MBG*2040: The Genetic Code and Translation
1/37
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
38 Terms
gene
The nucleotide sequence of the DNA.
mRNA transcript
The RNA copy of the template DNA strand of the gene.
protein
The purpose of translation: to decode the mRNA and make the functional product of the gene.
protein synthesis in prokaryotes
Transcription, translation and mRNA degradation often occur simultaneously - can happen synchronously because there is no nuclear envelope separating the processes.
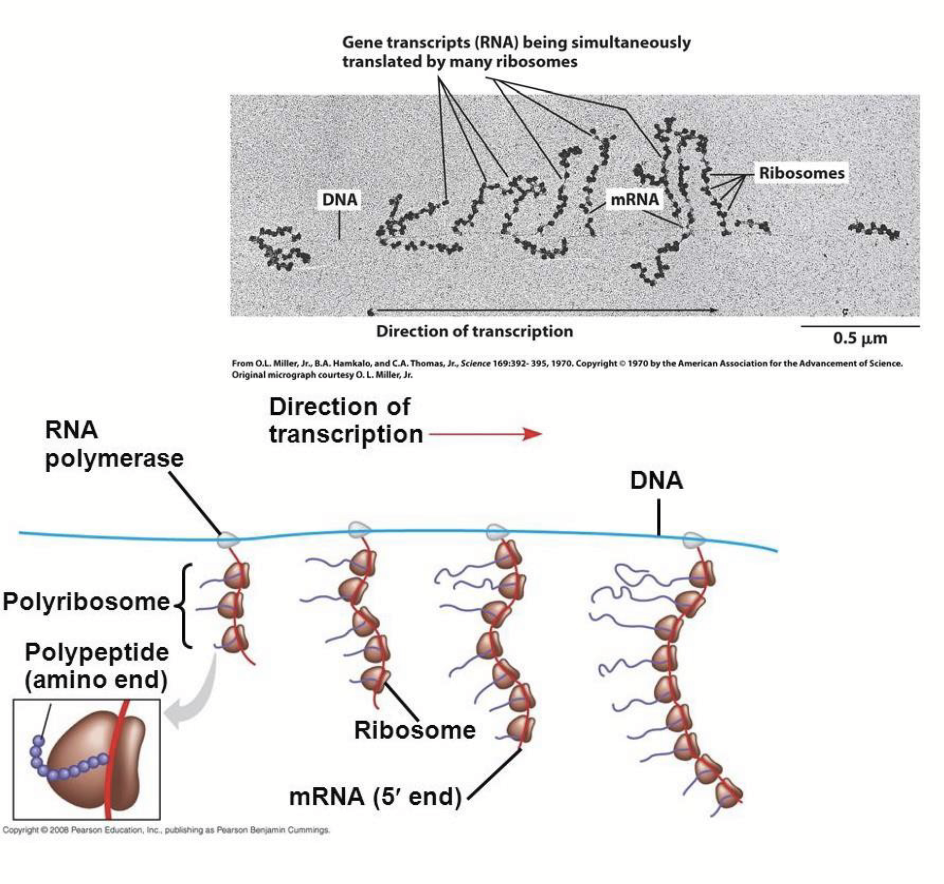
protein synthesis in eukaryotes
Transcription occurs in the nucleus and the mRNA translation occurs in the cytoplasm. Because of this, mRNA generally lasts longer.
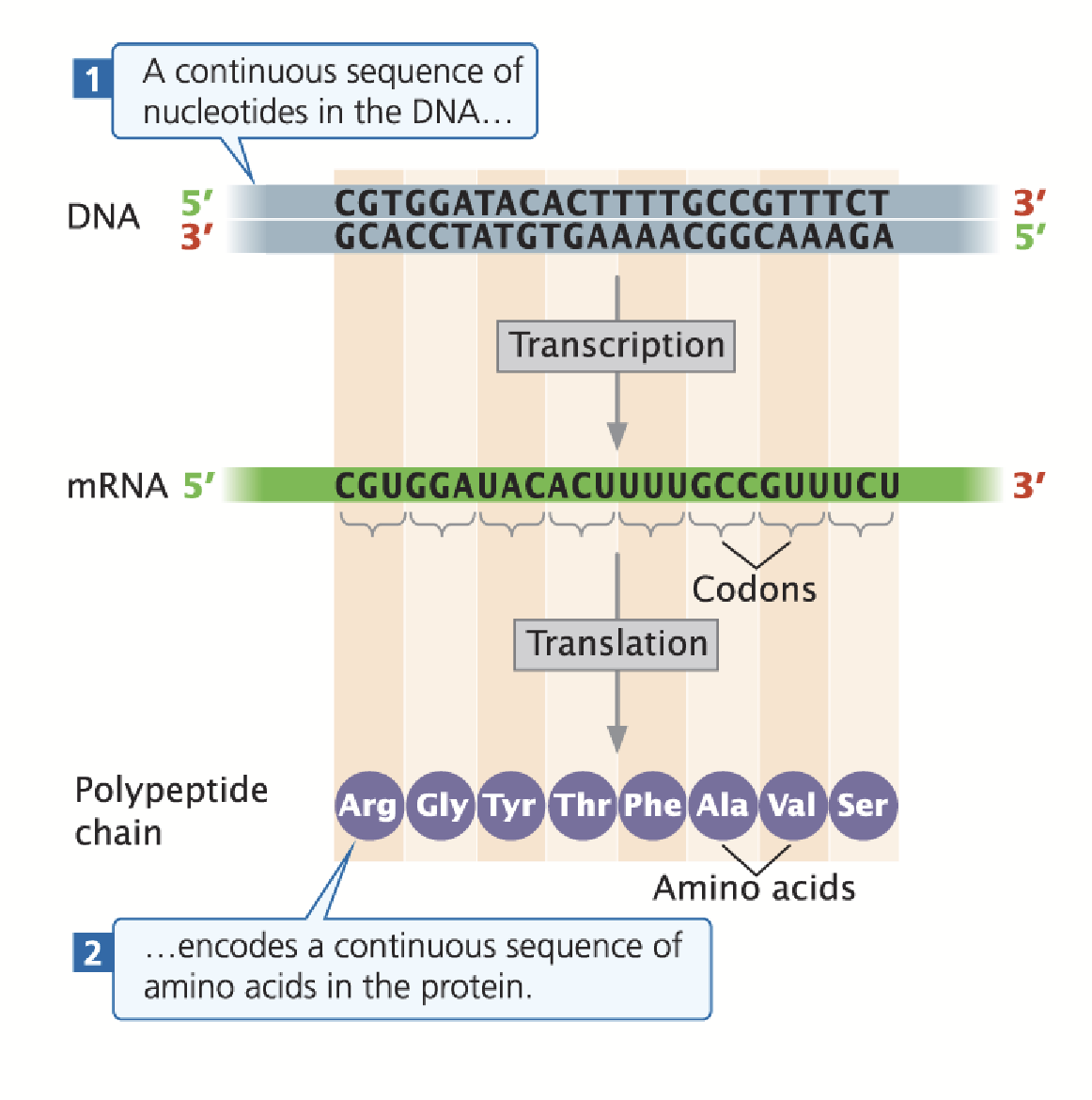
one gene, one colinear polypeptide
In prokaryotes, the sequence of base pair triplets in the coding region of a gene specify a colinear sequence of amino acids in its polypeptide product.
Beadle and Tatum
In the 1930s, through genetic analysis of nutritional mutants in the fungus Neurospora, these researchers discovered that one gene encoded one discrete polypeptide.
Charles Yanofsky and colleagues
In the 1960s, through mutational/biochemical analysis, they discovered that the sequence of nucleotide triplets in the trpA gene of E. coli corresponded to the sequence of amino acids in the TrpA protein.
We can predict what amino acids are in the protein!
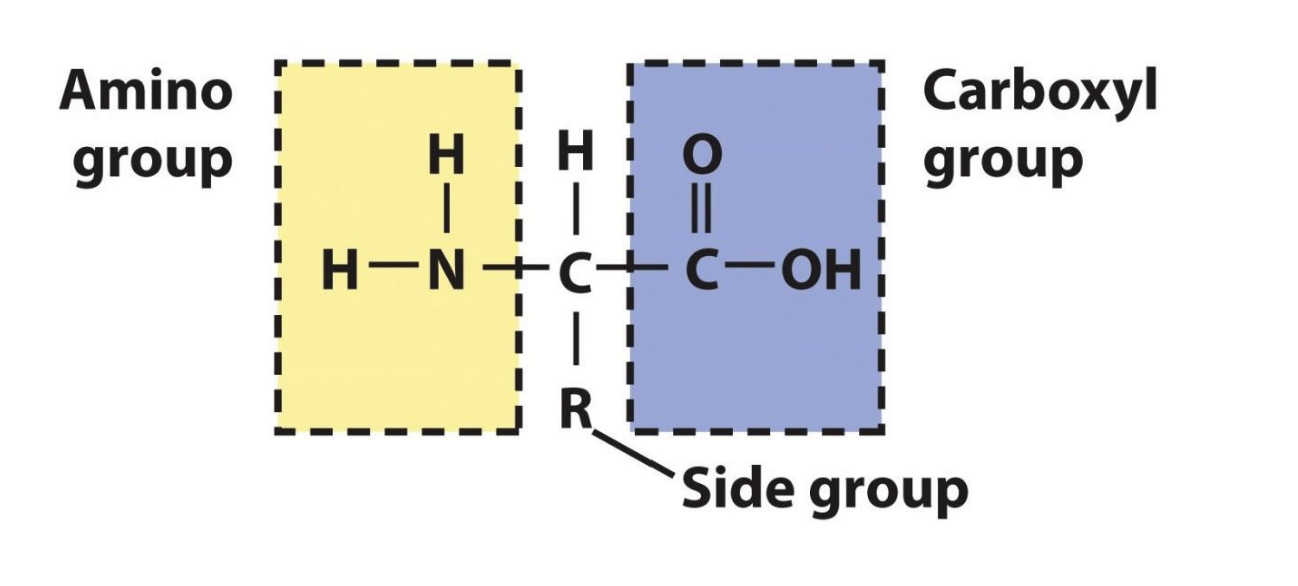
amino acids
Proteins are made of polypeptides; a polypeptide is a long chain of monomers, which have a free amino group (N-terminal), a free carboxyl group (C-terminal), and a side group (R).
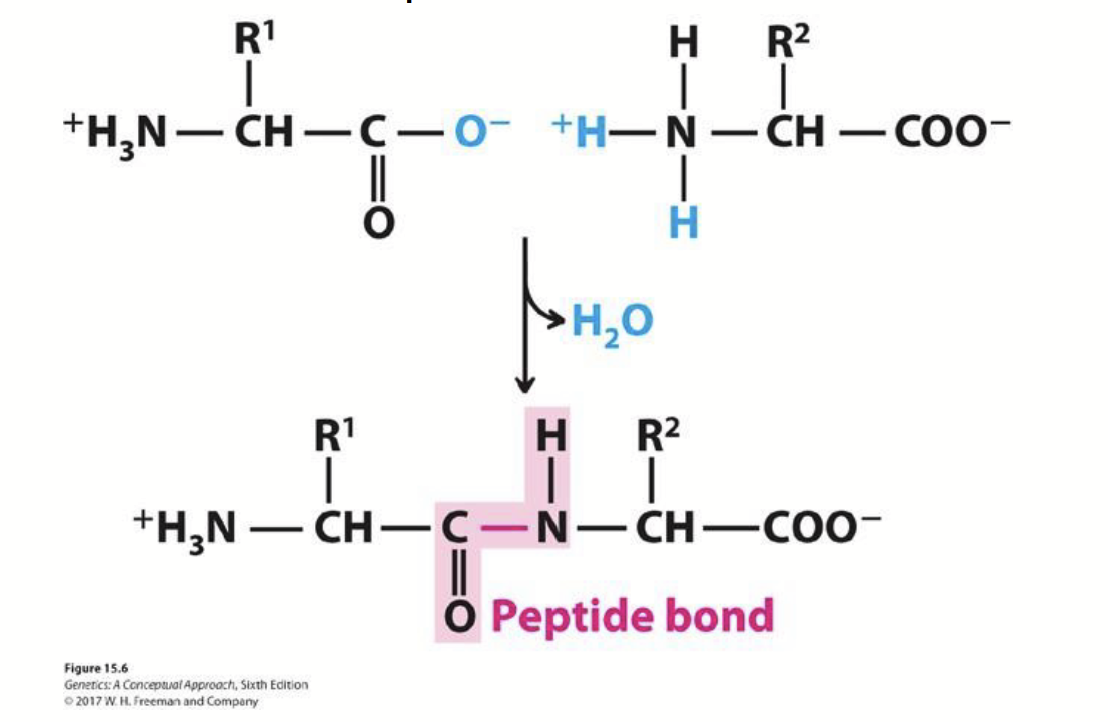
peptide bonds
How amino acids are joined. The carboxyl group of one amino acid is covalently attached to the amino group of the next amino acid with the expulsion of H2O.
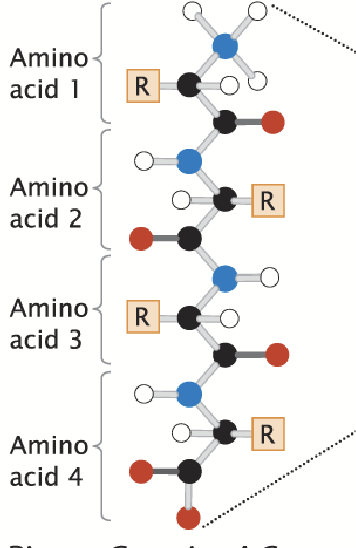
primary structure
The linear arrangement of amino acids; the sequence of amino acids in a protein.
The presence of the carboxyl (C terminus) and amino group (N terminus).
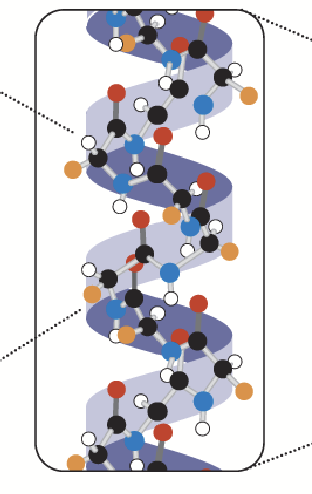
secondary structure
Determined by the spatial organization of amino acids; interactions between amino acids cause the primary structure to fold (e.g. alpha helix, beta sheets). Mostly determined by hydrogen bonding orientation between groups.
tertiary structure
Determined by the overall folding of the complete polypeptide; the secondary structure folds further (e.g. folded around alpha helix). Have combinations of secondary structures.
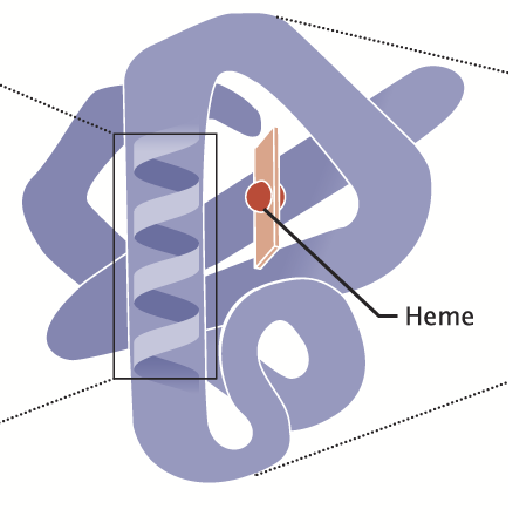
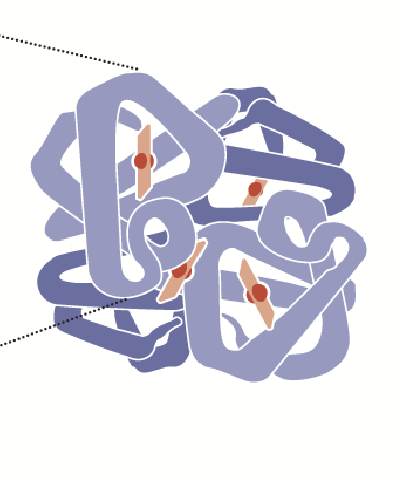
quaternary structure
In some proteins, more than one polypeptide interact to make a functional protein; two or more polypeptide chains may associate to create this structure.
what is the genetic code?
How many nucleotides are necessary to specify a single amino acid? Degenerate, some amino acids are specified by more than one codon.
Singlet code → since only 4 bases, only 4 codons specified… not enough for 20 amino acids.
Doublet code → would specify only 42=16 possible codons… not enough for 20 amino acids.
Triplet code → would specify 43=64 possible codons… sufficient for synthesis of the 20 amino acids if some amino acids were specified by more than one codon (which they are).
the genetic code
Composed of nucleotide triplets: 61 specify amino acids while 3 specify stop codons.
Degenerate: some amino acids are specified by more than one codon.
Comma-free.
Contains start and stop codons (non-sense).
Nearly universal!
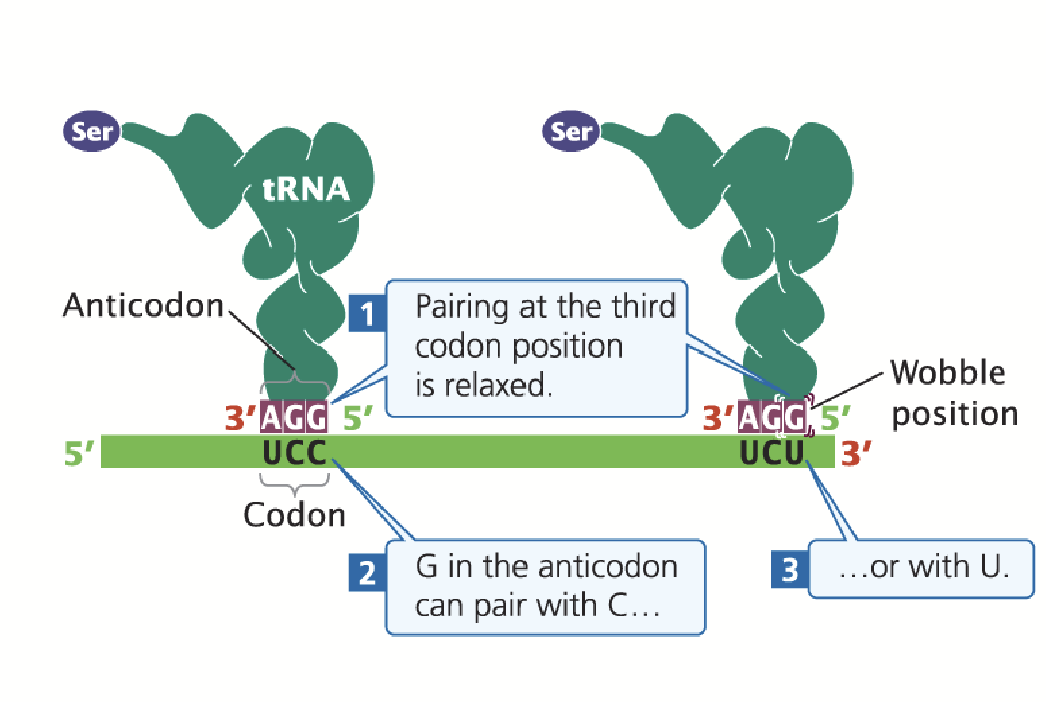
Wobble position
Base-pairing between mRNA codons and aminoacyl tRNAs is “anti-parallel”, but there is flexibility in binding at the third codon position (first anti-codon position).
Permits the tRNA anti-codon to bind to multiple codons in the mRNA.
Oftentimes, the base in the third codon position can be changed and still specify the same amino acid. This feature of the genetic code explains degeneracy.
tRNA modifications
tRNAs can be post-transcriptionally modified at the nucleotide level. Nucleotides can be substituted for different nucleotide derivatives.
e.g. Inosine (a rare base) is an adenine/guanine derivative, it can be produced via deamination (getting rid of an amino group on a base). It can base pair with either C, U, or A, giving the tRNA a lot more flexibility in terms of pairing partners.
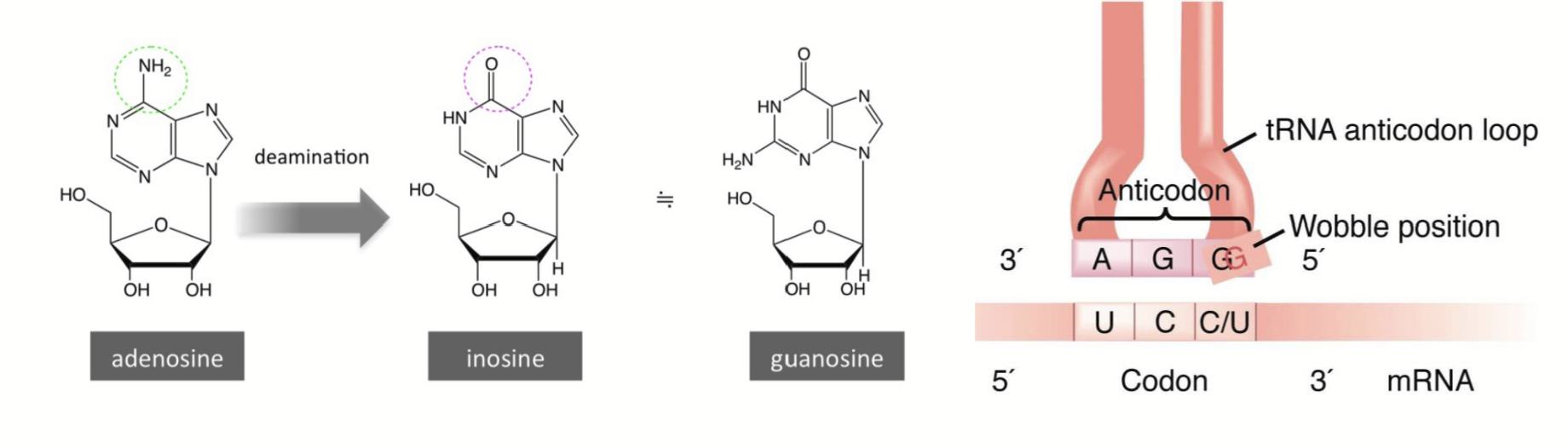
The Wobble Hypothesis
Nonstandard pairings could take place at the third position of a codon.
the macromolecules of translation
Ribosomes are made up of many proteins (>50) and ribosomal RNA (rRNA) molecules (3-4)
Amino acid activating enzymes (20)
tRNA molecules (40-60)
Soluble proteins (translation factors) involved in polypeptide chain initiation, elongation, and termination
ribosomes
Composed of proteins and several different rRNAs; composed of both a large and a small subunit that assemble. An “RNA machine” with key roles in protein synthesis, including the formation of peptide bonds between amino acids.
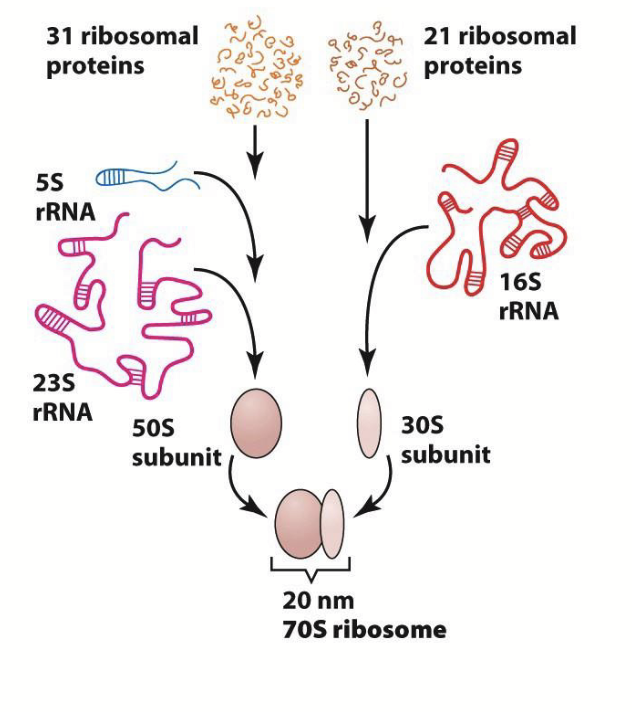
prokaryotic ribosome
31 ribosomal proteins and 21 ribosomal proteins.
31 ribosomal proteins + 5S rRNA + 23S rRNA = 50S subunit.
21 ribosomal proteins + 16S rRNA = 30S subunit.
50S subunit + 30S subunit = 20 nm 70S ribosome.
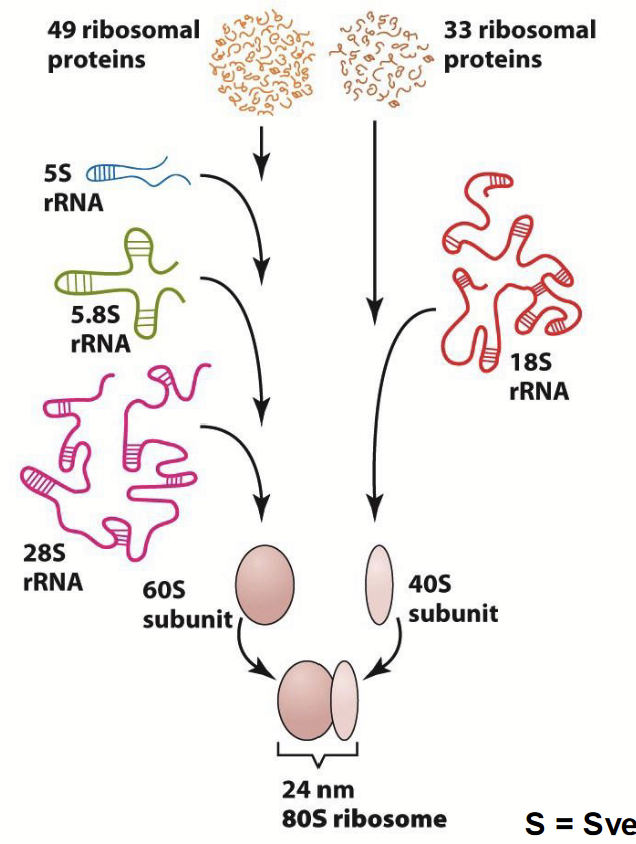
eukaryotic (mammalian) ribosome
49 ribosomal proteins and 33 ribosomal proteins.
49 ribosomal proteins + 5S rRNA + 5.8S rRNA + 28S rRNA = 60S subunit.
33 ribosomal proteins + 18S rRNA = 40S subunit.
60S subunit + 40S subunit = 24 nm 80S ribosome.
Svedberg units
S
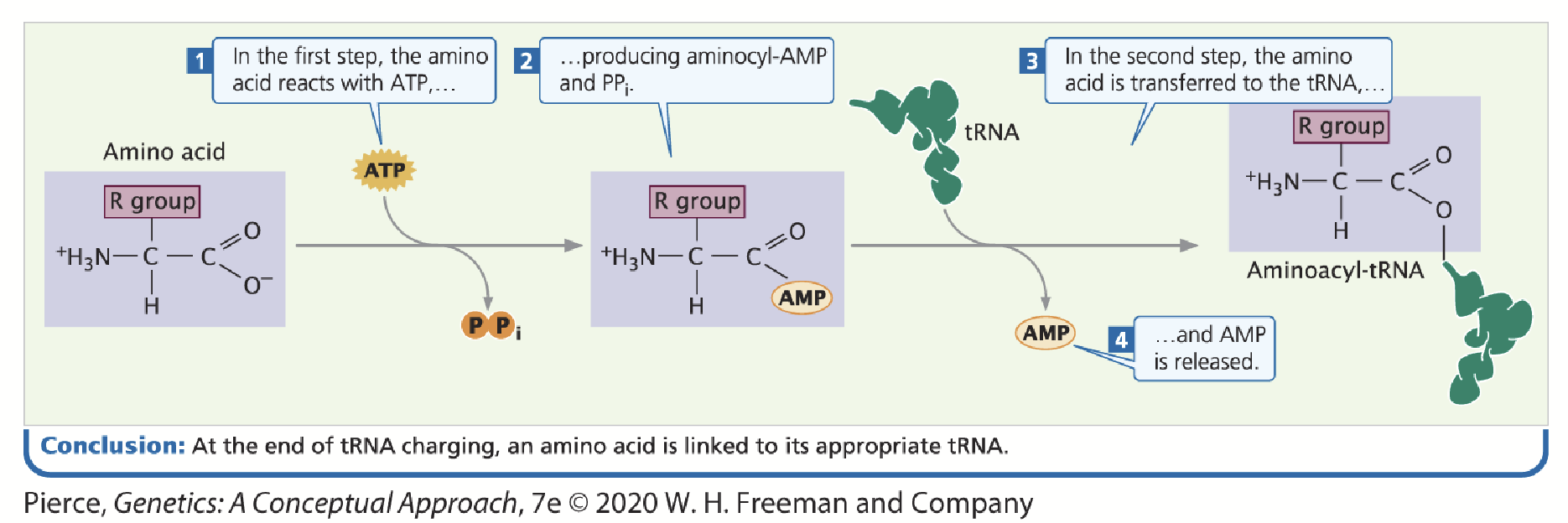
aminoacyl tRNA synthetase
Each amino acid has an enzyme that “charges” the tRNA with its specific amino acid (amino acid “activating” enzymes).
tRNA sequence is recognized by these enzymes; each enzyme is unique to each amino acid, recognizes amino acid based on their size, charge, and R group.
An amino acid becomes covalently attached to the appropriate tRNA in an ATP-dependent two-step reaction: 1) addition of AMP 2) addition of the tRNA.
aminoacyl tRNA/charged tRNA
When a tRNA’s respective amino acid is attached to its tRNA.
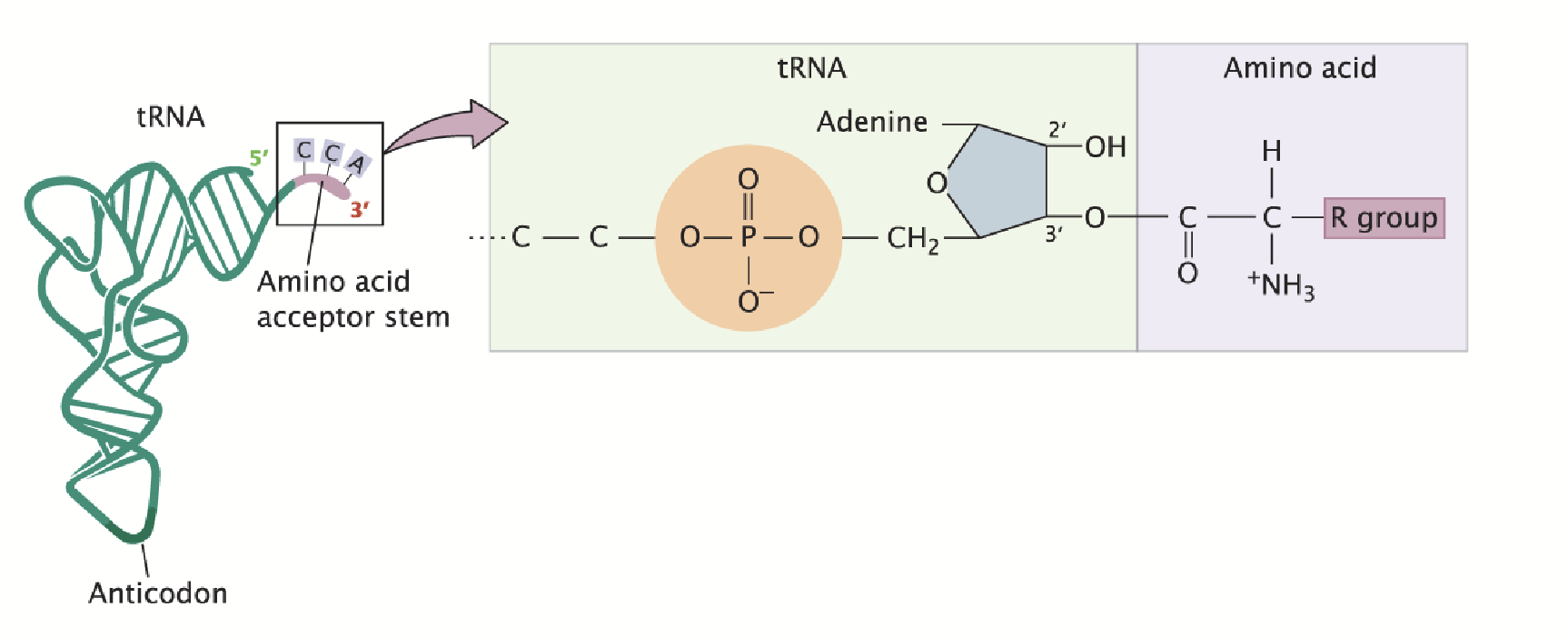
transfer RNAs (tRNAs)
The amino acid is covalently attached to the 3’ end.
Adapters between amino acids and the codons in mRNA. The anticodon base pairs with the codon of mRNA.
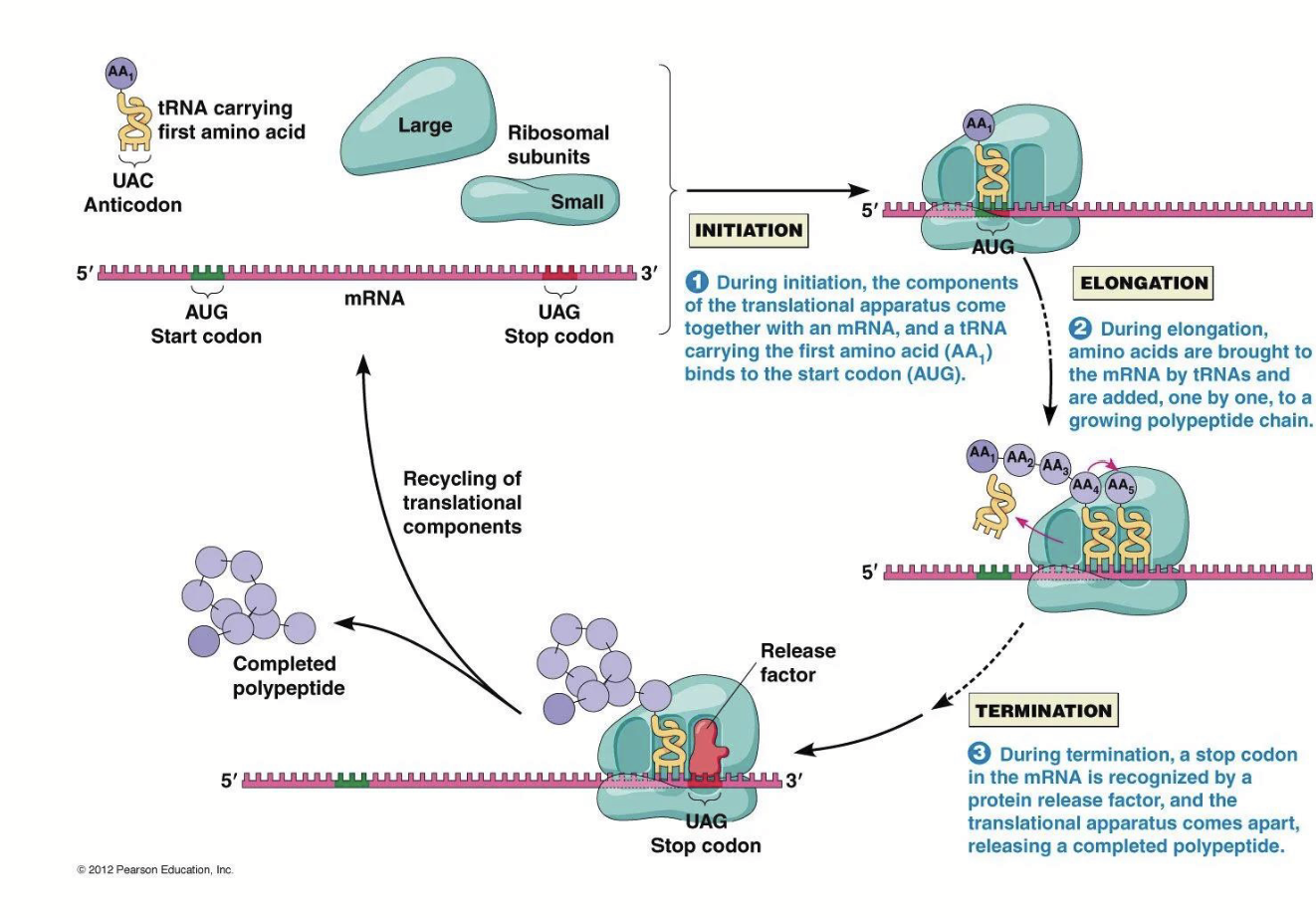
protein synthesis steps
Polypeptide Chain Initiation
Chain Elongation
Chain Termination
Basic steps are similar in prokaryotes and eukaryotes.
initiation in prokaryotes
mRNA, large and small ribosomal subunits, initiation factors (IF-3) and GTP are all required to form the initiation complex.
16S rRNA is a component of the 30S small ribosomal subunit and contains the complement to the Shine-Dalgarno (AGGAGG) sequence in the mRNA.
Pairing between the two sequences positions the ribosome near the AUG start codon.
IF-3 is required to inhibit large (50S) subunit from binding small (30S) subunit.
IF-1 and IF-2 position formylated (f)Met-tRNA over the start codon.
fMet-tRNA + 30S ribosome + IF2-GTP + IF-3 + IF-1 = 30S initiation complex.
GTP is hydrolyzed to GDP and the remaining IF factors dissociate. IF-3 dissociates, allowing the large 50S ribosomal unit to bind.
Complete complex = 70S initiation complex.
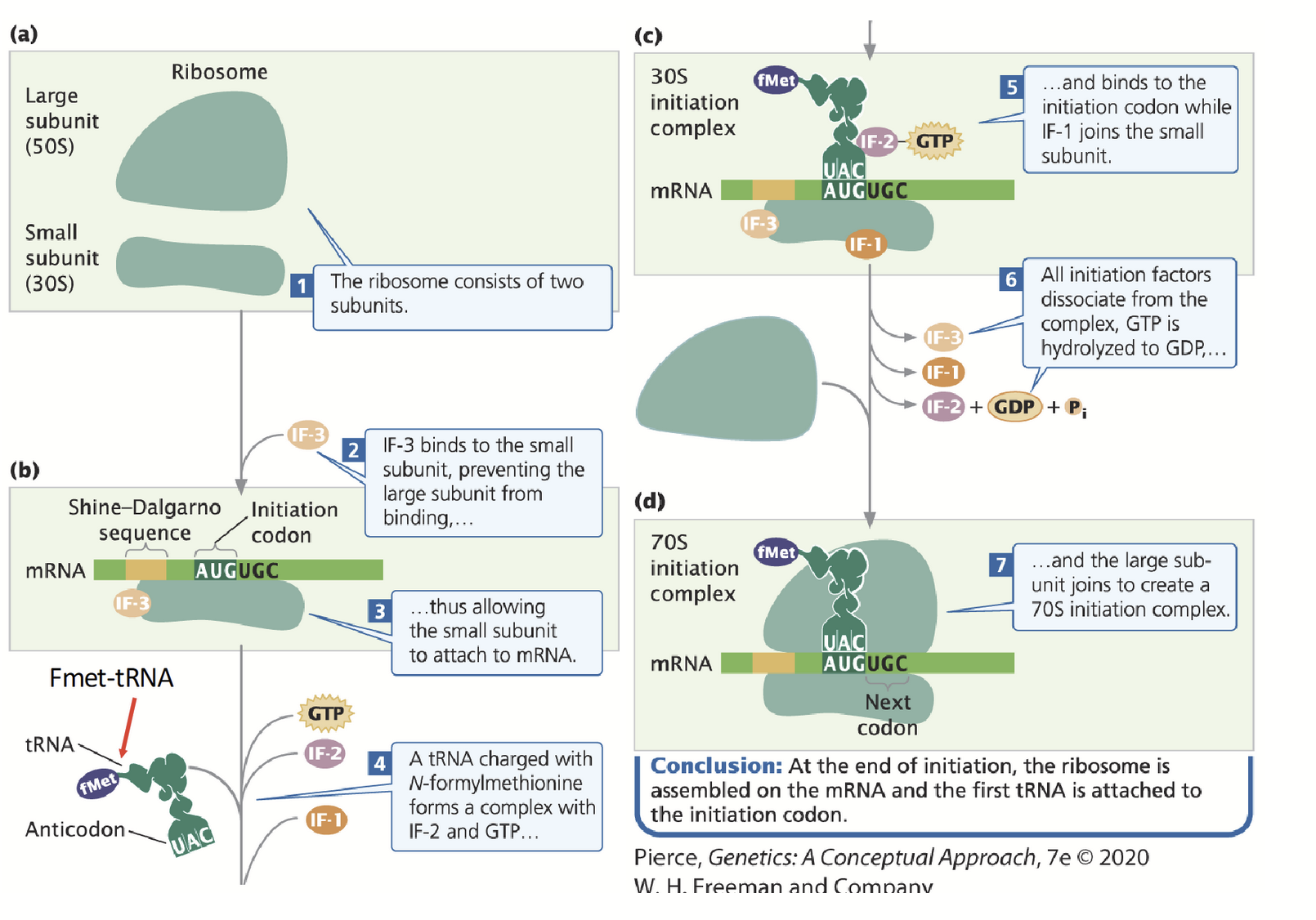
30S initiation complex
fMet-tRNA + 30S ribosome + IF2-GTP + IF-3 + IF-1
fMet-tRNA
Start codon in prokaryotes.
initiation in eukaryotes
The amino group of the methionine on the initiator tRNA is not formylated methionine (it is simply MET).
The initiation complex forms at the 5’ terminus of the mRNA (the 7-methylguanosine (7-MG) 5’ cap structure): no Shine-Dalgarno sequence.
Ribosome initiation complex scans inward for (usually) the first AUG initiation codon.
The Kozak sequence influences the efficiency of which AUG in the vicinity is used to start translation.
The poly(A) tail of the mRNA interacts with the mRNA 5’ 7-MG cap structure via a cap-binding protein complex (CBC) to promote translation initiation.
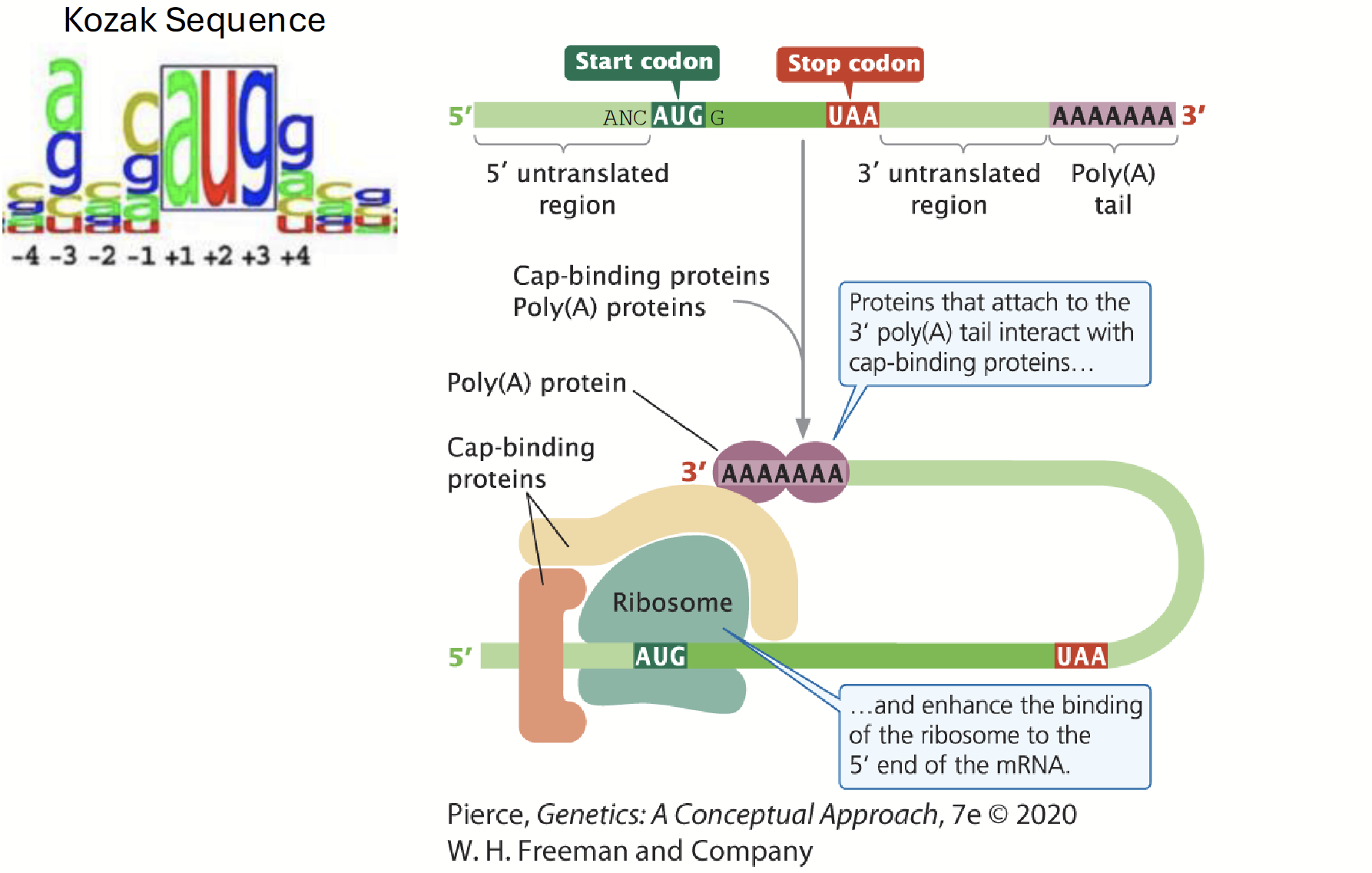
Kozak sequence
A consensus sequence that influences the efficiency of which AUG in the vicinity is used to start translation.
5’-C(A/G)NCAUGG-3’
elongation
A charged-tRNA binds to the A-site of the ribosome (requires complex formulation between the aminoacyl tRNA, elongation factor Tu plus GTP).
Once in the A site, GTP is hydrolyzed to GDP and is released along with EF-Tu.
rRNA in the large subunit catalyzes the formation of the peptide bond between the two amino acids.
Polypeptide chain is transferred to the amino acid in the A-site. As the chain grows, each new amino acid is added to the C-terminus of the previous one.
EF-G and GTP are needed to move the ribosome along the mRNA sequence; the tRNA does not move with the ribosome because it is bound by complementary base pairing.
The second amino-tRNA is now in the P-site (which contains the growing polypeptide chain).
F-Met/Met is now in the E-site and then moves into the cytoplasm to be recharged. A new aminoacyl-tRNA again binds in the A site (via EF-Tu and GTP) and the cycle is repeated.
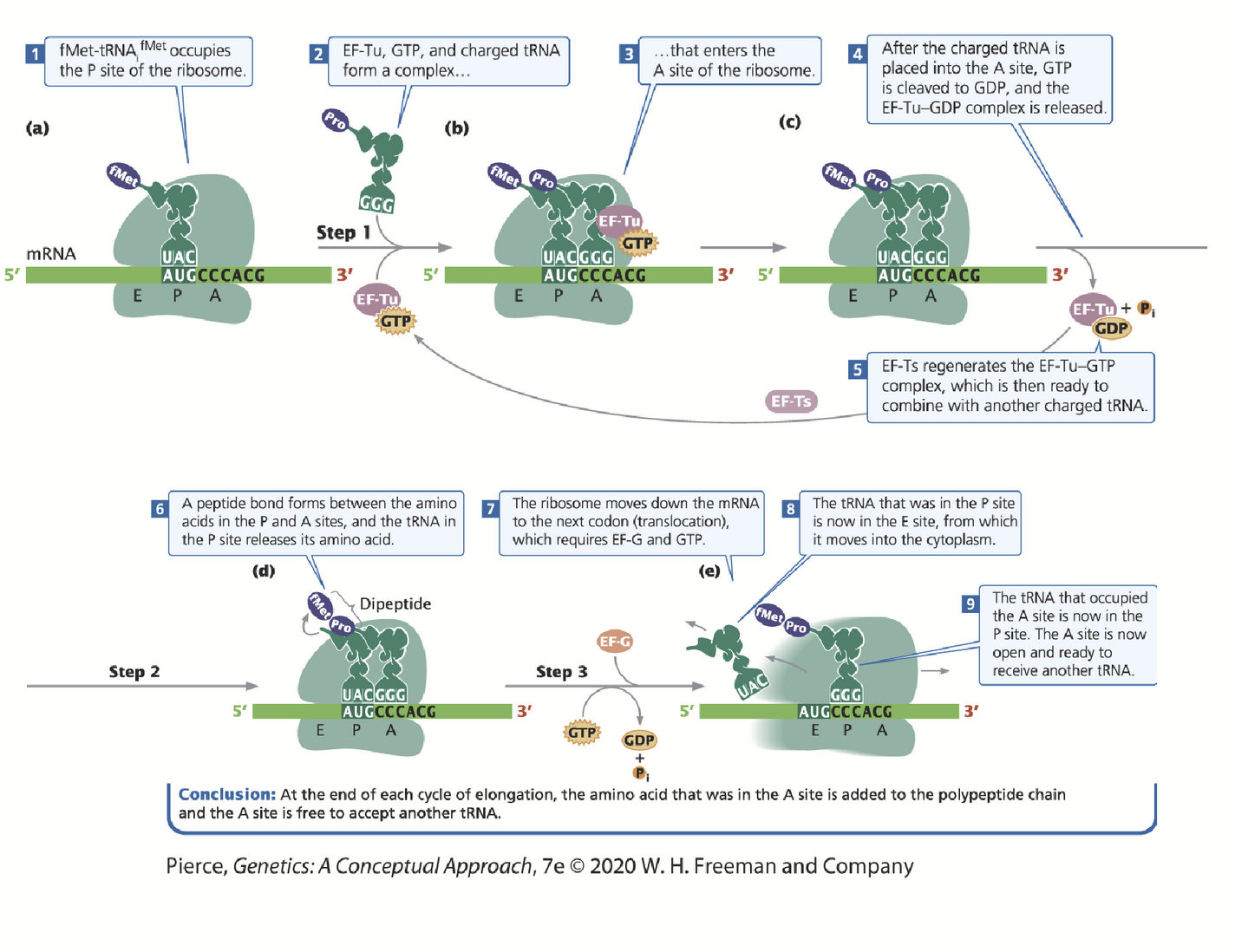
A site
Aminoacyl (acceptor) site. All amino acids (with the exception of f-Met or the first Met in eukaryotes) enters the A-site, moves to the P-site then E-site and then exits into the cytoplasm.
P site
Peptidyl site. Contains the growing polypeptide chain; first tRNA binds to the start codon here.
E site
Exit site.
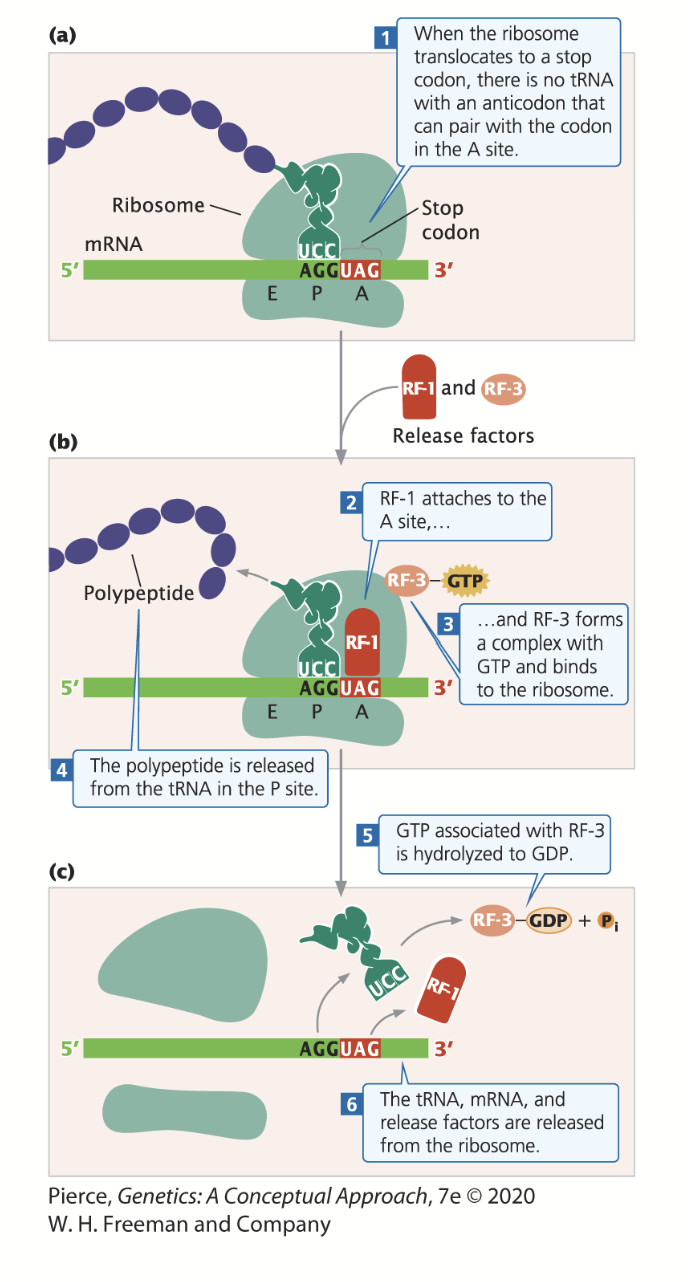
termination
Occurs in polypeptide chains when a chain-termination codon (stop codon) enters the A-site of the ribosome - there are no tRNAs that bind to stop codons (no amino acids that match the codons).
When a stop codon is encountered, a release factor (RF) binds to the A-site.
Release factor 1 (RF-1) recognizes UAG and UAA stop codons, while release factor 2 (RF-2) recognizes UAA and UGA stop codons.
RF-1 and RF-2 binding promotes the cleavage of the polypeptide chain from tRNA, releasing the chain and leading to termination. The binding of RF-3 and GTP to the ribosome assis in the dismantling of the entire complex.
Eukaryotes: eRF-1, eRF-2