.
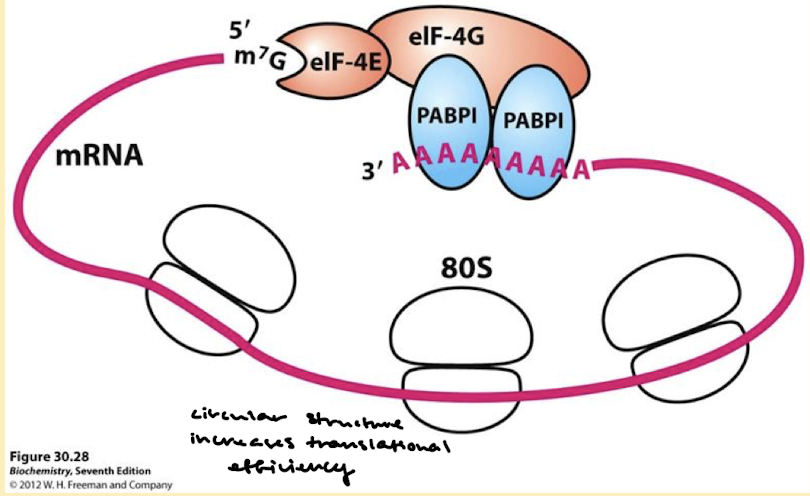
Why is translational efficiency improved by circular mRNA structure?
May facilitate the rebinding of the ribosomes following protein-synthesis termination.
Protect mRNA from exonucleolytic degradation.
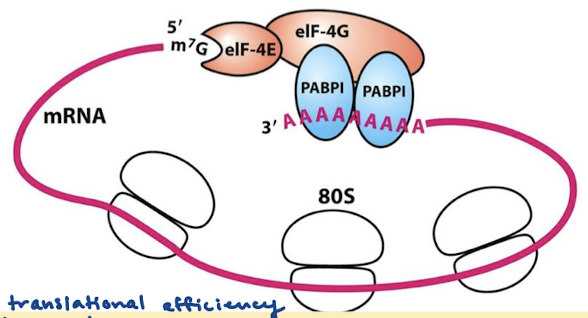
What is the structure of mRNA in cap-dependent initiation? What proteins are involved?
The mRNA is circular because of interactions between proteins that bind the 5’ cap (eIF-4E) and those that bind the pol A tail (PABPI). eIF-4G binds both proteins eIF-4E and PABPI.
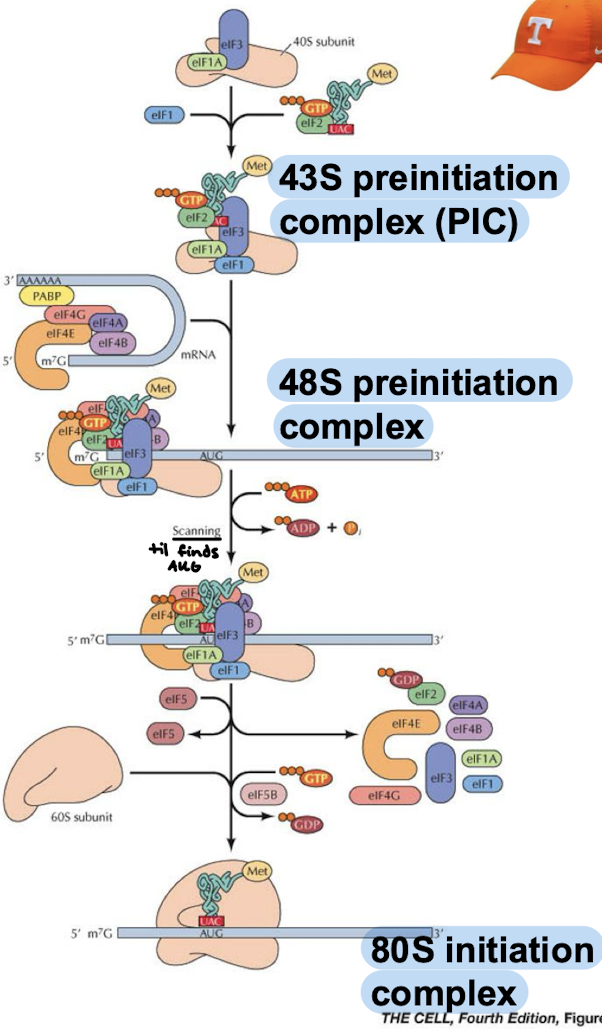
What are the eukaryotic counterparts of EF-Tu and EF-Ts? What are their functions?
EF-Tu = EF1alpha which brings the charged tRNA to the A site.
EF-Ts = EF1beta-gamma which facilitates the exchange of GDP for GTP of EF-Tu.
How is termination in eukaryotes carried out?
Carried out by a single release factor, eRF1.
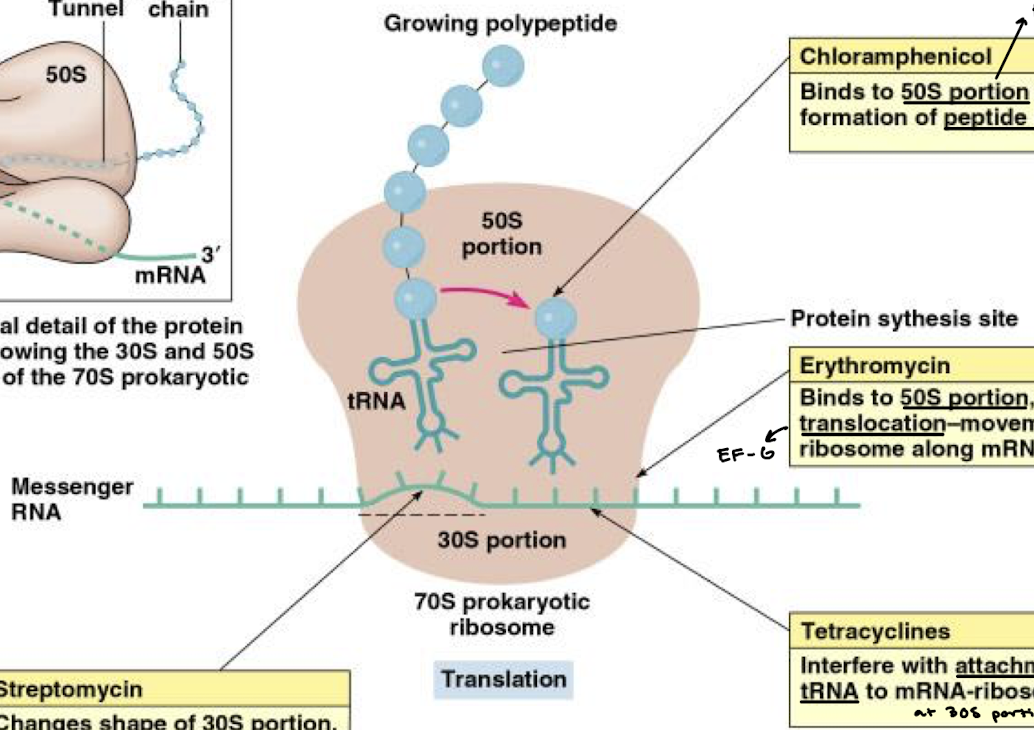
What is the function of antibiotic chloramphenicol in inhibition of bacterial protein synthesis?
Binds to the 50S portion (specifically the PTC) and inhibits formation of the peptide bond.
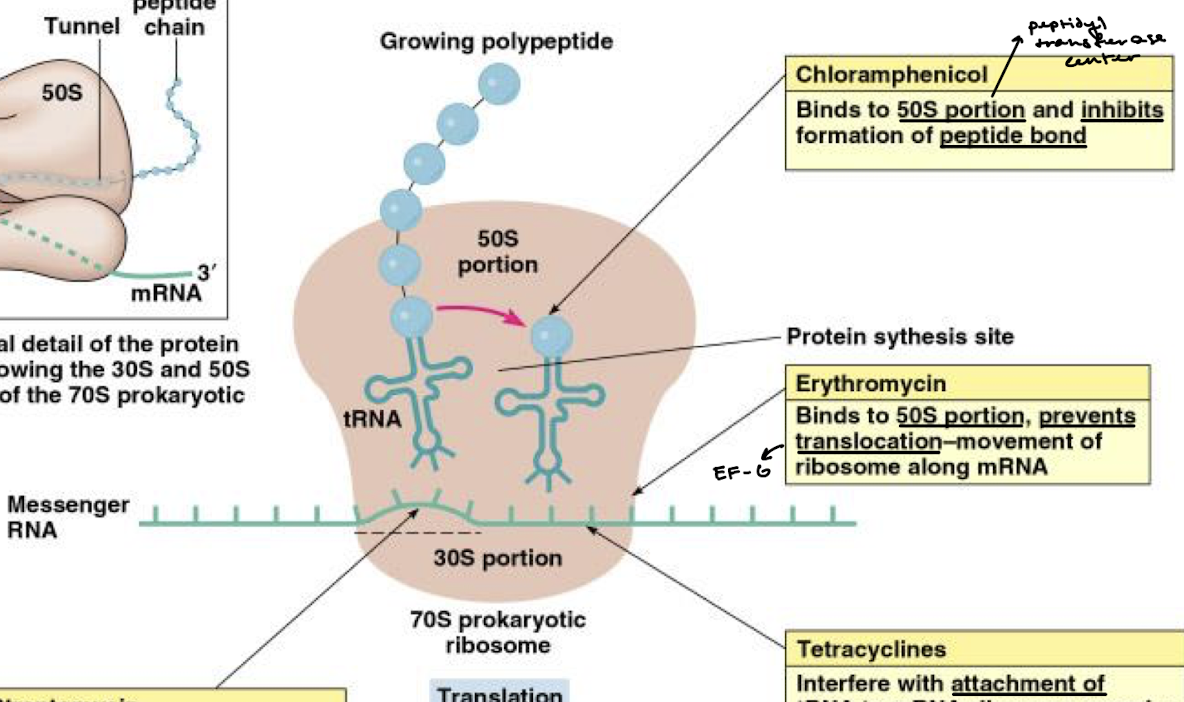
What is the function of erythromycin in inhibition of bacterial protein synthesis?
Binds to the 50S portion of the ribosome and prevents the translocation-movement of the ribosome along the mRNA (think EF-G).
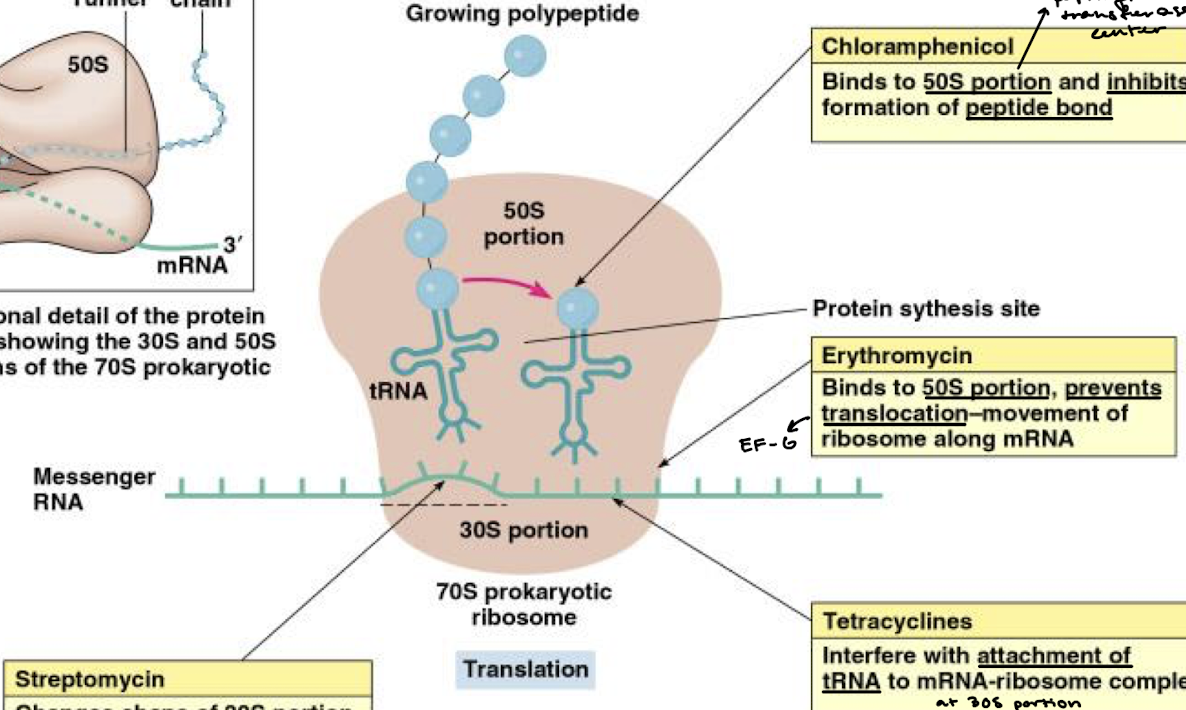
What is the function of tetracyclines in inhibition of bacterial protein synthesis?
They interfere with the attachment of tRNA to the mRNA-ribosome complex at the 30S portion.

What is the function of streptomycin in inhibition of bacterial protein synthesis?
Changes the shape of the 30S portion of the ribosome, causing the code on mRNA to be read incorrectly.
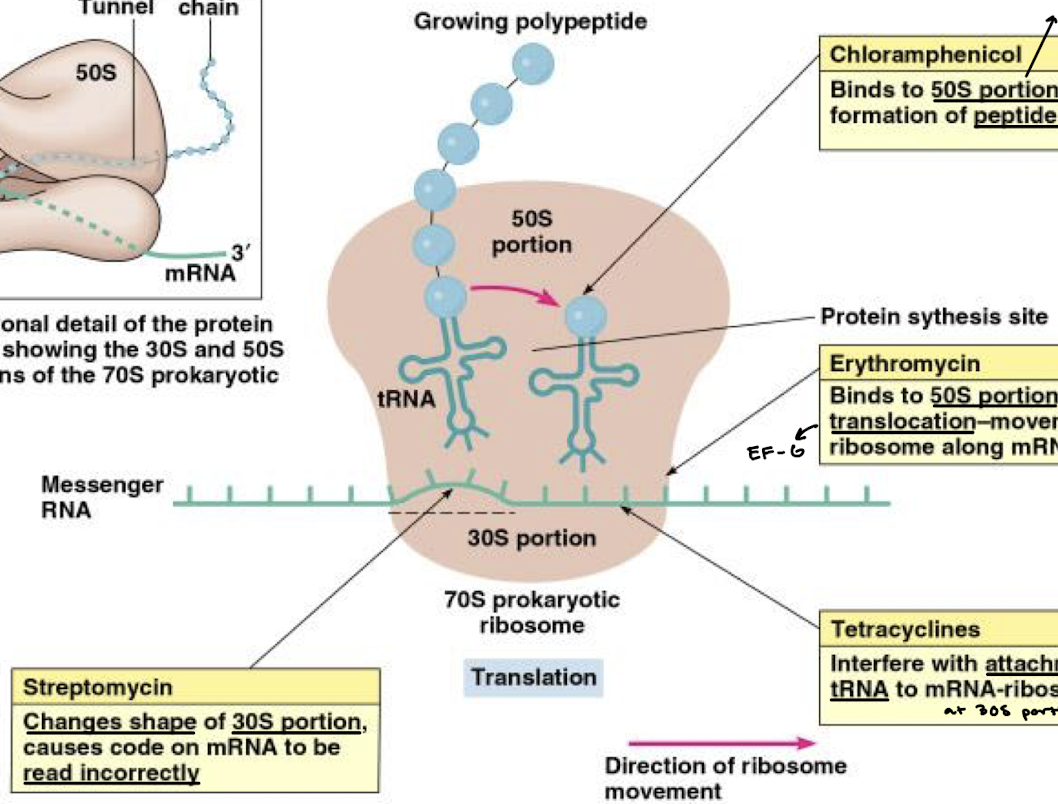
What is the function of cycloheximide in inhibition of protein synthesis?
Inhibits translocation in eukaryotes.
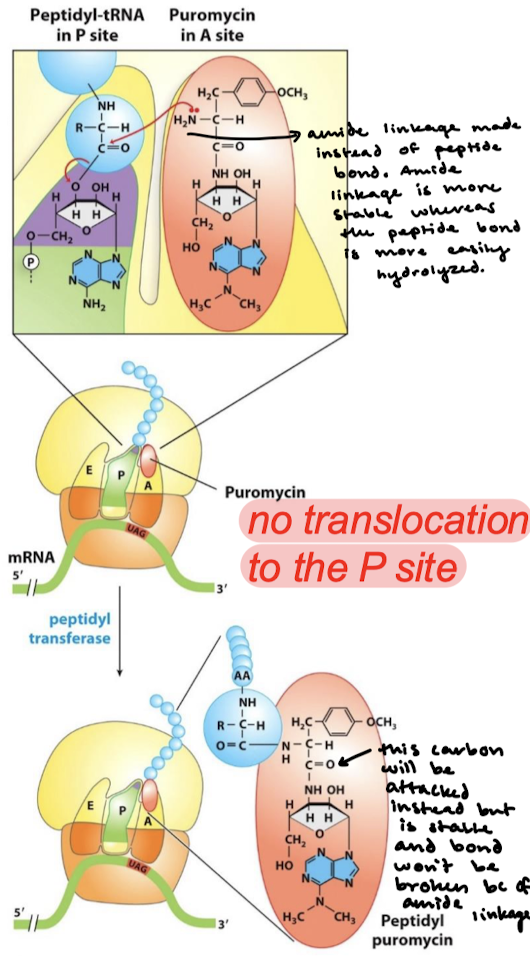
What organisms are affected by puromycin? How does puromycin act?
Either prokaryotes or eukaryotes. Puromycin resembles the aminoacyl terminus of an aminoacyl-tRNA. Its amino group joins the carbonyl group of the growing polypeptide chain to form an adduct that dissociates from the ribosome.
Why is the adduct formed by puromycin stable? Why won’t it be cleaved by the ribosome?
An aminoacyl-tRNA contains an ester linkage. Puromycin contains an amide linkage, which forms an amide linkage with the polypeptide chain instead fo a peptide bond. The amide linkage is more stable whereas the peptide bond is more easily hydrolyzed.
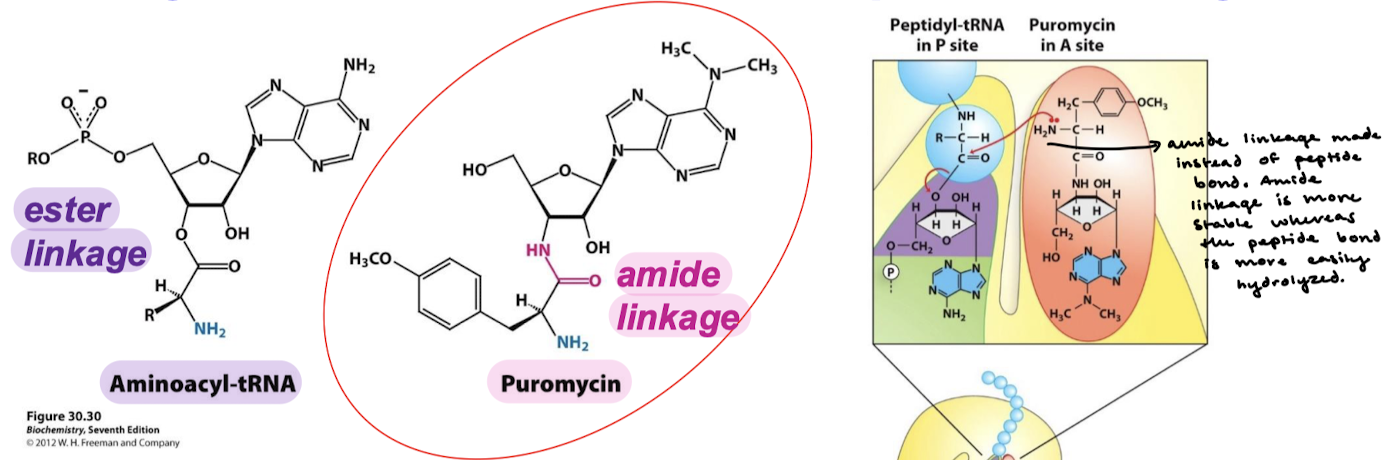
What happens after the amide linkage is formed instead of a peptide linkage when puromycin is acting?
There is no translocation to the P site because the carbonyl carbon that is typically nucleophilically attacked will be stable and the amide linkage will not be broken.
What is the effect of diphtheria toxin on eukaryote protein synthesis?
Diphteria is a respiratory infection and diphteria toxin is extremely lethal. The overall effect is halting of protein synthesis and killing of the cell.
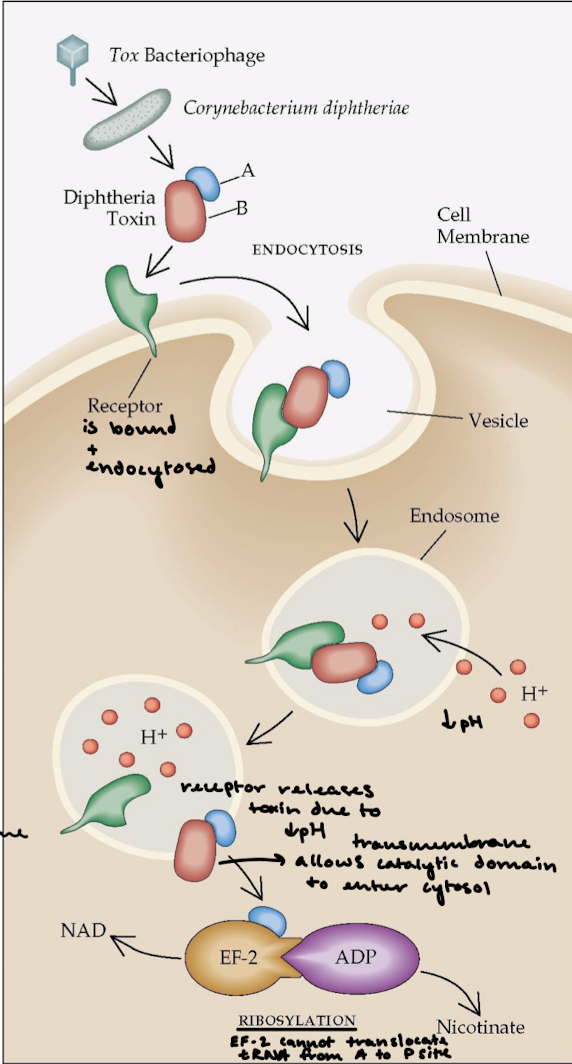
Describe the function of AB toxins.
A fragment = catalytic domain. B fragment = the transmembrane domain that binds the receptor for entrance into cell membrane and for catalytic domain to enter the cytosol.
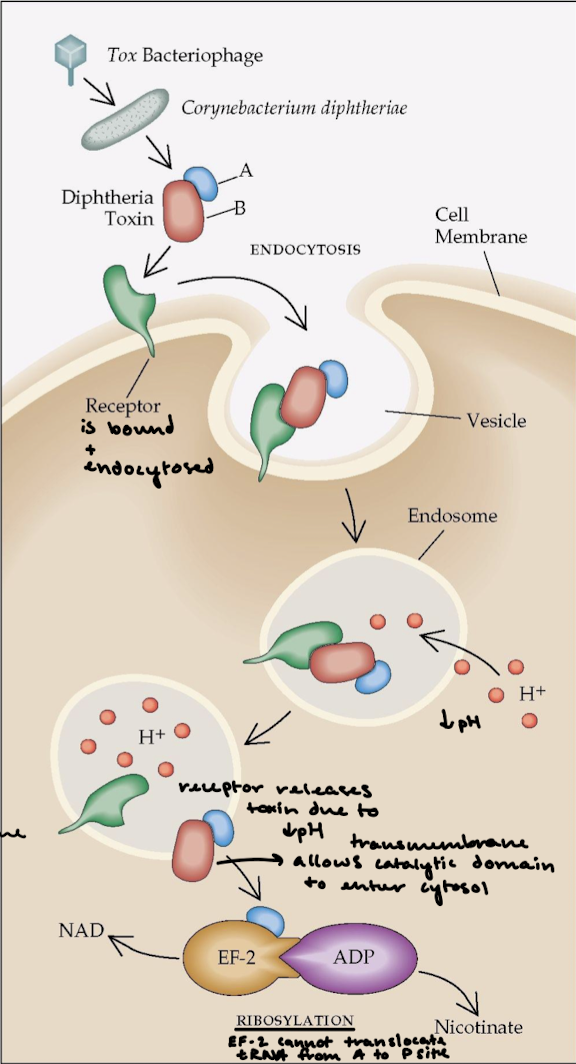
Describe what happens after the lysozome’s pH drops in the mechanism of diphtheria toxin.
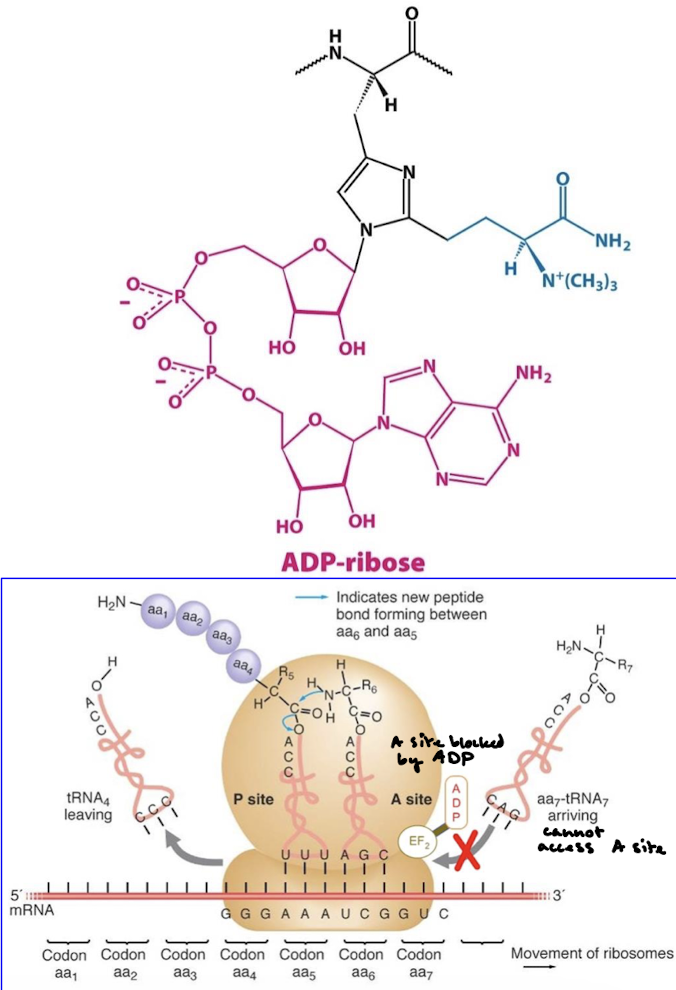
The receptor releases the toxin and the transmembrane domain allows the catalytic domain to enter the cytosol. The A subunit ADP-ribosylates eEF2, which is a translocase like EF-G. This halts protein synthesis and kills the cell.
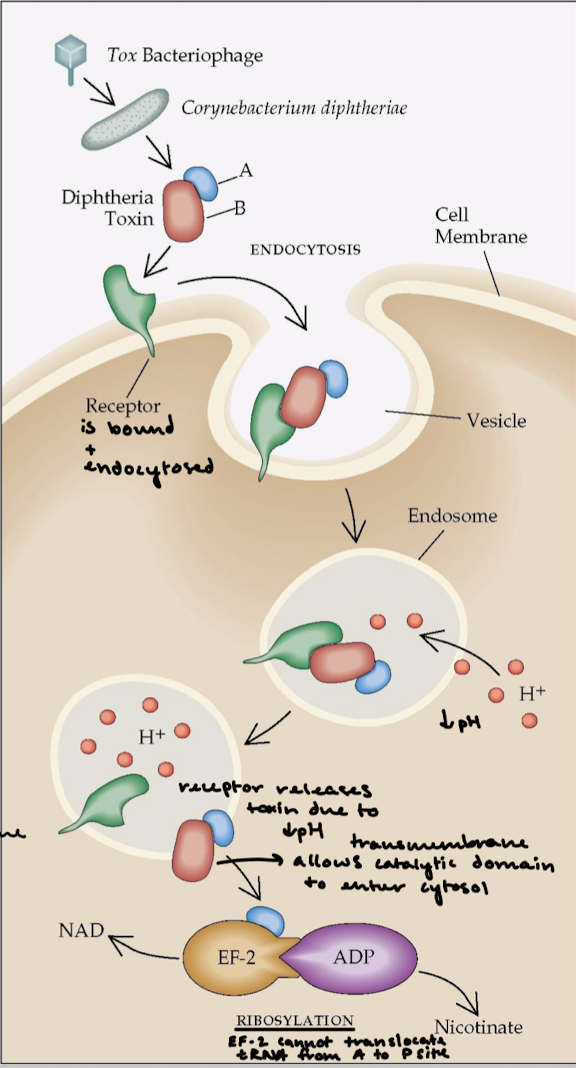
How can a single fragment A of the diphtheria toxin kill a cell?
eEF2 (translocase) contains diphthamide, an unusual amino acid that is formed by the post-translational modification of a histidine residue, which is important for maintaining the correct codon reading frame during translocation. The fragement’s NAD (nicotinamide adenine dinucleotide) glycohydralase activity will cleave ADP-ribose from NAD and covalently attaches ADP-ribose to diphthamide in eEF1. This blocks GTP binding to translocase.
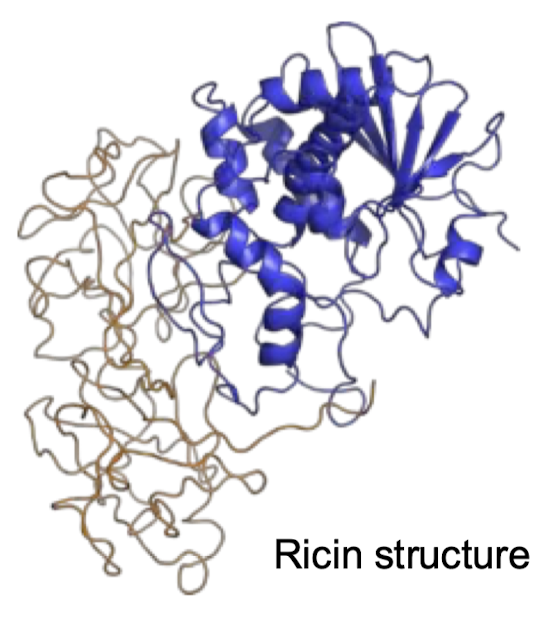
What organisms are affected by Ricin? Where is this toxin found? Describe the structure of Ricin including the domains and what connects them.
Ricin is a small (65 kDA) highly toxic protein found in the seeds of castor oil plant (Ricinus communis) that affects eukaryotes. 500 ug is lethal for an adult human. This is a heterodimeric protein, meaning a catalytic A chain is joined by a single disulfide bond to a B chain.
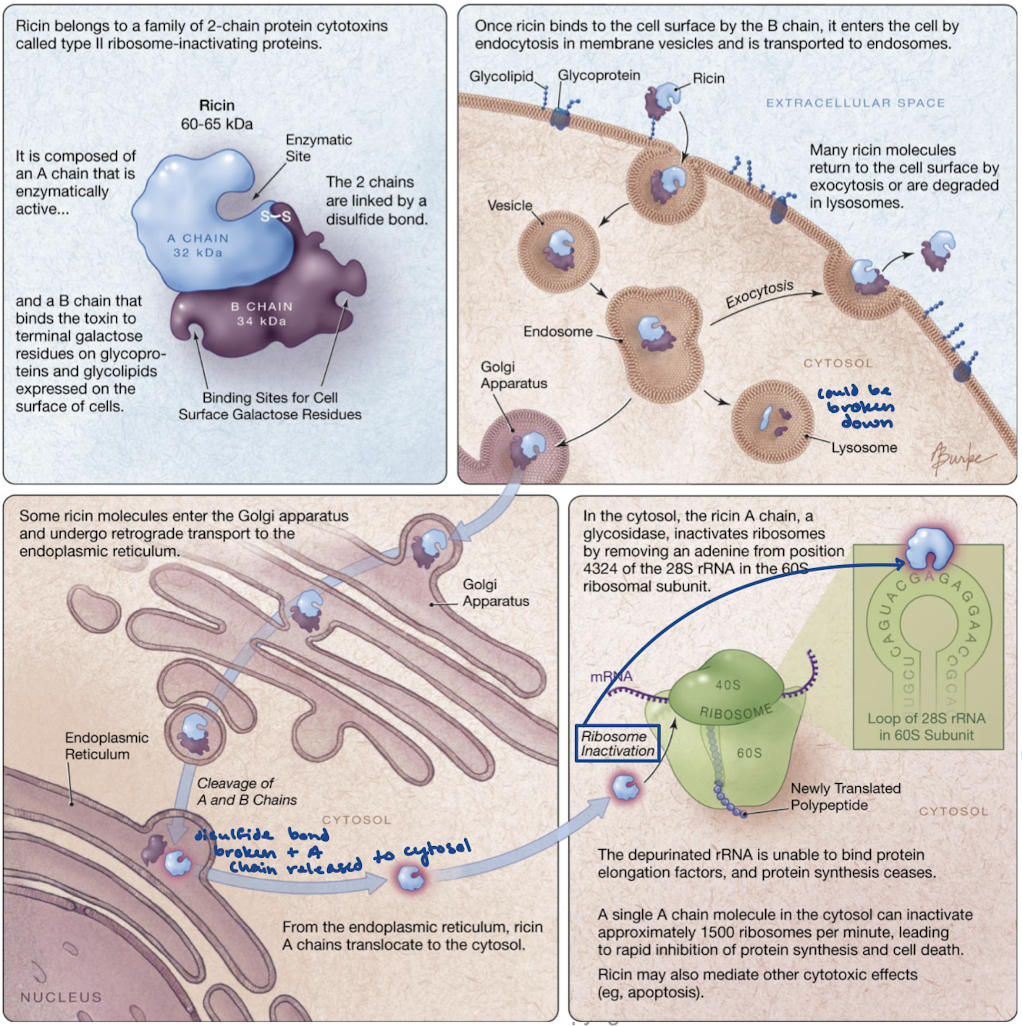
What is Ricin’s mechanism of action?
The B cahin binds the surface receptor and is endocytosed. It is either degraded in the lysosome or brought to the Golgi where it travels to the ER. The ER is where the disulfide bond is broken and the A chain is released to the cytosol. In the cytosol, the A chain cleaves adenine from a nucleotide in 28S rRNA (on the 60S subunit) that is crucial for binding EF-2. This blocks the ribosome from functioning and from protein synthesis.
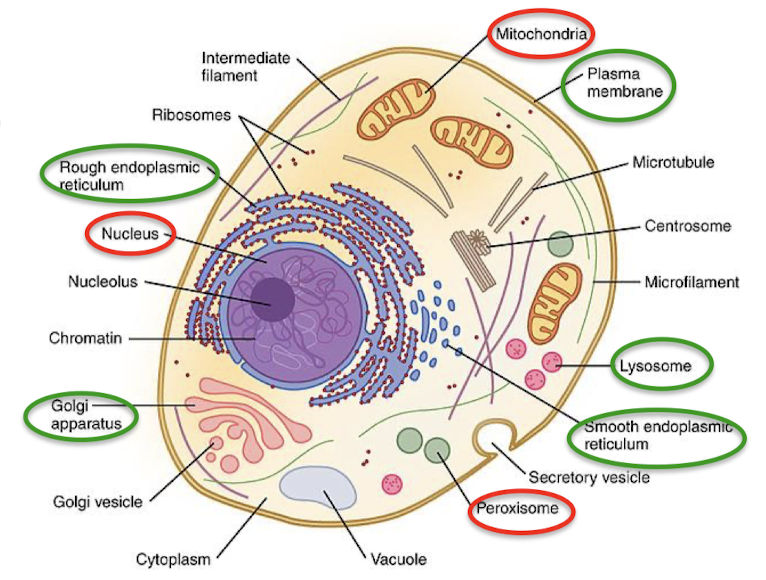
What is protein sorting (protein targeting) in eukaryotes? What are the two general mechanisms?
The process of directing protein to distinct organelles or out of the cell.
Completed proteins are synthesized in the cytoplasm and then delivered to the target intracellular location.
The secretory pathway directs proteins into the ER where they are inserted into the ER membrane co-translationally. Protein synthesis occurs on ribosomes bound to the ER.
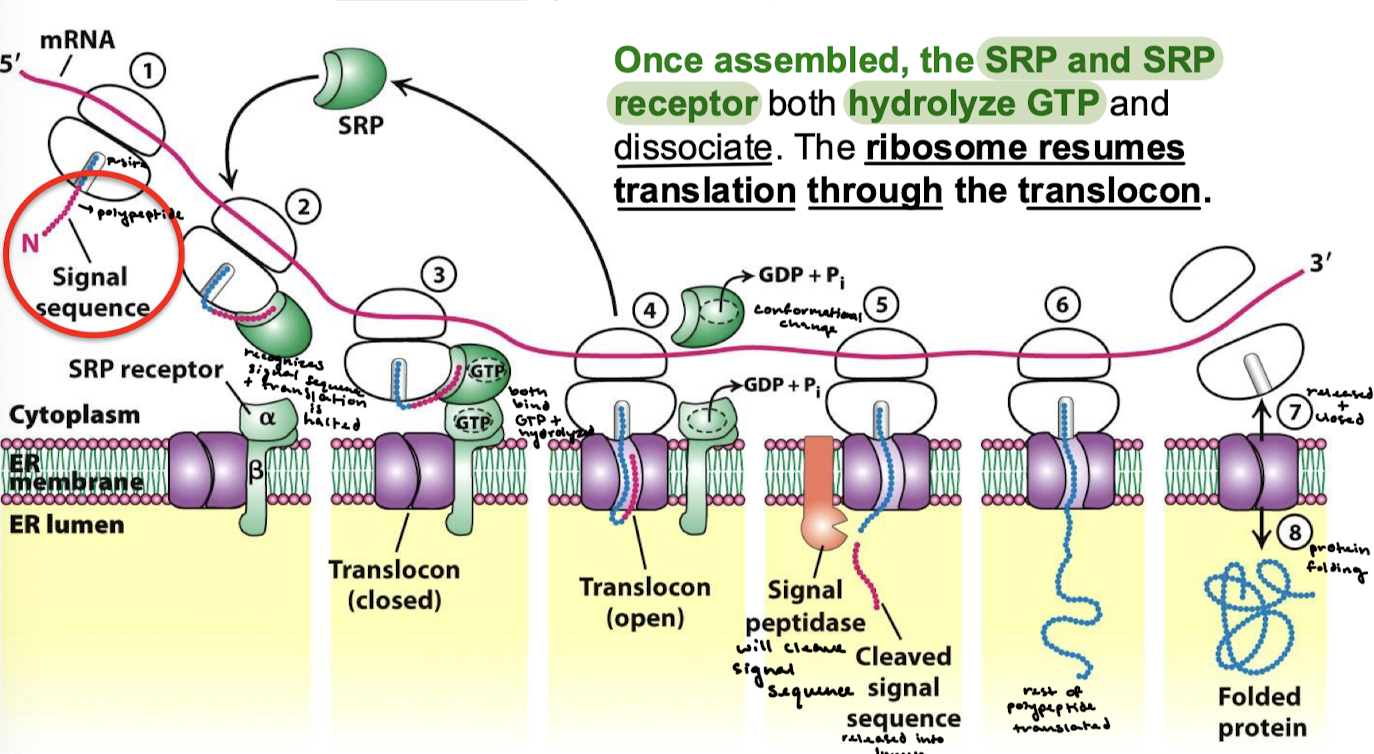
What are the components required for the cotranslational insertion of proteins into the ER? What is the function of each?
The signal sequence: identifies nascent protein as one that must cross the ER membrane.
The signal-recognition particle (SRP): Binds signal sequence as it exits the ribosome and directs complex to the ER.
The SRP receptor, an integral membrane protein with GTPase activity, binds the SRP-ribosome complex.
The translocon, a protein forming a channel for protein translocation, joins the complex.
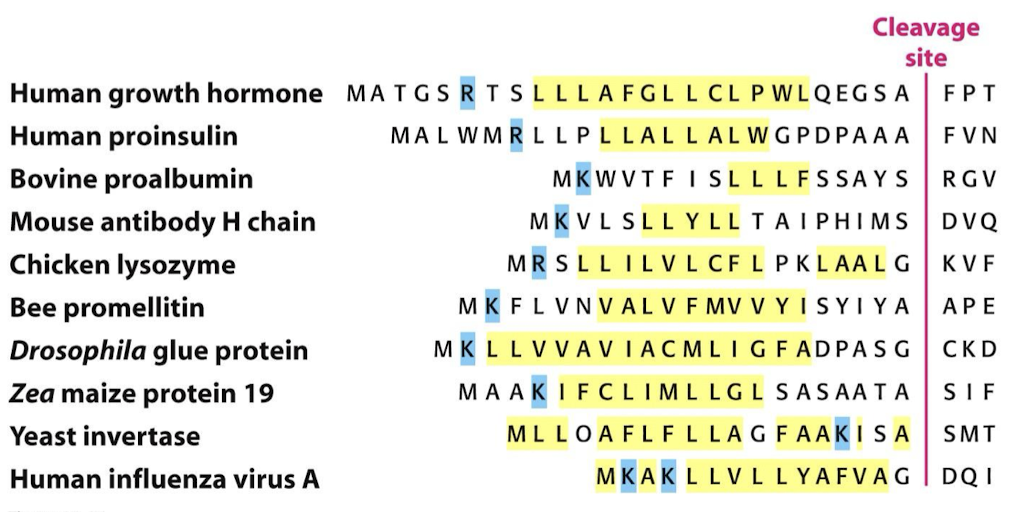
What amino acids make up the signal sequence? What removes the signal sequence?
9-12 hydrophobic amino acids with positively charged amino acids located at the N-terminus. A signal peptidase in the lumen of the ER will remove the signal sequence.
What activity does the SRP have? What does it bind? What happensn after binding?
A GTP-binding ribonucleoprotein with GTPase activity. It binds the signal sequence as it exits the ribosome and directs the complex to the ER. Binding of SRP to ribosome halts protein synthesis.
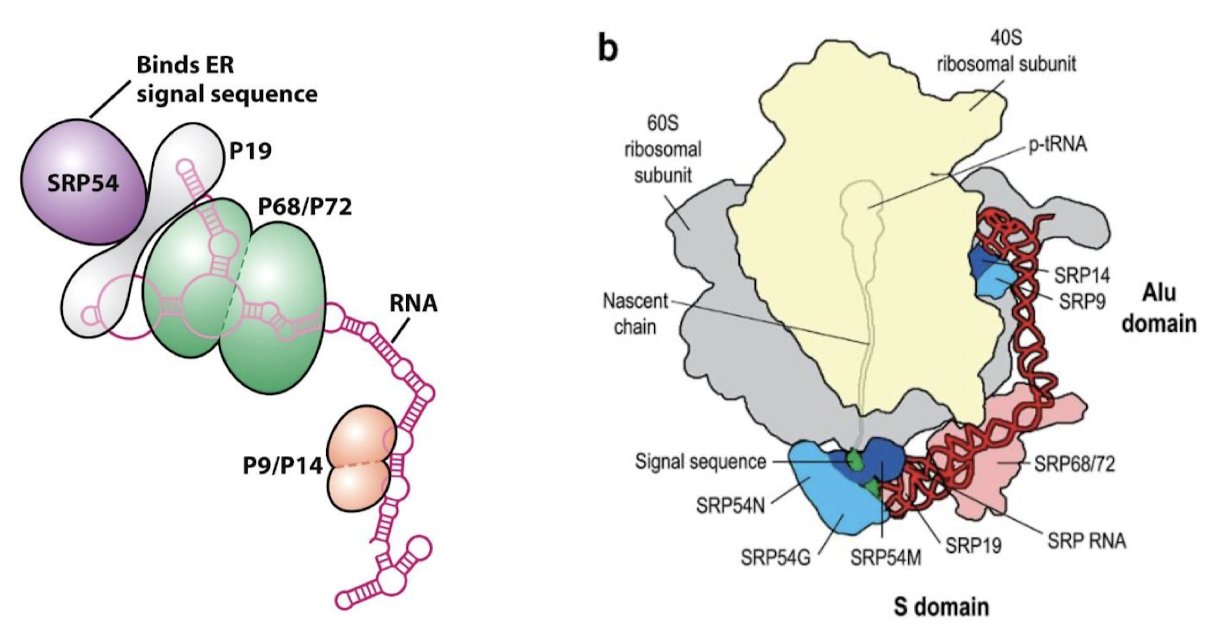
What activity does the SRP receptor have?
GTPase activity
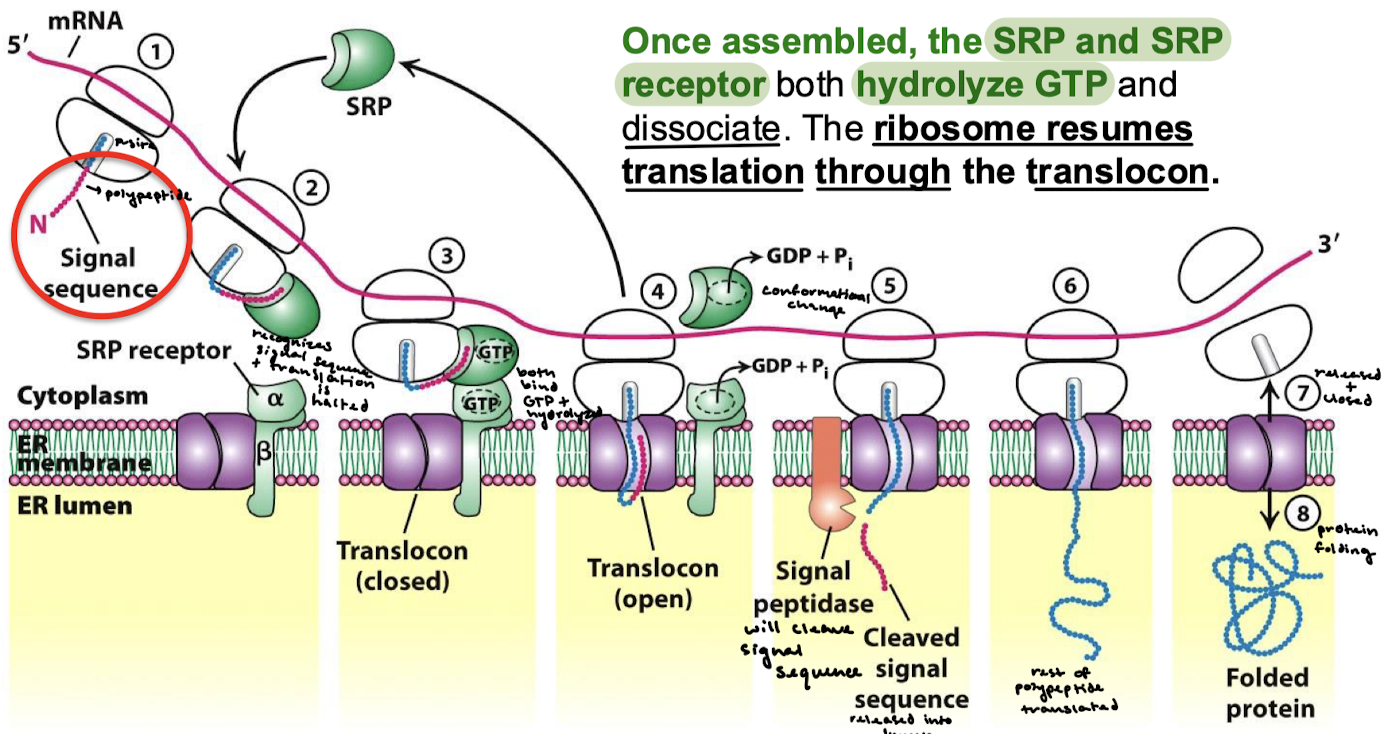
What happens once the SRP targeting complex is assembled?
The SRP and SRP receptor both hydrolyze GTP and dissociate. The ribosome resumes translation through the translocon.
Describe all eight steps for the SRP targeting cycle.
The N-terminal signal sequence is transcribed.
SRP recognizes the signal sequence, and translation is halted.
SRP binds the SRPR. GTP on both SRP and the receptor are hydrolyzed.
There is a conformational change and the translocon is opened.
The signal peptidase cleaves the signal sequence and releases it into the lumen.
The rest of the polypeptide is transcribed.
The ribosome is released, and the translocon closes.
Protein folding.
Both soluble proteins and transmembrane proteins targeted to the secretory pathway are delivered to the rough ER by SRP and undergo co-translational translocation. What are the main differences?
The signal sequence is cleaved in soluble proteins but not in transmembrane proteins.
How do transmembrane proteins get integrated by the translocon?
The first hydrophobic transmembrane domain binds SRP and does not have cleavage sites. Transmembrane domains exit the translocon laterally, and synthesis continues with the protein remaining within the ER membrane.